Abstract
The recognition and binding of cholesterol is an important feature of many eukaryotic, viral, and prokaryotic proteins, but the molecular details of such interactions are understood only for a few proteins. The pore-forming cholesterol-dependent cytolysins (CDCs) contribute to the pathogenic mechanisms of a large number of Gram-positive bacteria. Cholesterol dependence of the CDC mechanism is a hallmark of these toxins, yet the identity of the CDC cholesterol recognition motif has remained elusive. A detailed analysis of membrane interactive structures at the tip of perfringolysin O (PFO) domain 4 reveals that a threonine-leucine pair mediates CDC recognition of and binding to membrane cholesterol. This motif is conserved in all known CDCs and conservative changes in its sequence or order are not well tolerated. Thus, the Thr-Leu pair constitutes a common structural basis for mediating CDC-cholesterol recognition and binding, and defines a unique paradigm for membrane cholesterol recognition by surface-binding proteins.
Keywords: hemolysin, intermedilysin, sterol, toxin, pore
Membrane cholesterol is important to a variety of pathogenic processes that include virus fusion and budding (1) and the mechanisms of eukaryotic (2, 3) and prokaryotic toxins (4–7). Whether cholesterol is bound directly by these proteins as a receptor or it indirectly influences the binding or activity of the protein at the membrane surface remains unknown. The cholesterol-dependent cytolysins (CDCs) use cholesterol as their receptor at the membrane surface (7) and contribute to the pathogenesis of a large number of Gram-positive bacterial pathogens (8). The CDC–sterol interaction initiates a cascade of secondary and tertiary structural changes that lead to the formation of a large oligomeric complex, and ultimately a pore in the membrane of eukaryotic cells (9–13). Although significant progress has been made in understanding the assembly of the CDC pore complex, the structural basis for recognition and binding to cholesterol-rich membranes remains elusive.
Early studies with the Clostridium perfringens perfringolysin O (PFO) suggested that the highly conserved tryptophan-rich undecapeptide sequence at the base of domain 4 (14, 15) (Fig. S1) mediated the PFO–cholesterol interaction. However, recent studies by Soltani et al. (16) uncoupled cholesterol binding from the undecapeptide and showed that the membrane insertion of loops L1–L3 at the base of domain 4 was cholesterol dependent (Fig. S1). These observations are also consistent with a lack of conservation of the 3D structures of the undecapeptide in the closely related CDCs PFO (17) and Bacillus anthracis anthrolysin O (ALO) (18) (Fig. S1). These studies suggest the residues that comprise the cholesterol recognition motif are located within L1–L3 because these loops and the undecapeptide are the only structures at the tip of domain 4 exposed to the nonpolar bilayer core; the rest of the domain 4 surface is surrounded by water (19).
Cholesterol was thought to function as the sole CDC receptor until the discovery of intermedilysin (ILY), a CDC from Streptococcus intermedius. ILY was active only on human cells (20), a feature seemingly inconsistent with the “cholesterol as receptor” paradigm. Giddings et al. subsequently showed that ILY used human CD59 (hCD59) as its membrane receptor (21), but pore formation by ILY still required cholesterol (22). This enigma was partially resolved by Soltani et al. (23), who showed that ILY loops L1–L3 underwent a cholesterol-dependent membrane insertion after ILY bound to hCD59. Removal of membrane cholesterol prevented L1–L3 membrane insertion and appeared to trap ILY in an oligomeric prepore state, although the structural basis for this defect was unclear.
The structural motif that mediates the highly specific interaction of CDCs with cholesterol remains unknown, as does the role of cholesterol in the ILY mechanism. Herein, a detailed analysis reveals that a remarkably simple motif, composed of a threonine–leucine pair in loop L1, is conserved in all CDCs and is essential for specifically recognizing cholesterol in the membrane and initiating the cholesterol-dependent interaction of CDCs with membranes. These studies also resolve the basis for the cholesterol dependence of the ILY mechanism and provide a unifying structural basis for the cholesterol dependence of CDCs whether they bind directly to cholesterol or to a nonsterol receptor.
Results
Hemolytic and Binding Activity of PFO L1–L3 Alanine and Glycine Mutants.
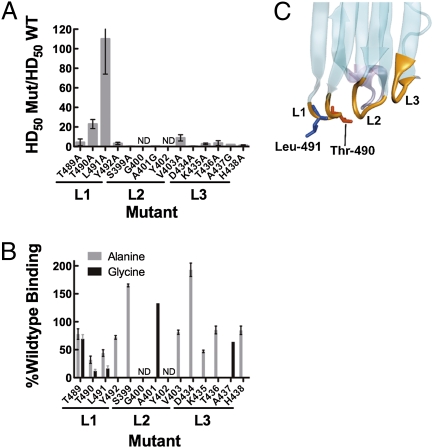
The domain 4 surface is largely surrounded by water; only loops L1–L3 and the undecapeptide at the tip of domain 4 interact with the bilayer (19). Because undecapeptide apparently played no role in cholesterol recognition (16), the search for the cholesterol recognition motif was focused on loops L1–L3. The residues comprising L1, L2, and L3 (Table S1) were subjected to scanning mutagenesis with alanine and/or glycine. Each mutant was screened for defects in hemolytic activity and in binding. Hemolytic activity was lost by mutating L491, and severely impaired by mutating T490 (Fig. 1A), and PFO binding to liposomes was greatly reduced by the same two mutations (Fig. 1B). We therefore focused our attention on T490 and L491 (Fig. 1C).
Fig. 1.
Hemolytic and binding activity of PFO L1–3 loop mutants. Residues within loops L1, L2, and L3 were systematically substituted with alanine and/or glycine and assayed for changes in hemolytic activity (A) The change in hemolytic activity is shown as the ratio of the HD50 (concentration of PFO or mutant required for 50% hemolysis, see Methods for details) for each mutant to that for wild-type PFO (HD50 = 0.34 nM). Hence, bar height is inversely correlated with activity (n = 4 for each mutant hemolytic analysis). Binding to cholesterol-rich liposomes was assessed by SPR. (B) Percent binding of wild-type PFO is calculated by the equation 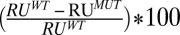 , where RUWT is the change in resonance units (RU) induced by passing 100 μL wild-type PFO (900 nM) and RUMUT is the change in resonance units for each mutant at the same concentration. In those cases where the mutant bound better than wild-type PFO, the percent change was calculated from
, where RUWT is the change in resonance units (RU) induced by passing 100 μL wild-type PFO (900 nM) and RUMUT is the change in resonance units for each mutant at the same concentration. In those cases where the mutant bound better than wild-type PFO, the percent change was calculated from 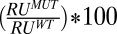 (n = 3 for each binding assay). Shown in C is the location of the Thr-490•Leu-491 pair in the lower half of PFO domain 4 (for the complete PFO structure refer to Fig. S1).
(n = 3 for each binding assay). Shown in C is the location of the Thr-490•Leu-491 pair in the lower half of PFO domain 4 (for the complete PFO structure refer to Fig. S1).
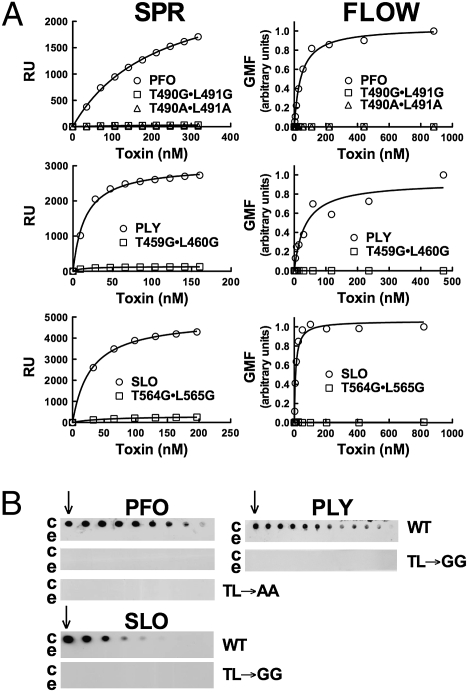
Double mutants were generated in which Thr-490 and Leu-491 were converted to either alanines or glycines. PFOT490A•L491A and PFOT490G•L491G retained less than 0.03% of the wild-type PFO hemolytic activity (Table 1). Similar results were observed when glycine was substituted for the analogous residues of streptolysin O (SLOT564G•L565G) and pneumolysin (PLYT459G•L460G) (Table 1), two PFO-like CDCs that bind directly to cholesterol-rich membranes.
Table 1.
Hemolytic activity of CDC derivatives containing mutations in the cholesterol-binding motif
| Toxin | HD50 (nM) | % WT activity |
| PFO | 0.34 ± 0.1 | 100 |
| PFOT490A•L491A | >1410 | <0.03 |
| PFOT490G•L491G | >1760 | <0.02 |
| SLO | 0.8 ± 0.1 | 100 |
| SLOT564G•L565G | >1640 | < 0.02 |
| PLY | 0.7 ± 0.3 | 100 |
| PLYT459G•L460G | >1890 | <0.02 |
| PFOT490S | 3.2 ± 0.7 | 11 |
| PFOL491I | 1.5 ± 0.6 | 22 |
| PFOL491V | 1.9 ± 0.1 | 18 |
| PFOT490S•L491I | 83 ± 6 | 0.4 |
| PFOT490S•L491V | 92 ± 2 | 0.3 |
| PFOT490L•L491T | >1760 | <0.02 |
Shown are the HD50 values for PFO, SLO, and PLY and their derivatives containing mutants in the cholesterol recognition motif.
The substitution of the Thr-Leu pair does not appear to affect the structure of PFO domain 4. Circular dichroism (Table S2) and tryptophan emission spectra (Fig. S2) (six of the seven tryptophan residues in PFO are in domain 4) of the mutants were not significantly different from native PFO. Furthermore, the sidechains of the four residues comprising loop L1 (Thr-489 to Tyr-492) extend into the aqueous solvent and do not form any predicted interactions with residues outside of loop L1, suggesting that their mutation is unlikely to perturb the structures of nearby loops or undecapeptide.
Thr–Leu Mutant Binding to Cholesterol-Rich Liposomes, Human Erythrocytes, and Immobilized Cholesterol.
Binding of PFOT490A•L491A, PFOT490G•L491G, SLOT564G•L565G, and PLYT459G•L460G to cholesterol-rich liposomes and human erythrocytes was examined by surface plasmon resonance (SPR) (liposomes) and flow cytometry (erythrocytes), respectively. These mutants lacked detectable binding to cholesterol-rich liposomes and to human erythrocytes (Fig. 2A). Direct binding to cholesterol immobilized on a PVDF membrane was not detected for any double mutants (Fig. 2B), consistent with the SPR and flow cytometry results. Taken together, these observations suggest that the loop L1 Thr–Leu pair mediates the recognition and binding of cholesterol at the membrane surface.
Fig. 2.
Alanine or glycine substitution of the Thr–Leu pair of PFO, PLY, and SLO abolishes binding. (A) Binding of PFO, PLY, SLO, and their derivatives to cholesterol-rich liposomes was measured by SPR (Left column). Binding of the same proteins to human RBCs was shown by flow cytometry (Right column). (B) A series of dot blots with twofold dilutions of cholesterol or epicholesterol (starting at 65 nmols, indicated by the arrow) probed with the same proteins and detected by antibody. Comparisons between binding of wild type and its derivatives can be made, but binding comparisons between CDCs are not valid due to differences in reactivity of the antibodies used for their detection. c, cholesterol; e, epicholesterol; TL→AA and TL→GG, the double alanine and double glycine mutants, respectively, of the Thr–Leu pair. The binding analyses are representative of three experiments. GMF, geometric mean fluorescence.
Loss of the cholesterol-dependent binding by PFOT490A•L491A could not be restored to a detectable level either by second site substitutions shown herein to increase binding of native PFO to liposomes (alanine substituted Ser-399 and Asp-434, Fig.1) (Fig. S3) or by substituting DOPC for POPC in liposomes containing 55 mol % cholesterol (Fig. S4), which is ≈25 mol % more cholesterol than is require for maximal binding of PFO to these liposomes (24, 25).
Structural Requirements of the CDC Cholesterol Recognition Motif.
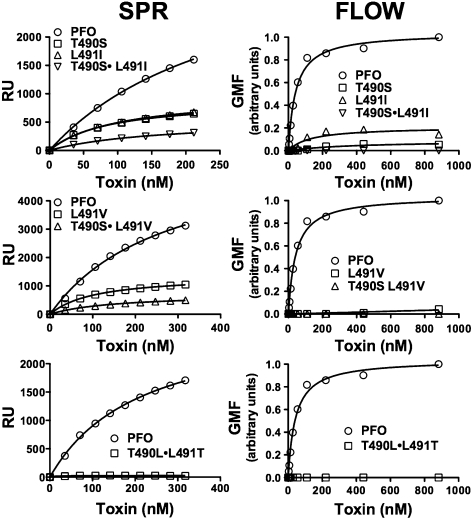
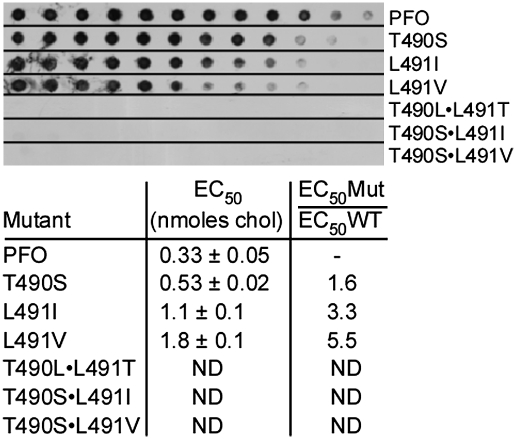
The Thr–Leu pair is conserved in all known CDCs, which suggests that even conservative substitutions are not tolerated. Substitution of serine for Thr-490 (PFOT490S), or of isoleucine or valine for Leu-491 (PFOL491I, PFOL491V), did not restore hemolytic activity to wild-type PFO levels, whereas double mutants PFOT490S•L491I and PFOT490S•L491V were reduced in activity by 240-fold or more (Table 1). Binding to cholesterol-rich liposomes and human erythrocytes was reduced for the single mutants and was virtually undetectable for the double mutants (Fig. 3). Similarly, binding of the single mutants to pure immobilized cholesterol was less than that observed for wild-type PFO, whereas binding was undetectable for the double mutants (Fig. 4).
Fig. 3.
Structural requirements of the cholesterol recognition motif. SPR analysis of binding for the various PFO mutants to cholesterol-rich liposomes is shown in the Left column. Flow cytometric analysis of binding to human erythrocytes is shown in the Right column. The SPR and flow cytometry results are representative of three or more experiments.
Fig. 4.
Binding of PFO mutants to immobilized cholesterol. The EC50 for PFOT490S, PFOL491I, and PFOL491V were compared to the EC50 for PFO. Upper panel is a representative dot blot that shows binding of each toxin to the various cholesterol concentrations (see Fig. 3 and Methods for details). In the table Below are the EC50 values and standard errors (n = 7) calculated from densitometric analysis of the dots and the fold increase in EC50 for each mutant. The double mutants PFOT490S•L491I, PFOT490S•L491V, and PFOT490L•L491T did not exhibit detectable binding to the immobilized cholesterol so an EC50 value was not determined (ND). The absolute concentrations of bound cholesterol on the PVDF membrane are not known; the values are used only to compare the relative binding of PFO and its derivatives (EC50Mut/EC50WT).
Scrambling the linear sequence by inverting their positions (PFOT490L•L491T) reduced hemolytic activity more than 5,000-fold (Table 1), whereas binding to cholesterol-rich liposomes and cells (Fig. 3) and to immobilized cholesterol was undetectable (Fig. 4). The structural arrangement of the Thr–Leu motif is therefore relatively inflexible, consistent with its conservation in all CDCs and its critical role in the specific recognition and binding of membrane cholesterol.
Role of the Thr–Leu Pair in the Intermedilysin Pore-Forming Mechanism.
Intermedilysin (ILY) first binds to its cellular receptor, hCD59 (21), and then undergoes a cholesterol-dependent insertion of its L1–L3 loops (16). Because hCD59 anchored ILY to the membrane, it was unclear why the ILY pore-forming mechanism remained sensitive to the cholesterol-dependent insertion of these loops. We recently showed, however, that ILY disengages from hCD59 during the prepore-to-pore transition (26). If the Thr–Leu pair is unable to initiate the cholesterol-dependent membrane interaction of loops L1–L3 we hypothesized that ILY would lose contact with the cell surface when it disengages from hCD59.
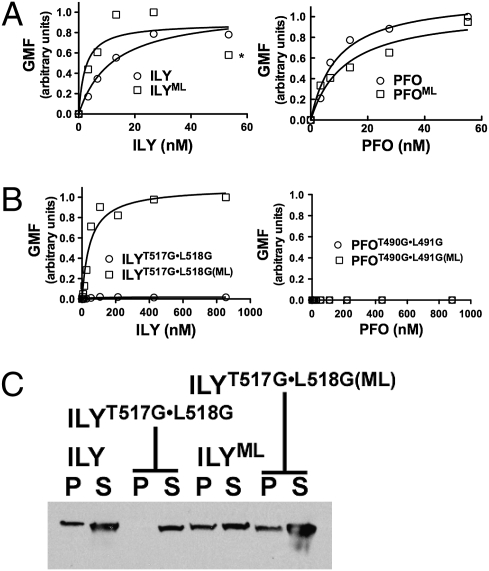
To test this hypothesis, we compared the membrane binding of native ILY and ILYT517G•L518G in two different states: a monomer-locked (ML) state that prevents disengagement of ILY from its receptor and the nonlocked state that allows ILY to undergo all conformational changes necessary to form a pore. A disulfide bond was introduced between β-strands 4 and 5 of the domain 3 core β-sheet, which prevents the disengagement of β5 from β4, a structural transition that is necessary to convert monomers into oligomers (11, 26). As predicted, ILYML and wild-type ILY bound to erythrocytes (Fig. 5A). Also, the monomer-locked version of ILYT517G•L518G (ILYT517G•L518G(ML)) remained cell bound because it cannot undergo the structural transitions that disengage it from hCD59 (Fig. 5B). Consistent with our hypothesis, binding of ILYT517G•L518G to erythrocytes was undetectable by flow cytometry (Fig. 5B). Hence, as ILYT517G•L518G disengages receptor it looses contact with the membrane if it is unable to initiate the cholesterol-dependent interaction of L1–L3. A parallel set of experiments was also carried out with PFO. PFO in its native or monomer-locked states binds to membrane (Fig. 5A). Since PFO requires the Thr–Leu pair to initiate contact with cholesterol-rich membranes, PFOT490G•L491G did not bind in either its monomer-locked or nonlocked state (Fig. 5B).
Fig. 5.
The Thr–Leu pair is necessary to maintain ILY-membrane contact during pore formation. Binding by flow cytometry of wild-type ILY and monomer-locked ILY (ILYML) to human erythrocytes (A) is compared with the Thr–Leu glycine-substituted mutant in nonlocked (ILYT517G•L518G) and monomer-locked background (ILYT517G•L518G(ML)) (B). Also shown are the corresponding analyses of nonlocked and monomer-locked versions of PFO and PFOT490G•L491G (A and B, Right). (C) SDS/PAGE and Western blot analysis showing ILY, ILYML, and ILYT517G•L518G(ML) are present in both pellet (P) and supernatant (S) fractions (due to receptor saturation with excess toxin) whereas the ILY double glycine mutant is found exclusively in the supernatant fraction. The flow cytometry and SDS/PAGE analyses are representative of three experiments. *ILYML-induced erythrocyte agglutination at this and higher concentrations, which depressed the fluorescence signal.
The flow cytometry studies were confirmed by SDS/PAGE analysis of the soluble and membrane-bound fractions of human erythrocytes treated with ILY and its derivatives. Consistent with the flow cytometry results ILY, ILYML, and ILYT517G•L518G(ML) were associated with the pellet and supernatant fractions due to receptor saturation with excess toxin (Fig. 5C). As predicted, ILYT517G•L518G was detected exclusively in the supernatant fraction.
Discussion
The strict requirement of CDC pore formation for membrane cholesterol has been known for over 50 years (27). Membranes that lack or are significantly depleted of cholesterol are not susceptible to the pore-forming activity of these toxins. For many years, cholesterol recognition was attributed to the highly conserved Trp-rich undecapeptide of the CDCs (14, 15), but recent studies by Soltani et al. (16) suggested that the cholesterol recognition motif resided within loops L1–L3, not the undecapeptide. The current studies have now discovered that only two residues, a Thr–Leu pair in Loop L1, are essential for CDC cholesterol recognition and binding in cholesterol-rich liposomes, in natural cell membranes, and in pure immobilized cholesterol. This motif is shown here to be critical for cholesterol recognition for four different CDCs, but its strict conservation in all CDC sequences suggests that the Thr–Leu pair functions similarly for all members of the family. The CDC cholesterol recognition motif is remarkably simple, but highly specific for cholesterol, and thus serves as a unique structural paradigm for cholesterol recognition motifs.
The Thr–Leu pair interacts specifically with cholesterol instead of preferentially inserting into cholesterol-rich domains. Inversion of the Thr–Leu sequence eliminated detectable binding, even though the free energy of partitioning Leu–Thr into the bilayer would be the same as for Thr–Leu. Also, conservative mutation of the Thr–Leu pair was not well tolerated, suggesting that the structure and orientation of the Thr–Leu sidechains are important factors in this recognition. It is unlikely that residues from the other two loops are involved in cholesterol recognition to any significant extent, although they likely contribute to the binding energy by partitioning their sidechains into the bilayer. Residues in L3 are not conserved, whereas L2 residue sidechains are either buried in the domain 4 core (Tyr-402 and Ala-404), lack a sidechain (Gly-401), or their substitution with alanine or glycine did not significantly affect binding (Ala-401 and Val-403). Ser-399 (L2) and Asp-434 (L3) increased binding of PFO, but placing these two mutants into the PFOT490A•L491A background did not restore any detectable binding. Also, it is unlikely that substitution of the TL pair affects the structure of loops L2 and L3 because the residues of loop L1 (Thr–Thr–Leu–Tyr) extend into the solvent and their sidechains do not form any predicted interactions with residues outside of L1, consistent with measurements that show no detectable perturbations of the domain 4 structure of these mutants. Hence, the Thr–Leu pair in the context of L1 appears to confer specificity for membrane cholesterol, whereas the subsequent membrane insertion of residues in loops 2 and 3 strengthen the interaction with the membrane.
The only other well-characterized sterol-binding motif is the eukaryotic cholesterol recognition amino acid consensus (CRAC) motif (reviewed in ref. 28). Significant structural and operational differences, however, exist between the CDC cholesterol recognition motif and the CRAC motif. The CRAC motif (L/V-X(1-5)-Y-X(1-5)-R/K) is typically located adjacent to a transmembrane α-helix that positions this sequence within the nonpolar core of the bilayer where it can form contacts along the length of the cholesterol molecule (29). In contrast, the CDC Thr–Leu pair must recognize specific features of cholesterol at or near the membrane surface to initiate the cholesterol-dependent interaction of the CDC with the cell. Previous studies have demonstrated that the 3-hydroxy headgroup of cholesterol in the 3-β stereoisomer configuration is required for recognition by the CDCs as well as a structurally intact A ring and iso-octyl hydrocarbon chain at C17 (30–33). Cholesterol is a planar molecule that packs parallel to the phospholipid acyl chains and adopts a nearly perpendicular orientation to the cell surface (34), with the 3-β-hydroxy group at the surface and the iso-octyl chain near the bilayer core. The 3-β-hydroxyl of cholesterol appears to serve as an important factor for stereo-specific binding of cholesterol, although it is unlikely to be the only component of the cholesterol headgroup to contribute to this interaction with the Thr–Leu pair. The stereo-specificity of the binding presumably results directly from the conformation of the Thr–Leu pair in the L1 loop and the consequent steric requirements of aligning the hydrogen bond donor(s) and acceptor(s) while simultaneously juxtaposing and maximizing multiple van der Waals contacts. It is also likely the structure of loop L1 and correct presentation of the Thr–Leu pair are necessary for cholesterol recognition. Thus, not all Thr–Leu pairs would be capable of mediating such an interaction. It is unlikely that the C and D rings and the iso-octyl hydrocarbon chain at C17 directly participate in CDC recognition because they reside far inside the bilayer core and are therefore not accessible to the Thr–Leu pair. Their structures, however, can affect the packing and orientation of the cholesterol molecule in the bilayer (34), which may indirectly affect the presentation of the cholesterol headgroup at the membrane surface.
Once the Thr–Leu pair recognizes cholesterol, the interaction of the CDC monomer with the membrane is furthered strengthened by the membrane insertion of additional residues of loops L1–L3 and residues of the undecapeptide (19, 35). The Thr–Leu interaction with cholesterol may also elicit rearrangement of the L1 polypeptide that then is transmitted throughout the protein. We previously observed that domain 4 binding corresponds to structural changes in PFO domain 3 that is located more than 70 Å above the membrane surface (11, 36). The change in the domain 3 structure leads to oligomerization of membrane bound monomers into the prepore, which then makes the transition to the β-barrel pore (11, 13).
ILY and the recently identified CDC from Gardnerella vaginalis, vaginolysin (VLY) do not bind directly to cholesterol but instead bind the protein receptor hCD59 (21, 37). Yet the pore-forming activity of both CDCs remains cholesterol dependent. Recent studies by Lachapelle et al. (26) showed that ILY disengages from hCD59 during the transition of oligomeric ILY from the prepore to pore state. We show here that the Thr–Leu pair of ILY must initiate a cholesterol-dependent interaction with the membrane, otherwise it loses contact with the membrane surface as individual monomers within the prepore complex disengage from the receptor during the prepore to pore transition. Because loops L1–L3 only insert in a cholesterol-dependent fashion after ILY binds its receptor hCD59 (16), it presumably orients loops L1–L3 and the undecapeptide near the membrane surface.
In conclusion, a remarkably simple structure composed of a threonine–leucine pair functions as the cholesterol recognition motif of the CDCs. The membrane binding of many other bacterial and eukaryotic toxins is cholesterol dependent, but their cholesterol recognition motif is unknown. Therefore it will be interesting to determine how widespread the Thr–Leu recognition is among other cholesterol-dependent membrane active proteins.
Methods
Bacterial Strains, Plasmids, and Chemicals.
The genes for PFO, SLO, and ILY were cloned into pTrcHisA (Invitrogen) as described previously (23). The gene for PLY was cloned into pQE-30 (a generous gift from R. Malley). All mutations in PFO were made in the cysteine-less background (PFOC459A). Mutations in other CDCs were generated in native SLO and PLY (both containing the native cysteine) and ILY (naturally cysteine-less) backgrounds. All chemicals and enzymes were obtained from Sigma, VWR, and Research Organics. All fluorescent probes were obtained from Molecular Probes (Invitrogen). Primary antibodies, anti-Xpress and anti-PLY (NCL-SPNm), were obtained from Invitrogen and Novocastra, respectively. Polyclonal anti-PFO antibody was affinity purified from hyperimmune rabbit serum. Secondary antibodies, goat anti-mouse labeled with fluorescein isothiocyanate (FITC) and goat anti-rabbit-FITC were obtained from Abcam and goat anti-mouse-HRP and goat anti-rabbit-HRP were obtained from Biorad. Sterols were obtained from Steraloids.
Generation and Purification of Toxin Derivatives.
Various amino acid substitutions were made in PFO, SLO, PLY, and ILY using PCR QuikChange mutagenesis (Stratagene) and verified by DNA sequencing analysis performed by the Oklahoma Medical Research Foundation Core DNA Sequencing Facility. The expression and purification of recombinant toxins and derivatives from Escherichia coli were carried out as described (23). Purified protein was dialyzed into buffer [100 mM NaCl, 50 mM HEPES; (pH 7.5)] overnight at 4 °C and stored in 50 μM TCEP (except for monomer-locked derivatives) and 10% (vol/vol) sterile glycerol at −80 °C.
Hemolytic Activity and Trypsin Sensitivity of Toxins and Their Derivatives.
The pore-forming activity of each toxin and derivative was measured using a hemolytic titration assay as previously described (13). The trypsin sensitivity of each mutant was examined as previously described (16) and compared to that of native toxin. No significant difference in cleavage patterns was noted in the various mutants and wild-type CDCs.
Liposome Preparation.
Liposomes containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; Avanti Polar Lipids) or 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and cholesterol at a molar ratio of 45:55 were prepared as described (13).
SPR Analysis.
SPR was measured with a BIAcore 3000 system (Oklahoma Medical Research Foundation) using a L1 sensor chip (BIAcore). To prepare the L1 chip with liposomes, 10 μL of 20 mM CHAPS was injected at a flow rate of 10 μL/min. Liposomes (0.5 mM final lipid concentration) were then injected at the same flow rate for 15 min. After injection of liposomes, 50 mM NaOH was injected for 3 min, followed by injection of 0.1 mg/mL BSA for 1 min. All injections were performed at 25 °C. The L1 chip was regenerated and stripped of liposomes by repeated injections of 20 mM CHAPS and 50 mM NaOH until original resonance units (RU) readings were reached. No loss of sensor chip binding capacity resulted from regeneration.
Binding analyses were performed in HBS [100 mM NaCl, 50 mM HEPES (pH 7.5)] at 25 °C. Nine consecutive 50-μL injections of PFO or derivatives (100 ng per injection), SLO or derivatives (25 ng per injection), or PLY or derivatives (25 ng per injection) were passed over the liposome-coated chip at a flow rate of 10 μL/min. No change in the RU was observed due to buffer.
Flow Cytometry.
Twofold serial dilutions of the various toxins or their derivatives were incubated with washed erythrocytes (1 × 106 cells) in PBS for 1 h at 4 °C (reaction volume 100 μL) to minimize cell lysis. Primary antibody (anti-Xpress or anti-PLY) was added 1:100 in 50 μL PBS containing 0.1% (wt/vol) BSA and incubated 30 min at 4 °C. Secondary antibody (goat anti-mouse-FITC) was added 1:100 in 50 μL PBS containing 0.1% (wt/vol) BSA and incubated 30 min at 4 °C. Cells were pelleted (14,000 × g for 10 min) and washed twice with ice-cold PBS. Samples were then brought to a final volume of 500 μL with ice-cold PBS and analyzed by a FACSCalibur flow cytometer (University of Oklahoma Health Sciences Center), gating on both live and dead cells. The emission wavelength was 530 nm, and the excitation was 488 nm with a bandpass of 30 nm.
SDS/PAGE and Western Blot.
ILY or its derivatives (142 nM) were incubated with huRBCs at 37 °C for 45 min, and each sample was centrifuged at 15,000 rpm for 15 min to separate cell-bound and soluble ILY. Each pellet was solubilized with 25% (wt/vol) SDS, boiled for 5 min and the proteins separated on a 10% SDS/PAGE gel (125 V for 70 min), and transferred to nitrocellulose. The blot was incubated for 1 h with blot wash [20 mM Tris-HCl (pH 8.0), 0.3 M NaCl, 0.5% (vol/vol) Tween 20] made 3% (wt/vol) in dry nonfat milk and 7.7 mM in sodium azide (blocking buffer), and incubated with anti-Xpress primary antibody overnight at room temperature. Blots were washed three times with blot wash and incubated with goat anti-mouse-HRP for 45 min, washed three more times, and treated with ECL Western Blotting Detection kit (Amersham). Blots were exposed to X-ray film (Life Science Products) to detect the ILY bands.
Cholesterol Dot Blot Analysis.
Binding of the CDCs and their derivatives to pure cholesterol was determined by dot blot. A PVDF membrane was wetted with 100% methanol and then washed in 25 mL PBS buffer before placing it into a Hybri-dot Manifold apparatus (Bethesda Research Labs) to which cholesterol and epicholesterol stocks (in 100% ethanol) were serially diluted (in PBS) from 65 nmol to 16 pmol in each set of 12 wells. After 30 min the sterol solution was pulled through the membrane by vacuum. The membrane was removed from the manifold and blocked in blocking buffer (as for the Western blot, above) for 1 h. Each set of 12 wells was cut into strips and each strip was incubated with the various CDCs or their derivatives (5 μg/mL in a total of 10 mL blocking buffer) for 1 h at room temperature. Blots were washed with blot wash (25 mL) three times and incubated with primary antibody (1:1,000 in blocking buffer) for 30 min at room temperature and then removed by washing the blot three times with 25 mL of blot wash each time. Secondary antibody conjugated to horseradish peroxidase was added to the membrane (1:2,000 in blocking buffer), incubated for 15 min at room temperature, and unbound antibody was removed as done for the primary antibody. Bound toxin was visualized by developing the blot as done for the Western blot. To quantify binding, the film was scanned and the optical density of the dots determined using Image J analysis software (38). The EC50 (effective concentration of cholesterol for 50% binding of each toxin) was determined using Prism software (GraphPad Software, Inc.).
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health National Institute of Allergy and Infectious Diseases (AI037657) and the Robert A. Welch Foundation (Chair Grant BE-0017). The technical assistance of P. Parrish, J. Henthorn's assistance with flow cytometry, P. Mehta-D'Souza's assistance with SPR, and K. Rodgers and N. Rahman with circular dichroism are appreciated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911581107/DCSupplemental.
References
- 1.Kielian M, Chatterjee PK, Gibbons DL, Lu YE. Specific roles for lipids in virus fusion and exit. Examples from the alphaviruses. Subcell Biochem. 2000;34:409–455. doi: 10.1007/0-306-46824-7_11. [DOI] [PubMed] [Google Scholar]
- 2.Ishitsuka R, Kobayashi T. Cholesterol and lipid/protein ratio control the oligomerization of a sphingomyelin-specific toxin, lysenin. Biochemistry. 2007;46:1495–1502. doi: 10.1021/bi061290k. [DOI] [PubMed] [Google Scholar]
- 3.Barlic A, et al. Lipid phase coexistence favors membrane insertion of equinatoxin-II, a pore-forming toxin from Actinia equina. J Biol Chem. 2004;279:34209–34216. doi: 10.1074/jbc.M313817200. [DOI] [PubMed] [Google Scholar]
- 4.Giesemann T, et al. Cholesterol-dependent pore formation of Clostridium difficile toxin A. J Biol Chem. 2006;281:10808–10815. doi: 10.1074/jbc.M512720200. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay K, Bhattacharyya D, Banerjee KK. Vibrio cholerae hemolysin. Implication of amphiphilicity and lipid-induced conformational change for its pore-forming activity. Eur J Biochem. 2002;269:4351–4358. doi: 10.1046/j.1432-1033.2002.03137.x. [DOI] [PubMed] [Google Scholar]
- 6.Martín C, et al. Membrane restructuring by Bordetella pertussis adenylate cyclase toxin, a member of the RTX toxin family. J Bacteriol. 2004;186:3760–3765. doi: 10.1128/JB.186.12.3760-3765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alouf JE, Billington SJ, Jost BH. Repertoire and general features of the cholesterol-dependent cytolysins. In: Alouf JE, Popoff MR, editors. Bacterial Toxins: A Comprehensive Sourcebook. 3rd Ed. London: Academic Press; 2005. pp. 643–658. [Google Scholar]
- 8.Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tweten RK, Harris RW, Sims PJ. Isolation of a tryptic fragment from Clostridium perfringens θ-toxin that contains sites for membrane binding and self-aggregation. J Biol Chem. 1991;266:12449–12454. [PubMed] [Google Scholar]
- 10.Heuck AP, Hotze EM, Tweten RK, Johnson AE. Mechanism of membrane insertion of a multimeric beta-barrel protein: Perfringolysin O creates a pore using ordered and coupled conformational changes. Mol Cell. 2000;6:1233–1242. doi: 10.1016/s1097-2765(00)00119-2. [DOI] [PubMed] [Google Scholar]
- 11.Ramachandran R, Tweten RK, Johnson AE. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit β-strand alignment. Nat Struct Mol Biol. 2004;11:697–705. doi: 10.1038/nsmb793. [DOI] [PubMed] [Google Scholar]
- 12.Shatursky O, et al. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: A novel paradigm for pore-forming toxins. Cell. 1999;99:293–299. doi: 10.1016/s0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- 13.Shepard LA, et al. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: An α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs T, et al. The conserved undecapeptide shared by thiol-activated cytolysins is involved in membrane binding. FEBS Lett. 1999;459:463–466. doi: 10.1016/s0014-5793(99)01297-1. [DOI] [PubMed] [Google Scholar]
- 15.Sekino-Suzuki N, Nakamura M, Mitsui KI, Ohno-Iwashita Y. Contribution of individual tryptophan residues to the structure and activity of theta-toxin (perfringolysin O), a cholesterol-binding cytolysin. Eur J Biochem. 1996;241:941–947. doi: 10.1111/j.1432-1033.1996.00941.x. [DOI] [PubMed] [Google Scholar]
- 16.Soltani CE, Hotze EM, Johnson AE, Tweten RK. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc Natl Acad Sci USA. 2007;104:20226–20231. doi: 10.1073/pnas.0708104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 18.Bourdeau RW, et al. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J Biol Chem. 2009;284:14645–14656. doi: 10.1074/jbc.M807631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran R, Heuck AP, Tweten RK, Johnson AE. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat Struct Biol. 2002;9:823–827. doi: 10.1038/nsb855. [DOI] [PubMed] [Google Scholar]
- 20.Nagamune H, et al. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect Immun. 1996;64:3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giddings KS, Zhao J, Sims PJ, Tweten RK. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat Struct Mol Biol. 2004;12:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- 22.Giddings KS, Johnson AE, Tweten RK. Redefining cholesterol's role in the mechanism of the cholesterol-dependent cytolysins. Proc Natl Acad Sci USA. 2003;100:11315–11320. doi: 10.1073/pnas.2033520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soltani CE, Hotze EM, Johnson AE, Tweten RK. Specific protein-membrane contacts are required for prepore and pore assembly by a cholesterol-dependent cytolysin. J Biol Chem. 2007;282:15709–15716. doi: 10.1074/jbc.M701173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan JJ, Tweten RK, Johnson AE, Heuck AP. Cholesterol exposure at the membrane surface is necessary and sufficient to trigger perfringolysin O binding. Biochemistry. 2009;48:3977–3987. doi: 10.1021/bi9002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson LD, Johnson AE, London E. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: Insights into the origin of perfringolysin O-lipid raft interaction. J Biol Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- 26.LaChapelle S, Tweten RK, Hotze EM. Intermedilysin-receptor interactions during assembly of the pore complex: Assembly intermediates increase host cell susceptibility to complement-mediated lysis. J Biol Chem. 2009;284:12719–12726. doi: 10.1074/jbc.M900772200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard JG, Wallace KR, Wright GP. The inhibitory effects of cholesterol and related sterols on haemolysis by streptolysin O. Br J Exp Pathol. 1953;34:174–180. [PMC free article] [PubMed] [Google Scholar]
- 28.Epand RF, et al. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45:6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamin N, et al. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol Endocrinol. 2005;19:588–594. doi: 10.1210/me.2004-0308. [DOI] [PubMed] [Google Scholar]
- 30.Prigent D, Alouf JE. Interaction of streptolysin O with sterols. Biochim Biophys Acta. 1976;433:422–428. doi: 10.1016/0005-2736(76)90511-3. [DOI] [PubMed] [Google Scholar]
- 31.Zitzer A, Westover EJ, Covey DF, Palmer M. Differential interaction of the two cholesterol-dependent, membrane-damaging toxins, streptolysin O and Vibrio cholerae cytolysin, with enantiomeric cholesterol. FEBS Lett. 2003;553:229–231. doi: 10.1016/s0014-5793(03)01023-8. [DOI] [PubMed] [Google Scholar]
- 32.Geva M, Izhaky D, Mickus DE, Rychnovsky SD, Addadi L. Stereoselective recognition of monolayers of cholesterol, ent-cholesterol, and epicholesterol by an antibody. Chembiochem. 2001;2:265–271. doi: 10.1002/1439-7633(20010401)2:4<265::AID-CBIC265>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 33.Izhaky D, Addadi L. Stereoselective interactions of a specialized antibody with cholesterol and epicholesterol monolayers. Chem Eur J. 2000;6:869–874. doi: 10.1002/(sici)1521-3765(20000303)6:5<869::aid-chem869>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 34.Olsen BN, Schlesinger PH, Baker NA. Perturbations of membrane structure by cholesterol and cholesterol derivatives are determined by sterol orientation. J Am Chem Soc. 2009;131:4854–4865. doi: 10.1021/ja8095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heuck AP, Tweten RK, Johnson AE. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J Biol Chem. 2003;278:31218–31225. doi: 10.1074/jbc.M303151200. [DOI] [PubMed] [Google Scholar]
- 36.Ramachandran R, Tweten RK, Johnson AE. The domains of a cholesterol-dependent cytolysin undergo a major FRET-detected rearrangement during pore formation. Proc Natl Acad Sci USA. 2005;102:7139–7144. doi: 10.1073/pnas.0500556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelber SE, Aguilar JL, Lewis KL, Ratner AJ. Functional and phylogenetic characterization of Vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J Bacteriol. 2008;190:3896–3903. doi: 10.1128/JB.01965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.