Abstract
Cholera toxin is encoded in the genome of CTXφ, a lysogenic filamentous phage of Vibrio cholerae. CTXφ variants contribute to the genetic diversity of cholera epidemic strains. It has been shown that the El Tor variant of CTXφ hijacks XerC and XerD, two host-encoded tyrosine recombinases that normally function to resolve chromosome dimers, to integrate at dif1, the dimer resolution site of the larger of the two V. cholerae chromosomes. However, the exact mechanism of integration of CTXφ and the rules governing its integration remained puzzling, with phage variants integrated at either or both dimer resolution sites of the two V. cholerae chromosomes. We designed a genetic system to determine experimentally the tropism of integration of CTXφ and thus define rules of compatibility between phage variants and dimer resolution sites. We then showed in vitro how these rules are explained by the direct integration of the single-stranded phage genome into the double-stranded bacterial genome. Finally, we showed how the evolution of phage attachment and chromosome dimer resolution sites contributes to the generation of genetic diversity among cholera epidemic strains.
Keywords: lysogenic conversion, site-specific recombination
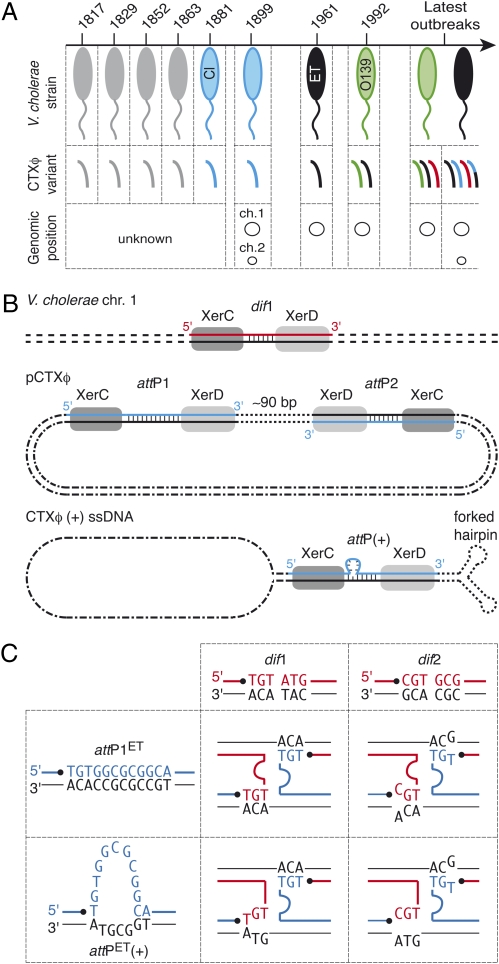
Cholera toxin, which is responsible for the deadly diarrhea associated with the disease of the same name, is one of the most significant virulence factors of Vibrio cholerae (1). It is encoded in the genome of a lysogenic filamentous bacteriophage, CTXφ (2). Different CTXφ variants exist, the two main ones being classified as “classical” and “El Tor,” according to the biotype of the hosts in which they originally were identified (Fig. 1A) (3). The existence of several different CTXφ variants and their integration in variable copy numbers on the first, the second, or both V. cholerae chromosomes contribute to the genetic diversity of cholera epidemic strains (Fig. 1A) (4).
Fig. 1.
Lysogenic conversion of V. cholerae strains by CTXφ. (A) CTXφ variants found in cholera epidemic strains. V. cholerae strains: gray, unknown; blue, classical; black, El Tor; green, O139. CTXφ variants: gray, unknown; blue, classical; black, El Tor; green, O139; red, G; black and blue, El Tor and classical hybrid. (B) Scheme of Xer recombination sites. dif1, dimer resolution site of the N16961 chromosome 1; attP1 and attP2, Xer sites found in the replicative form of CTXφ (pCTXφ); attP(+), the recombination site created by the folding of the (+) ssDNA genome of CTXφ. The two DNA strands of each site are drawn. The strand cleaved by XerC is shown in color. Vertical bars indicate bases present in the overlap region of each site. (C) Schemes of the Watson–Crick bp interactions that could stabilize the strand exchange catalyzed by XerC between the overlap regions of the two chromosome dimer resolution sites of N16961 and the two putative attachment sites of El Tor variants of CTXφ. dif1, chromosome 1 dimer resolution site; dif2, chromosome 2 dimer resolution site; attP1ET, attP1 found in CTXφ El Tor variants; attP(+)ET, attachment site unmasked by the folding of the (+) ssDNA genome of El Tor variants of CTXφ. The strands cleaved by XerC on dimer resolution and phage attachment sites are shown in red and in blue, respectively. Pairing interactions are indicated by the proximity of the bases.
In contrast to most other lysogenic phages, such as phage λ, CTXφ does not encode its own integration machinery. Instead, it has been shown that the El Tor variant of CTXφ hijacks XerC and XerD, two host-encoded tyrosine recombinases that normally function to resolve chromosome dimers (5), to integrate at dif1, the dimer resolution site of the larger of the two V. cholerae chromosomes (6). Xer recombination sites consist of binding sites for XerC and XerD, separated by a 6-bp to 8-bp overlap region; strand exchanges occur at the border of this region (7). The replicative form of CTXφ harbors two putative Xer recombination sites in inverted orientations, attP1 and attP2 (pCTXφ; Fig. 1B). This observation led to the proposal of two integration models (8, 9). The first model predicts that recombination occurs between dif1 and attP1 as the result of an unknown architectural function played by attP2. The second model relies on the formation of a forked hairpin by the ∼150-bp region encompassing attP1 and attP2 in the (+) ssDNA genome of the phage, which unmasks a putative phage attachment site, attP(+) [CTXφ (+) ssDNA; Fig. 1B]. Both models rely on the exchange of a single pair of strands catalyzed by XerC and on the conversion of the resulting Holliday junction into product by repair and/or replication.
According to both models, no integration should occur at dif2, the dimer resolution site of the second V. cholerae chromosome, because no Watson–Crick bp interactions could stabilize the exchange of strands catalyzed by XerC (Fig. 1C). Nevertheless, the classical variant of CTXφ was found integrated at dif2 in classical strains (3) and in recent El Tor isolates (4, 10–14). The mode of dissemination of this variant is of particular interest, because it allows the production of an elevated amount of cholera toxin (15, 16) that seems to be implicated in a high proportion of severe infections associated with classical strains (17). It was proposed that this variant recently invaded the genome of El Tor strains through chitin-induced competence and homologous recombination (18). However, the transformation efficiency was very low, in the order of 10−6 to 10−4. In addition, such a mechanism does not explain how CTXφ initially achieved dif2 integration or the diversity of combinations of the different genetic elements that were found integrated at the two chromosome dimer resolution sites of V. cholerae strains (4).
In this study, we designed a sensitive assay to monitor the efficiency with which CTXφ integrates at dif1 and dif2 in V. cholerae. Using this assay, we demonstrated the specificity of integration of the El Tor variant of CTXφ harbored by strain N16961. In contrast, the classical variant of CTXφ harbored by strain 569B efficiently integrated at both dif1 and dif2. We found that the altered integration behavior of the classical phage is caused by two base changes in the overlap region of attP2 that allow V. cholerae XerCD to recombine the classical ssDNA attP(+) region with dif1 and dif2 in vitro. These results further support the ssDNA integration model and allow the definition of rules of compatibility between phage attachment and dimer resolution sites that explain the tropism of integration of the different CTXφ variants. Based on these rules, we designed a phage attachment site that exclusively targets dif2. We also explained how O1 and O139 V. cholerae strains with altered dimer resolution sites can escape lysogenic conversion by the most common variants of CTXφ and showed how a new variant of CTXφ has evolved that can integrate into the genome of these particular strains. Taken together, these results suggest that lysogenic conversion by CTXφ is the primary mode of acquisition of the cholera toxin genes, which, along with the evolution of phage attachment and chromosome dimer resolution sites, contributes to the generation of genetic diversity among cholera epidemic strains.
Results
Design of a Sensitive Assay to Monitor the Efficiency and Specificity of CTXφ Integration.
Previous studies that addressed the mechanism of integration of CTXφ used Southern and PCR techniques to determine the location of each integration event. These techniques are robust. However, because the integration of CTXφ at dif2 might be a very rare event, we sought to design a time- and cost-effective method to monitor its frequency of occurrence. To this aim, we inserted dif2 in the coding region of the Escherichia coli lacZ gene in such a manner that the produced peptide might retain its β-galactosidase activity. We then engineered an N16961 El Tor strain in which the endogenous lacZ gene was deleted and in which dif2 was replaced by the lacZ::dif2 allele (Table 1). These cells turn blue in the presence of X-gal because the β-galactosidase they produce is active. dif2 integration events disrupt the lacZ ORF, thereby abolishing β-galactosidase production. Thus we could screen for such events simply by plating cells on X-gal media.
Table 1.
In vivo integration of RSET and RSCl
| Phage machinery | attP | dif | Chromosome | % integration | Screened colonies |
| Classical | Classical | dif1 | 1 | 36.4 | 466 |
| Classical | Classical | dif2 | 2 | 4.0 | 451 |
| El Tor | El Tor | dif1 | 1 | 100.0 | 454 |
| El Tor | El Tor | dif2 | 2 | <0.1 | 1099 |
| El Tor | Classical | dif1 | 1 | 100.0 | 675 |
| El Tor | Classical | dif2 | 2 | 36.3 | 1556 |
| Classical | El Tor | dif1 | 1 | 91.7 | 743 |
| Classical | El Tor | dif2 | 2 | <0.1 | 883 |
| El Tor | El Tor | dif1 | 2 | 100.0 | 875 |
| El Tor | El Tor | dif2 | 1 | <0.1 | 1070 |
Data were obtained from at least three independent experiments.
The two chromosomes harbored by classical strains possess the same dif2 dimer resolution site (CP000626.1). This situation could have favored dif2 integration because of the absence of the preferential attachment site of CTXφ in the bacterial genome. To mimic this situation, we used as a background for our assays an N16961 El Tor strain in which dif1 and the integrated copies of CTXφ surrounding it had been deleted (Table S1). Finally, we engineered a lacZ−/dif2− N16961 El Tor strain in which dif1 was replaced by a functional E. coli lacZ::dif1 gene to monitor the efficiency with which CTXφ integrates at dif1 (Table S1).
The Variant of CTXφ Found in the 569B Classical Strain Integrates at Both dif1 and dif2.
Studies of the mechanism of integration of the classical variant of CTXφ were complicated because no phage production was observed in any of the strains harboring it (3, 11). To circumvent this difficulty, we used conjugation to deliver circular DNA molecules carrying the replication and integration region of the phage found in the 569B classical strain, hereafter referred to as “RSCl,” directly into V. cholerae cells. To this aim, we cloned RSCl in a conjugative vector that carries a chloramphenical resistance gene but cannot replicate autonomously in V. cholerae (Table S2). We also cloned the replication and integration region of RS1, a truncated derivative of CTXφ found in the N16961 El Tor strain (19), hereafter referred to as “RSET” (Table S2). The attachment site of RS1 is identical to the attachment site of the El Tor variants of CTXφ. In addition, RS1 efficiently and specifically integrates at dif1, making it a perfect control for our experiments. Conjugation of the RSET and RSCl vectors gave rise to similar numbers of CmR colonies. RSET integrated in 100% of the colonies of dif1+/dif2− cells and in no colonies of dif1−/dif2+ cells, confirming its highly specific integration at dif1 (Table 1). In contrast, RSCl integrated in 36.4% of the colonies of dif1+/dif2− cells and in 4% of the colonies of dif1−/dif2+ cells, indicating that it efficiently targets both dif1 and dif2 (Table 1). The integration specificity was not linked to the genomic context of the dimer resolution sites, because RSET integrated as efficiently when dif1 was located on chromosome 2 as when it was on chromosome 1 (Table 1). Finally, integration was suppressed entirely in xerC− cells and did not require homologous recombination (Table S3).
Two Bases Determine the Capacity of RSCl to Integrate at Both dif1 and dif2.
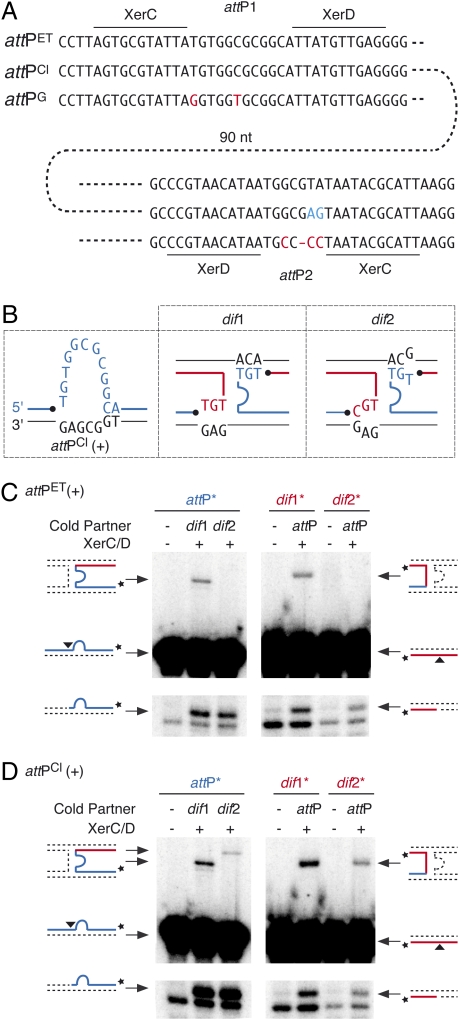
We next investigated which differences in the sequences of RSCl and RSET were involved in their different integration behaviors. Because these differences were numerous, we decided to restrict the number of positions tested by searching for CTXφ residues that would be specifically conserved among classical or El Tor isolates in the available genomic sequences of toxigenic V. cholerae strains. During this search, we identified three categories of CTXφ attachment regions, two of which seemed specifically linked to classical and El Tor isolates (Fig. 2A and Table S4). Indeed, the attachment region of classical variants of CTXφ, attPCl, differs from the attachment region of El Tor and O139 variants, attPET, by two residues in the overlap region between the XerC- and XerD-binding sites of attP2 (Fig. 2A, blue residues). Introduction of the classical residues in the attachment region of RSET led to its efficient integration at both dif1 and dif2 (Table 1), and introduction of the El Tor residues in the attachment region of RSCl abolished dif2 integration (Table 1), demonstrating that the respective relaxation and specific integration behaviors of RSCl and RSET are determined solely by a difference of sequence at these two positions.
Fig. 2.
The (+) ssDNA of classical variants of CTXφ recombines with both dif1 and dif2. (A) The three categories of phage attachment regions found in CTXφ variants. Residues specifically conserved in classical and G variants of the phage are shown in blue and red, respectively. (B) Schemes of Watson–Crick bp interactions that could stabilize the strand exchange catalyzed by XerC between the overlap regions of the two dimer resolution sites of N16961 and the attachment site found in the (+) ssDNA genome of classical variants of CTXφ. The legend is as in Fig. 1C. (C) V. cholerae XerCD-mediated recombination of attPET(+) with dif1 and dif2. A short radioactively labeled attP substrate was reacted with a longer cold dimer resolution substrate (Left Panels), and a short radioactively labeled dimer resolution substrate was reacted with a longer cold attP substrate (Right Panels). Schemes of substrate and products are indicated on the side of each panel. A black triangle indicates the position of cleavage of V. cholerae XerC. A star indicates the position of the radioactive label of the probe. (D) V. cholerae XerCD-mediated recombination of attPCl(+) with dif1 and dif2. Schemes of the products and of the substrate are indicated as in Fig. 2B.
XerCD Recombines attPCl(+) with dif1 and dif2 in Vitro.
The T-to-G transversion found in the overlap region of classical attP2 allows the recovery of one Watson–Crick bp interaction on one side of the reaction between the (+) ssDNA of attPCl and dif2 (Fig. 2B, red strand). Perfect Watson–Crick pairing is not recovered on the other side but is replaced by a TG wobble bp (Fig. 2B, blue strand). Likewise, one Watson–Crick bp is replaced by a TG wobble bp on one side of the reaction between the stem of the hairpin formed by the (+) ssDNA of attPCl and dif1 (Fig. 2C, red strand). This finding prompted us to check whether V. cholerae XerC and XerD could recombine the (+) ssDNA of attPCl with dif1 and dif2 in vitro. We did so using purified V. cholerae XerC and XerD proteins and annealed synthetic oligonucleotides that mimic dif1, dif2, and the stems of the hairpins created by attPCl and attPET (Table S5).
Three steps can be defined in the strand exchange reaction performed by tyrosine recombinases: first, a single strand in each of the two recombining sites is cleaved by one recombinase, generating two 3′-phosphorotyrosyl recombinase/DNA covalent intermediates; the liberated 5′-hydroxyl extremities then are exchanged; finally, they attack the phosphorotyrosyl bond of the partner site to form phosphodiester bonds. Cleavage of each of the two recombining strands and their subsequent ligation to the opposite partner strand can be monitored when the strand is labeled at its 3′ extremity: Strand cleavage leads to the apparition of a shorter migration product on a sequencing gel; ligation to a partner strand harboring a longer extension on the 5′ side of the XerC-binding site leads to the creation of a longer recombinant product. Ligation products were detected when attPET(+) was reacted against dif1 (Fig. 2C, Upper) and when attPCl(+) was reacted against dif1 or dif2 (Fig. 2D, Upper). Thus, in the absence of any other host or phage factors, XerC and XerD can promote the exchange of one pair of strands when one Watson–Crick bp is replaced by a TG wobble bp on one side of the recombining complex. However, no ligation product was detected between attPET(+) and dif2; Watson–Crick pairing is lost on both sides of this reaction (Fig. 2C, Upper).
Design of a dif2-Specific Phage Variant.
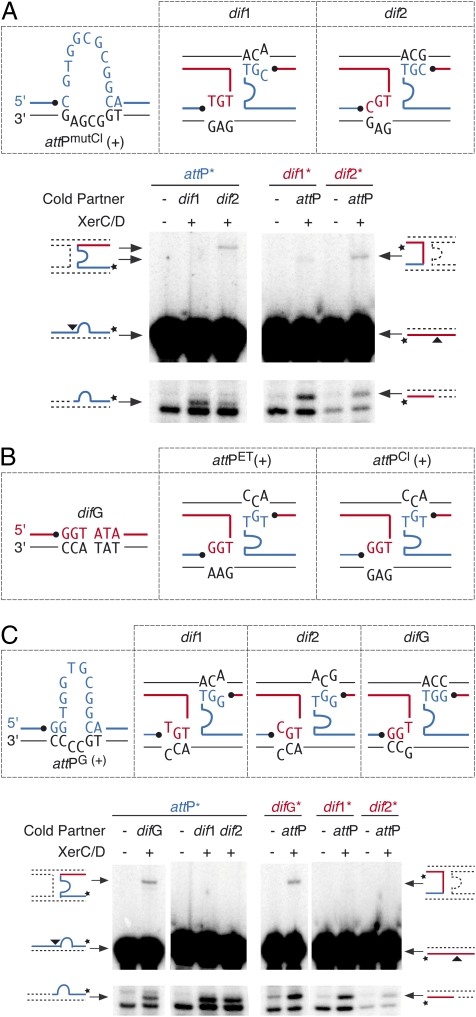
To demonstrate further that the specificity of integration of the different variants of CTXφ is governed by the ability to establish bp interactions that stabilize strand exchanges, we designed a phage that, based on the ssDNA integration model, should integrate specifically at dif2. To this aim, the thymine immediately downstream of the site of cleavage of XerC was replaced by a cytosine in attPCl(+) [attPmutCl(+); Fig. 3A]. Because of this substitution, perfect Watson–Crick pairing is lost on both sides of the reaction with dif1 (Fig. 3A). However, it is re-established on both sides of the reaction with dif2 (Fig. 3A). As expected, we observed a normal number of XerC-mediated strand exchanges between attPmutCl(+) and dif2 in vitro, whereas recombination with dif1 was barely detectable (Fig. 3A). In vivo, this process resulted in the fully specific integration of a vector carrying the attPmutlCl(+) attachment site at dif2 (attPmutCl; Table 2).
Fig. 3.
Homology determinants implicated in lysogenic conversion. (A) Design of a variant of CTXφ specifically integrating at dif2. The legend is as in Figs. 1C and 2C. (B) Scheme showing the possible pairing interactions between difG and attPET(+) or attPCl(+). The legend is as in Fig. 1C. (C) V. cholerae XerCD-mediated recombination of attPG(+) with dif1, dif2, and difG. A scheme of the possible pairing interactions is shown above the gels. The legend is as in Figs. 1C and 2C.
Table 2.
Integration compatibility of CTXφ attP(+) sites and of dif sites
| Phage machinery | attP | dif | % integration | Screened colonies |
| El Tor | mutCl | dif1* | <0.1 | 1256 |
| El Tor | mutCl | dif2† | 30.4 | 1449 |
| El Tor | El Tor | difG* | <0.1 | 1335 |
| Classical | Classical | difG* | <0.1 | 1399 |
| El Tor | G | difG* | 1.1 | 2110 |
| El Tor | G | dif1* | <0.3 | 307 |
| El Tor | G | dif2† | <0.4 | 271 |
Data were obtained from at least three independent experiments.
*On chromosome 1.
†On chromosome 2.
Rules of Compatibility Drive the Evolution Between Phage Attachment and Dimer Resolution Sites.
We next investigated the sequence of the dimer resolution sites of natural CTXφ- V. cholerae strains. We found that the first chromosome of many recently isolated O1 El Tor, O139 and non-O1 non-O139 strains harbors a dimer resolution site with an overlap region different from that of dif1 and dif2; we called this site “difG” (Fig. 3B and Table S6). RSET and RSCl did not integrate at difG in vivo (Table 2). This result was expected, because no bp interactions could stabilize strand exchanges between difG and attPET(+) or attPCl(+) (Fig. 3B). However, the stem of the hairpin created by the folding of the third category of attachment sites that was identified during our database searches (attPG; Fig. 2A) seemed suitable for integration with difG (Fig. 3C). In contrast, attPG(+) did not seem suitable for integration at dif1 or dif2. Correspondingly, V. cholerae XerCD specifically recombined attPG(+) with difG in vitro (Fig. 3C), and a phage harboring attPG specifically integrated at difG in vivo (Table 2) independently of the chromosomal context of the site (Table S7).
Discussion
Previous work on the El Tor variant of CTXφ suggested that it specifically integrates at dif1 (6). In this context, the integration of CTXφ at dif2 in classical strains (3) and in recent El Tor isolates (4, 10–14) was puzzling. It was observed recently that El Tor strains can acquire the dif2-integrated classical copy of CTXφ via chitin-induced competence and homologous recombination (18). However, such a mechanism left open the questions of how dif2 integration could be achieved initially and the frequency at which this integration could occur. In addition, it did not explain the diversity of combinations of the different genetic elements that were found integrated at the chromosome dimer resolution sites loci of V. cholerae strains (4). To address these points, we designed a sensitive assay to monitor the specificity and efficiency of dif1 and dif2 integration events. We then cloned the replication and integration region of the variant of CTXφ found in the 569B classical strain into a vector harboring a conditional origin of replication; this vector was delivered into N16961 El Tor cells by conjugation. Surprisingly, we observed that this vector efficiently integrated at both dif1 and dif2 independently of the chromosomal context of the sites (Table 1). This result explains how the classical variant of CTXφ could integrate in the genome of classical strains even if their two chromosomes share the same dif2 dimer resolution site (CP000626.1) and why it was found integrated at dif1 on the first chromosome of the El Tor BX330286 strain although such a configuration had not been observed previously in any other strain (4). In contrast, the N16961 El Tor variant of CTXφ specifically targeted dif1 (Table 1). Likewise, we observed that classical and El Tor variants of CTXφ could not integrate at difG, the dimer resolution site on the larger chromosome of many nontoxigenic strains, but that difG is targeted specifically by a third type of phage that was isolated recently (Table 2).
CTXφ integration is irreversible (6, 9), and the secretion of new phage particles relies on the production of its ssDNA genome by rolling circle replication across tandemly integrated copies (20, 21). However, we observed that RSCl can integrate in tandem copies that should ensure such a production (Fig. S1). Correspondingly, the sequencing of the genome of several V. cholerae strains revealed the presence of tandem copies of the classical phage (El Tor strains BX330286 and B33) (4). In addition, it was observed that classical copies of the phage coexist with intact El Tor copies and/or with the RS1 element in other strains (4, 14); such coexistence also could ensure the production of classical CTXφ virions. Interestingly, the conjunct production of El Tor and classical phages could favor the emergence of hybrid phages, a phenomenon that has been observed recently (Fig. 1A) (4, 13). Taken together, these observations suggest that lysogenic conversion by CTXφ is the primary mode of acquisition of the cholera toxin genes; this mode of acquisition, along with the evolution of phage attachment and chromosome dimer resolution sites, contributes to the generation of genetic diversity among cholera epidemic strains.
Our results further indicate that the relaxation and specific integration behaviors of the classical and the El Tor variants of CTXφ are determined solely by a difference in sequence at two positions in the seven-bp overlap region of attP2, immediately 3′ to the position at which XerC should cleave (Fig. 1B). Two models have been proposed for CTXφ integration at dif1 (8, 9). In the first model, XerCD would catalyze the formation of a Holliday junction between the host dimer resolution sites and the dsDNA form of attP1, which is found on the replicative form of the phage (8). In this model, attP2 is thought to play a structural role in stabilizing the synapse and/or the exchange; this assumption makes it difficult to explain how mutations in its overlap region could influence the specificity of integration. In contrast, the base immediately 3′ to the XerC cleavage site in the overlap region of attP2 contributes to the formation of bp interactions during the strand exchange predicted by the ssDNA integration model (Fig. 1C) (9). Here, we show that all of the combinations between (+) ssDNA phage attachment sites and chromosome dimer resolution sites in which Watson–Crick bp interactions could stabilize the exchange of strands catalyzed by XerC [attPET(+)xdif1, attPmutCl(+)xdif2, and attPG(+)xdifG] were recombined in vitro and promoted integration in vivo. No in vitro recombination and no in vivo integration were detected for combinations in which no Watson–Crick bp interactions could be formed [attPET(+)xdif2, attPmutCl(+)xdif1, attPG(+)xdif1, and attPG(+)xdif2]. In the remaining two combinations we tested [attPCl(+)xdif1 and attPCl(+)xdif2], proper Watson–Crick bps could form only on one side of the recombination complex, but nevertheless in vitro recombination and in vivo integration were observed. To stabilize the exchange of strands, tyrosine recombinases normally require one Watson–Crick bp interaction on each side of the recombination complex immediately after the position of cleavage of the recombinases (22–31). The only notable exception to this rule is the integrase of some bacteroides conjugative transposons (32). In this context, it does not seem fortuitous that a TG wobble bp replaces the missing Watson–Crick bp in reactions between attPCl(+) and dif1 or dif2 (Fig. 2B). Taken together, these results give considerable support to the ssDNA integration model and allow the definition of rules of compatibility between phage attachment and dimer resolution sites that dictate the possibility for lysogenic conversion.
Finally, in four of the five effective combinations of attachment and dimer resolution sites we tested [attPET(+)xdif1, attPCl(+)xdif1, attPCl(+)xdif2, and attPmutCl(+)xdif2], the efficiency of integration correlated with the efficiency of recombination observed in vitro. Furthermore, the in vitro recombination efficiency of these four combinations (Fig. 2 C and D and Fig. 3A, Upper) correlated with the stability and/or frequency of formation of the two cleaved substrates involved in the strand exchange (Fig. 2 C and D and Fig. 3A, Lower). For instance, the lower efficiency of the recombination of attPmutCl(+)xdif2 when compared with attPET(+)xdif1, attPCl(+)xdif1, and attPCl(+)xdif2 is explained by the lower stability of the dif2/XerC and attPmutCl/XerC covalent complexes compared with the dif1/XerC, attPET/XerC, and attPCl/XerC covalent complexes (with respective mean frequencies of 0.15 ± 0.08%, 0.16 ± 0.08%, 0.46 ± 0.18%, 0.41 ± 0.12%, and 1.19 ± 0.65% out of at least four independent experiments). The relatively low number of dif2/XerC covalent intermediates fits with previous results that indicated that the control of recombination is more stringent at dif2 than at dif1 (5). The relatively low number of attPmutCl/XerC covalent intermediates further suggests that the presence of a cytosine immediately 5′ to the XerC cleavage site is detrimental to XerC cleavage and/or to the stability of the cleaved complex. In contrast, the frequency with which a vector carrying the attPG attachment region integrated at difG was low when compared with the efficiency of the attPG(+)xdifG recombination reaction in vitro (Table 2 and Fig. 3B). This finding suggests that factors other than the efficiency with which strand exchanges are performed govern CTXφ lysogeny. Such factors also could explain why integration was less efficient for vectors harboring the classical replication machinery than for vectors harboring the El Tor replication machinery (Tables 1 and 2). Some factors could play a role in the production and/or stabilization of the (+) ssDNA genome of the phage. Others might play a role in the conversion of the Holliday junction intermediate into fully recombinant products by repair and/or replication. The observation that attPG and RSCl are detrimental to some of these factors should help unravel these other important aspects of CTXφ integration.
Methods
Strains and Plasmids.
Relevant strains and plasmids are described in Tables S1 and S2, respectively. Both E. coli and V. cholerae cells were grown in LB at 37 °C with shaking (220 rpm). Unless otherwise indicated, cognate antibiotics were used at the following concentrations: streptomycin, 100 μg/mL; spectinomycin, 100 μg/mL; chloramphenicol, 34 μg/mL for E. coli and 3 μg/mL for V. cholerae; and rifampicin, 100 μg/mL for E. coli and 2 μg/mL for V. cholerae. All V. cholerae reporter strains were constructed by allele exchange methods using derivatives of suicide vectors carrying either sacB or rpsL as a counter selectable marker (33, 34). Engineered strains were confirmed by PCR and sequencing. For long storage, cells were maintained at –70 °C in LB containing 20% glycerol. Classical and El Tor phage replication and integration machinery (RS elements) were amplified using genomic DNA of V. cholerae strains 569B or N16961, respectively, as templates. The amplicons were cloned into the suicide vector pSW23T (30). Plasmids carrying hybrid phage variants were engineered by inverse PCR. The recombinant suicide vectors carrying the functional lacZ::dif allele flanking by the chromosomal fragments of V. cholerae were constructed by cloning the 28-bp dif site in the natural ClaI site of the E. coli lacZ gene.
In Vivo Integration Assay.
For conjugation both donor [diaminopimelic acid (DAP) auxotroph E. coli)] and recipient (V. cholerae) strains were grown separately in LB to an OD of ∼0.3 at 600 nm. Bacteria were pelleted by centrifugation, washed, and mixed in 25% of the initial volume in fresh LB supplemented with 0.3 mM DAP. The mixture then was deposited on a sterile filter paper covering an LB-agar plate supplemented with DAP. After 4 h of incubation at 37 °C, bacterial cells were resuspended and plated on LB plates containing X-gal isopropyl β-D-1-thiogalactopyranoside (IPTG) and cognate antibiotics. Transconjugants carrying integrated or replicative forms of phage machinery were monitored after 36 h of growth at 37 °C.
Protein Purification.
The XerC and XerD ORFs were amplified by PCR from N16961 genome and cloned into the pTYB-11 (New England Biolabs) expression vector using SapI and PstI restriction sites. Proteins were produced at 30 °C in BL21Gold cells (Stratagene). XerD-producing cells were grown for 2 h in the presence of 0.1 mM IPTG. XerC-producing cells were grown for 2 h in the presence of 0.2% glucose and 0.5 mM IPTG. Cells were collected and resuspended in buffer A (25 mM TrisHCl, pH8/1 M NaCl/10% glycerol), frozen in dry ice, and lysed with a Carver press. Lysates were centrifuged for 1 h at 25,000 × g. The supernatants were loaded on chitin bead columns and washed extensively with buffer A. Intein tag cleavage was performed in buffer A adjusted to 0.5 M NaCl and supplemented with 50 mM DTT at 7 °C for 16 h. Untagged XerC and XerD were eluted, and small aliquots were frozen and stored at −70 °C. Protein concentrations were evaluated by the Bradford methods using BSA as standard.
In Vitro Recombination Assays.
Synthetic oligos used to mimic dif1, dif2, difG, attPET, attPCl, attPmutCl, and attPG are shown in Table S5. Recombination reactions were performed in a 20-μL volume, in the presence of 25 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, 0.1 μg/mL BSA, 40% glycerol, and 5 nM each of the cold and radioactively labeled recombination substrates. XerC and XerD were used at 150 nM and 100 nM final concentrations, respectively. Reactions were incubated for 3 h at 37 °C, ethanol precipitated, and analyzed by PAGE using a 10% acrylamide-urea gel. Dried gels were exposed to phosphor screen. Signals were detected using a Typhoon instrument and quantitated using the IQT 7.0 software (GE Healthcare).
Supplementary Material
Acknowledgments
We thank R.K. Bhadra and D. Mazel for the kind gift of V. cholerae strains and C. Possoz for helpful discussions. This work was supported by the Fondation pour la Recherche Médicale (Equipe 2007), the European Molecular Biology Organization (YIP 2006), and the Centre National pour la Recherche Scientifique (ATIP+).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online awww.pnas.org/cgi/content/full/0910212107/DCSupplemental.
See Commentary on page 3951.
References
- 1.De SN. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature. 1959;183:1533–1534. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- 2.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 3.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype Vibrio cholerae: Functional phage genes but dysfunctional phage genomes. J Bacteriol. 2000;182:6992–6998. doi: 10.1128/jb.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun J, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Val M-E, et al. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 2008;4:e1000201. doi: 10.1371/journal.pgen.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 7.Barre F-X, Sherratt DJS. Xer site-specific recombination: Promoting chromosome segregation. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Vol. 1. Washington, D.C.: ASM Press; 2002. pp. 149–161. [Google Scholar]
- 8.McLeod SM, Waldor MK. Characterization of XerC- and XerD-dependent CTX phage integration in Vibrio cholerae. Mol Microbiol. 2004;54:935–947. doi: 10.1111/j.1365-2958.2004.04309.x. [DOI] [PubMed] [Google Scholar]
- 9.Val M-E, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Das B, Halder K, Pal P, Bhadra RK. Small chromosomal integration site of classical CTX prophage in Mozambique Vibrio cholerae O1 biotype El Tor strain. Arch Microbiol. 2007;188:677–683. doi: 10.1007/s00203-007-0275-0. [DOI] [PubMed] [Google Scholar]
- 11.Faruque SM, et al. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci USA. 2007;104:5151–5156. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ledón T, et al. El Tor and Calcutta CTXPhi precursors coexisting with intact CTXPhi copies in Vibrio cholerae O139 isolates. Res Microbiol. 2008;159:81–87. doi: 10.1016/j.resmic.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Minh NB, et al. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical type cholera toxin B in Vietnam 2007-2008. J Clin Microbiol. 2009 doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raychoudhuri A, et al. Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis. 2009;15:131–132. doi: 10.3201/eid1501.080543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung DT, Mekalanos JJ. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci USA. 2005;102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mekalanos JJ. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 17.Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udden SM, et al. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci USA. 2008;105:11951–11956. doi: 10.1073/pnas.0805560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldor MK, Rubin EJ, Pearson GD, Kimsey H, Mekalanos JJ. Regulation, replication, and integration functions of the Vibrio cholerae CTXphi are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 20.Davis BM, Waldor MK. CTXphi contains a hybrid genome derived from tandemly integrated elements. Proc Natl Acad Sci USA. 2000;97:8572–8577. doi: 10.1073/pnas.140109997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyer KE, Kimsey HH, Waldor MK. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXphi. Mol Microbiol. 2001;41:311–323. doi: 10.1046/j.1365-2958.2001.02517.x. [DOI] [PubMed] [Google Scholar]
- 22.Arciszewska L, Grainge I, Sherratt D. Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J. 1995;14:2651–2660. doi: 10.1002/j.1460-2075.1995.tb07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunes-Düby SE, Yu D, Landy A. Sensing homology at the strand-swapping step in lambda excisive recombination. J Mol Biol. 1997;272:493–508. doi: 10.1006/jmbi.1997.1260. [DOI] [PubMed] [Google Scholar]
- 24.Nunes-Düby SE, Azaro MA, Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site-specific recombination. Curr Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 25.Lee SY, Landy A. The efficiency of mispaired ligations by lambda integrase is extremely sensitive to context. J Mol Biol. 2004;342:1647–1658. doi: 10.1016/j.jmb.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhu XD, Pan G, Luetke K, Sadowski PD. Homology requirements for ligation and strand exchange by the FLP recombinase. J Biol Chem. 1995;270:11646–11653. doi: 10.1074/jbc.270.19.11646. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Jayaram M. Role of partner homology in DNA recombination. Complementary base pairing orients the 5′-hydroxyl for strand joining during Flp site-specific recombination. J Biol Chem. 1995;270:4042–4052. doi: 10.1074/jbc.270.8.4042. [DOI] [PubMed] [Google Scholar]
- 28.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 30.Demarre G, et al. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol. 2005;156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Barabas O, et al. Mechanism of IS200/IS605 family DNA transposases: Activation and transposon-directed target site selection. Cell. 2008;132:208–220. doi: 10.1016/j.cell.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malanowska K, Yoneji S, Salyers AA, Gardner JF. CTnDOT integrase performs ordered homology-dependent and homology-independent strand exchanges. Nucleic Acids Res. 2007;35:5861–5873. doi: 10.1093/nar/gkm637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.