Abstract
Analysis of chromosomal aberrations is used to determine the prognosis of neuroblastomas (NBs) and to aid treatment decisions. MYCN amplification (MNA) alone is an incomplete poor prognostic factor, and chromosome 11q status has recently been included in risk classification. We analyzed 165 NB tumors using high-density SNP microarrays and specifically compared the high-risk groups defined by MNA (n = 37) and 11q-deletion (n = 21). Median patient age at diagnosis was 21 months for MNA tumors and 42 months for 11q-deletion tumors, and median survival time after diagnosis was 16 months for MNA and 40 months for 11q deletion. Overall survival (at 8 years) was ∼35% in both groups. MNA and 11q deletion were almost mutually exclusive; only one case harbored both aberrations. The numbers of segmental aberrations differed significantly; the MNA group had a median of four aberrations, whereas the 11q-deletion group had 12. The high frequency of chromosomal breaks in the 11q-deletion group is suggestive of a chromosomal instability phenotype gene located in 11q; one such gene, H2AFX, is located in 11q23.3 (within the 11q-deletion region). Furthermore, in the groups with segmental aberrations without MNA or 11q deletion, the tumors with 17q gain have worse prognosis than those with segmental aberrations without 17q gain, which have a favorable outcome. This study has implications for therapy in different risk groups and stresses that genome-wide microarray analyses should be included in clinical management to fully evaluate risk, aid diagnosis, and guide treatment.
Keywords: comparative genomic hybridization/SNP array, chromosomal aberration, H2AFX, MYCN amplification, pediatric tumor
Neuroblastoma (NB), a tumor of the sympathetic nervous system, is the most common extracranial solid tumor of childhood. Clinical manifestations vary from aggressive malignant growth to spontaneous regression. Tumors with near-triploid karyotypes with numerical aberrations (i.e., whole-chromosome gains and losses) have a good prognosis, whereas tumors with near-diploid or near-tetraploid karyotypes and segmental rearrangements, including deletions of parts of chromosome arms 1p or 11q, gain of 17q, and amplification of the MYCN proto-oncogene, have a poor prognosis (1–3). Children under age 1 year with NB generally present with a localized tumor and have an excellent outcome, whereas older children more often have aggressive NB with a poor prognosis despite intensive treatment (4). MYCN status, 11q23 allele status, and tumor ploidy recently have been included in the International Neuroblastoma Risk Group (INRG) classification system, after a review of the INRG database, which contains 8,800 patients with NB (5).
Genomic microarrays, either comparative genomic hybridization (CGH) or SNP arrays, are good tools for analyzing chromosomal rearrangements in tumors. Several studies have analyzed chromosomal breaks in NB on a genome-wide scale (2, 6–14). Recently, a study using array-based CGH to investigate nearly 500 NB tumors revealed two genetic classes of NB apparently related to different mechanisms of instability (15). Whole chromosome changes without segmental alterations were associated with an excellent outcome, even in patients age >18 months or in an advanced disease stage. The other class included tumors with segmental chromosomal alterations in which 1p, 3p, and 11q deletions and 1q, 2p, and 17q gains were identified as statistically significant prognostic factors; the authors suggested that any segmental alteration is associated with an increased risk of relapse. MYCN amplification and 1p and 11q deletion identified patients with particularly aggressive relapse and decreased overall survival.
We previously presented an analysis of 92 NB tumors performed with SNP arrays from Affymetrix, which provided both copy-number and allele-specific information at a resolution of 10–12 kb (16). We extended this study to include a total of 165 Swedish NB tumors. Here we highlight the features and differences in the larger number of NB tumors of all clinical stages, with emphasis on the 11q-deletion phenotype, which exhibits a poor prognostic chromosome instability phenotype with late onset of disease.
Results
Overall Genomic Data.
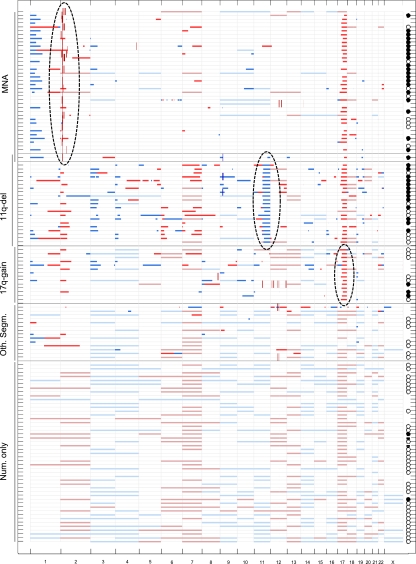
The genomic features of MYCN amplification, 11q deletion, and 17q gain represent segmental aberrations known to be involved in unfavorable NBs; thus, the presence/absence of these features was considered in the grouping of tumors. Of the 165 tumors analyzed, 37 displayed MYCN amplification (without 11q deletion), 21 displayed 11q deletion (without MYCN amplification), 1 displayed both MYCN amplification and 11q deletion, 14 displayed 17q gain (without MYCN amplification or 11q gain), and 14 displayed other segmental aberrations (involving neither MYCN amplification, 11q deletion, nor 17q gain). In addition, 47 cases displayed only numerical changes, whereas 31 cases displayed a flat profile (Table 1). It is evident that MYCN amplification and 11q deletion represent two different groups of aggressive NBs. It also is clear that 17q gain occurs at high frequency in both of these groups, but that 17q gain is sometimes present without MYCN amplification or 11q deletion. A large group of NBs, generally with favorable outcomes, exhibit only numerical aberrations, with gains and/or losses of whole chromosomes. In several cases, we detected neither segmental nor numerical aberrations, but found a normal-looking, so-called “flat” profile. This overall view provides the basis for our grouping of tumors (Fig. S1). The data for all of the genomic profiles except the flat profile group are summarized in Fig. 1. Fig. 1 also includes outcome data for patients who are dead of disease (filled circles to the right), those with an overall survival of at least 5 years (open circles), and those who died from surgical complications (horizontal line).
Table 1.
Genomic profiles in the analyzed tumors (n = 165)
| Group | Cases, n |
| MNA | 37 |
| MNA + 11q-del | 1 |
| 11q-del | 21 |
| 17q-gain (no MNA, no 11q-del) | 14 |
| Other segmental aberrations | 14 |
| Numerical only (whole chromosome loss and/or gain only) | 47 |
| Flat profile | 31 |
Fig. 1.
Summary of SNP array data of gains and losses for 134 NB cases. Horizontal lines show segmental loss (clear blue) and gain (clear red) and whole chromosome loss (pale blue) and gain (pale red). Short vertical lines show amplification (brown) or homozygous loss (dark blue). The genomic profile group is indicated to the left. The inclusion features for the three high-risk groups—MYCN amplification, 11q deletion, and 17q gain—are indicated by dotted ovals. Note that only one case has both MYCN amplification and 11q deletion. Patients who have died of disease are indicated to the right by filled black circles, whereas patients who are still alive and have an overall survival of at least 5 years are indicated by open circles. Two cases in the numerical-only group are represented by a horizontal line; these patients died of surgical complications. Cases with a flat profile (n = 31) are not shown. See Materials and Methods for definitions of the genomic groups.
Comparison of Segmental Aberrations in the Genomic Profile Groups.
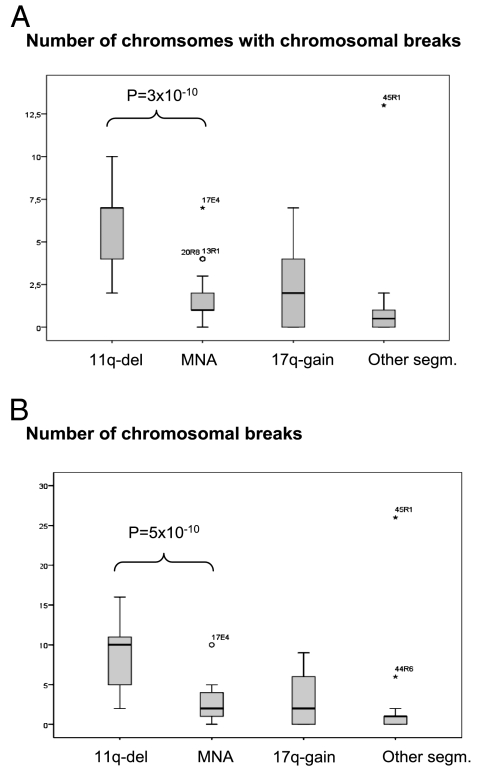
All chromosome breakpoints were recorded for all cases. Breakpoints for the MNA group (n = 298, 37 tumors) and the 11q-deletion group (n = 248, 21 tumors) are plotted in Fig. S2. In Fig. 2, the data are corrected for ascertainment bias, with breaks associated with MYCN amplification and 11q deletion omitted for MNA cases and 11q-deletion cases, respectively, as well as for 17q gain. Tumors with 11q deletion contained significantly more chromosomes with breaks compared with MYCN-amplified tumors (Fig. 2A; median of one chromosome with breaks in MNA versus seven chromosomes in 11q-deleted; P = 3 × 10−10, adjusted for ascertainment bias) and significantly more chromosomal breaks overall (Fig. 2B; median value of 4 breaks in MNA vs. 12 breaks in 11q-deleted; adjusted P value = 5 × 10−10). The 17q-gain group had a median value of four breaks, and the other-segmental group had a median of one break.
Fig. 2.
Boxplots showing that the 11q-deletion tumors have significantly more chromosomal breaks than the other groups. (A) Number of chromosomes with chromosomal breaks for each case. (B) Number of chromosomal breaks per case. In the boxplots, the upper and lower hinges of the box represent the 75th and 25th percentiles, respectively; whiskers indicate the highest and lowest values that are not outliers or extreme values; the thick horizontal line represents the median; open circles represent outliers; and asterisks represent extremes. The groups were compared after adjustment for ascertainment biases. The level of significance for the difference between the 11q-deleted and MYCN-amplified groups is indicated in both figures.
The relative distribution of breaks differed significantly (P < 0.001) between the two groups. The breakpoints in the MNA group focused on 1p and 6q, in addition to the 17q breakpoint seen in both groups. In contrast, the breakpoints in the tumors with 11q deletion were spread more evenly over the genome part from the concentration on 17q.
Correlation of Genomic and Clinical Data.
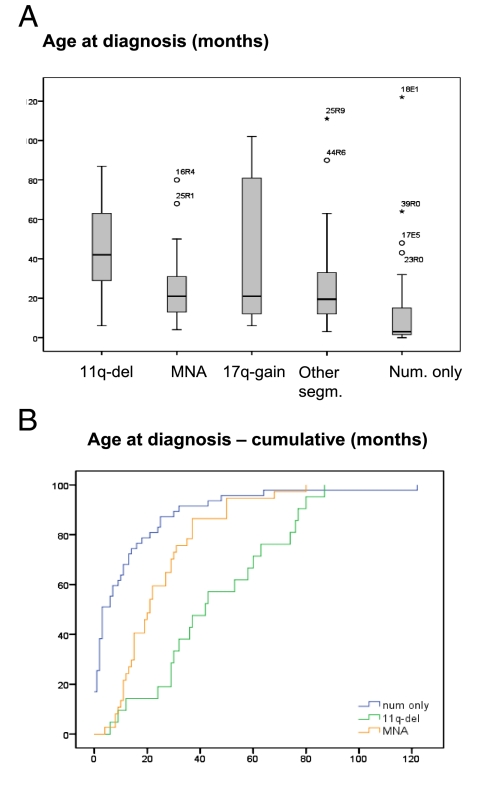
A significant difference in age at diagnosis based on genomic profile was seen (Fig. 3). The median age at diagnosis was 3 months in the numerical-only group, 21 months in the MYCN-amplified and 17q-gain groups, and 42 months in the 11q-deletion group (P = 0.002, MNA vs. 11q-del).
Fig. 3.
Age at diagnosis of the 165 NB patients by genomic profile group, showing a significant difference in age at diagnosis between groups of MYCN-amplified cases versus 11q-deleted cases. (A) Boxplot showing age at diagnosis by group. (B) Cumulative age at diagnosis of disease.
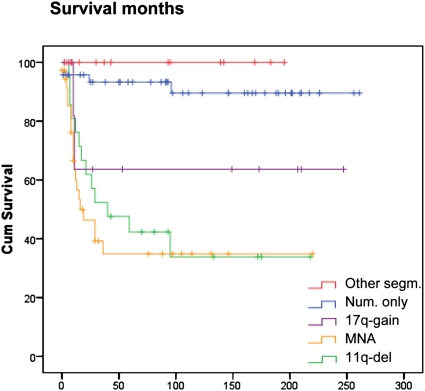
In terms of overall survival, the MYCN-amplified and the 11q-deleted tumors both had a very poor prognosis. The median survival after diagnosis was 16 months for MYCN-amplified group and 40 months for the 11q-deletion group (P = 0.002; MNA vs. 11q-del), but overall survival at 8 years was ∼35% for both groups (Fig. 4). This is in contrast to the numerical-only group, in which only four cases had a fatal outcome, two due to surgical complications. The 17q-gain group had an overall survival of ∼60%. No patients in the other-segmental group died of disease.
Fig. 4.
Kaplan-Meier overall survival for patients with tumors with different genomic profiles. The tumors are grouped as follows: the other segmental group (red line), the numerical-only group (blue line), the 17q-gain group (violet line), the MYCN-amplification group (yellow line), and the 11q-deletion group (green line). The single case with both MYCN amplification and 11q deletion was omitted from the analysis. See Materials and Methods for definitions of the genomic profile groups.
Chromosomal Breaks in 11q-Deleted Tumors Relative to H2AFX.
The high frequency of chromosomal breaks in 11q-deleted tumors prompted us to analyze the position of the deletion breakpoint relative to the known chromosomal instability gene H2AFX located in 11q. Of the 21 tumors with 11q deletion (22 tumors when the single case with both MNA and 11q deletion was included), none had an interstitial deletion, and thus all had deletions extending from the breakpoint to the q-terminal of chromosome 11. A breakpoint cluster region was noted at positions 67–73 Mb from pter (15 cases; Fig. S3). The most distal breakpoint was at position 110 Mb from pter, which is proximal to the H2AFX locus at position 118 Mb from pter. Interestingly, one case demonstrated a small rearrangement located in the H2AFX region (Fig. S3, open arrow).
Analysis of H2AFX Expression.
Real-time RT-PCR was used to compare H2AFX expression in 11q-deleted tumors and tumors with intact chromosome 11. NB cell lines with and without 11q deletion were analyzed as well. The 11q-deleted tumors and cell lines had significantly less H2AFX transcripts than the tumors and cell lines with intact chromosome 11 (P = 0.04, fold change = 3.4 and P = 0.01, fold change = 2.4, respectively). The 11q-deleted tumors also had less H2AFX expression than the MYCN-amplified tumors (P = 0.02, fold change = 8.4) (Fig. S4).
Discussion
Ever since MYCN was identified as an oncogene and found to be highly amplified in NB tumors in the 1980s (17), it has been used as a diagnostic and prognostic marker (mainly in FISH analysis) for identifying this particular group of high-risk NB tumors. In addition, it has long been known that there are other high-risk NB groups, that is, non–MYCN-amplified NB cases (18). The use of whole-genome–based approaches, such as multiplex ligation–dependent probe amplification (MLPA), SNP arrays, and CGH, has identified new risk groups, and it is now clear that the 11q-deletion group represents a distinct high-risk group within the non–MYCN-amplified cases (16, 19).
We have analyzed 165 NB tumors from a cohort of Swedish patients using SNP arrays. These represent all NB cases in Sweden between 1980 and 2008 for which fresh or fresh-frozen tumor material was available. Clinical data on all patients were obtained from the Swedish Children’s Cancer Registry. A quick inspection of the SNP data (Fig. 1) and a comparison of the different genomic profile groups show that the MYCN-amplified and 11q-deletion groups stand out as unfavorable, with a large number of patients who have died of disease. This contrasts with the numerical-only group, for example, in which only 2 out of 41 patients have died of disease. Our data and previous data clearly show that the MYCN-amplified and the 11q-deletion groups are high-risk groups. Our data also clearly show that the two groups have significantly different genomic profiles (Fig. 1); whereas MYCN-amplified cases involve the MYCN amplification and often 1p deletion and 17q gain, but very few other segmental aberrations, the 11q-deletion cases generally present with many more segmental aberrations. The two groups also are almost mutually exclusive with respect to MYCN amplification and 11q deletion, with only one case exhibiting both of these features. Furthermore, in the 11q-deletion cases, 17q gain is a very common feature, but the segmental loss of several other chromosomes are seen as well. These intriguing similarities and differences between the MYCN-amplified and the 11q-deleted cases led us to scrutinize these features.
A comprehensive breakpoint analysis revealed that the 11q-deleted tumors have a significantly higher frequency of chromosomal breaks in tumor cells, with more chromosomes exhibiting chromosomal breaks (Fig. 2). Obviously, all of the MYCN-amplified tumors and the 11q-deletion tumors exhibit a cluster of breaks in distal 2p (site of the MYCN gene) and 11q, respectively. The distribution of the breaks on other chromosomes also appears to differ as well, with the MYCN profile showing a break pattern focusing on 1p and 6q, along with 17q and a few other positions, and the 11q breaks demonstrating a more “shotgun”-like distribution pattern (Fig. S2).
The MYCN-amplified and 11q-deleted groups were similar in terms of overall survival (Fig. 4). The overall survival after diagnosis was ∼35% for both groups after 8 years of follow-up, although the 11q-deleted group displayed a somewhat later slope. In contrast, there was a large difference between the MYCN-amplified group and the 11q-deleted group in terms of age at diagnosis (Fig. 3), with a median age at diagnosis of 21 months in the MYCN-amplified group and 42 months in the 11q-deleted group. In comparison, the numerical-only group, which had a very favorable outcome, with only 2 out of 41 patients dying of disease, had a median age at diagnosis of only 6 months. The other-segmental group had an excellent prognosis; no patients in this group have died of disease. The MYCN-amplified and 11q-deletion groups are significantly associated with poor prognosis. In the groups with segmental aberrations without MYCN amplification or 11q deletion, the tumors with 17q gain have a worse prognosis than those without 17q gain.
It is obvious that the 11q-deleted tumors constitute a distinct group of unfavorable NB tumors with distinct features that differ from those of the other important high-risk NB group, the MYCN-amplified tumors. These features are summarized in Table 2. Although both groups have features of high-risk tumors and similar adverse outcomes, they are mutually exclusive, having either MYCN amplification or 11q deletion. In our study, 37 cases had MYCN amplification, 21 cases had 11q deletion, and only 1 case had both of these features. We previously reported that the size of the shortest region of overlap (SRO) of 1p deletion differs between these two groups (16). With these expanded data, we are able to show that the chromosome 1p SRO of deletions is in positions 17–32 Mb (from 1pter) in the MYCN-amplified tumors and in positions 0–10.4 Mb in the 11q-deletion tumors, with no overlap between the two SROs (16). Thus, as has been suggested earlier (20, 21), it is possible that the elusive 1p tumor suppressor gene could be two different genes, one more proximal (17–32 Mb), associated with MYCN-amplified tumors, and one more distal (0–10.4), associated with 11q-deleted tumors.
Table 2.
Features of the 11q-del and MNA phenotypes
| MNA group | 11q-del group |
| Amplification of MYCN | No amplification of MYCN* |
| No 11q deletion* | 11q deletion |
| Few segmental aberrations (median, 4) | Many segmental aberrations (median, 12) |
| Chromosomal breaks on few other chromosomes in addition to chromosome 2 (location of MYCN) | Chromosomal breaks on several chromosomes in addition to chromosome 11 |
| Break distribution ”more focused” | Break distribution ”more shotgun-like” |
| 1p deletion region more proximal on 1p (consensus in 17–32 Mb) | 1p deletion region more distal on 1p (consensus in 0–10.4 Mb) |
| Poor prognosis; 8-year survival ∼35% | Poor prognosis; 8-year survival ∼35% |
| Median age at diagnosis 21 months | Median age at diagnosis 42 months |
| Median survival from diagnosis 16 months | Median survival from diagnosis 40 months |
| Significantly higher H2AFX expression† | Significantly lower H2AFX expression† |
Only rare cases of both MNA and 11q-deletion in the same tumor.
In NB primary tumors and cell lines.
Which genes could be involved in the distinctly unfavorable 11q-deletion NB tumor group? The high frequency of breaks in the 11q-deleted tumors, even though the patients are so young, led us to consider a chromosomal instability (CIN) phenotype gene for this type of tumor. One or more important genes on chromosome 11q that are lost in the 11q-deleted tumors could be responsible for this CIN. The other allele could be inactivated by mutations or epigenetic means, or else the gene could be subject to haploinsufficiency. One candidate gene is the H2AFX gene, which resides in the chromosome region 11q23.2-q23.3, at position 118.469.799–118.471.369 Mb, a region generally lost in tumors with 11q deletion (Fig. S3). H2AFX encodes a core histone H2A variant that constitutes ∼2%–25% of the cellular H2A and is randomly incorporated into nucleosomes. H2AFX is phosphorylated in chromatin flanking DNA double-strand breaks (DSBs), and H2AFX-deficient mammalian cells have defective chromosomal DSB repair, marked ionizing radiation sensitivity, and increased genomic instability (22, 23). The loss of one H2AFX allele has been shown to compromise genomic stability and to enhance the susceptibility to cancer in the absence of p53 (23). We hypothesize that the increased number of chromosomal breaks seen in tumors with 11q deletion can be explained in part by the loss of one copy of the H2AFX gene. Real-time RT-PCR analysis supports this concept, in that NB tumors and cell lines with 11q deletion have significantly less H2AFX gene transcripts than those with intact chromosome 11 (Fig. S4).
In our study, 19% of tumors had a flat profile with no aberrations. In most cases, this likely reflects the presence of normal tissue in the sample. The flat profile was seen mostly in tumors that had been diagnosed and resected during the 1980s and 1990s, probably reflecting the fact that these samples were contaminated with normal cells to a greater extent than more recent samples.
This study underscores the biological heterogeneity of NB. The vast differences in tumor genetics between the two high-risk NB groups discussed here raises the question of whether these two groups should receive the same type of high-risk treatment. In particular, if the hypothesis of a CIN phenotype gene is correct, then treatment of these tumors may benefit from the more radical surgical removal of all tumor cells, because the remaining tumor has a particularly high capacity to mutate. Moreover, our findings underscore the fact that whole genome–based detection approaches are essential to the clinical diagnosis of NB tumors. This could be accomplished using the MLPA technique with a large set of probes, but preferably would be done using dense genome-wide microarray analyses with CGH or, as in the present study, dense SNP arrays. FISH analyses with a limited number of probes are insufficient for a complete evaluation of a patient’s genomic profile and thus his or her risk, diagnosis, and further treatment.
Materials and Methods
Tumor Material and Microarray Experiments.
Microarray analyses of 92 NB tumors using Affymetrix human 250K gene mapping arrays have been reported previously (16). These analyses included the majority of cases from Sweden, together with a group of tumors from other countries. In this study, all available tumors from the Swedish NB Registry have been analyzed. These include 80 cases from the previous study and an additional 85 cases analyzed more recently using the same criteria, giving a total of 165 tumors for this study (Table S1). All tumors were staged according to International Neuroblastoma Staging System (INSS) (24) and INRG criteria (25) (Table S1). Ethical permission was granted by the local ethics committee. For real-time RT PCR analysis, 36 NB tumors were used (eleven 11q-deleted tumors and 25 tumors with intact chromosome 11, of which 9 were MYCN-amplified, 10 were from the other-structural group, and 6 were from the numerical-only group). In addition, seven NB cell lines without 11q-deletion (SK-N-BE (2), SK-N-SH, SHSY5Y, IMR-32, SK-N-F1, NB69, and SHEP) and three NB cell lines with 11q deletion (SK-N-AS, Kelly, and SK-N-DZ) was used.
Data Analysis.
Primary data analysis was performed using GDAS software (Affymetrix), and further statistical studies were performed using CNAG (Copy Number Analyzer for Affymetrix GeneChip Mapping arrays) version 3.0 (Genome Laboratory, Tokyo University; http://www.genome.umin.jp) (26, 27). All cases of chromosomal gain, loss, or amplification were scored for both segmental and numerical aberrations, including detailed information about the breakpoint positions when applicable. Tumors were divided into groups in essentially the same way as others have done previously (15, 19), with some minor modifications. The groups (detailed in Fig. S1) were (i) the MNA group, comprising cases with the MYCN amplification; (ii) the 11q-deletion group, comprising cases with 11q deletion; (iii) the 17q-gain group, comprising cases without MYCN amplification and without 11q deletion but with 17q gain; (iv) the other segmental group, comprising cases with segmental aberrations other than MYCN amplification, 11q deletion, or 17q gain; (v) the numerical-only group, comprising cases with no segmental aberrations but with numerical aberrations (i.e., whole chromosome gains and/or losses only); and (vi) the flat profile group, comprising cases with neither segmental nor numerical changes. In the series of Swedish NB tumors studied here, only one case had both MYCN amplification and 11q deletion.
Regions of gains and losses were recorded for each case and used as input data for a graphical presentation of all cases (Fig. 1) in a program written in MATLAB version 7.6.0 (MathWorks). For statistical purposes, a chromosomal break in a tumor was defined as a clear change in gene dose level in the genomic profile. In some cases, MYCN amplification was more complex, exhibiting a group of peaks around the MYCN locus. In these cases, MYCN amplification was counted as one genetic aberration event. Moreover, in rare cases, a single chromosome had a fragmentation pattern with more than 50 breaks, for example. To prevent single cases of this type from skewing the statistics, we set a permitted maximum of five breaks per chromosome. To avoid ascertainment bias in statistical analyses and comparisons of groups with MYCN amplification, 11q deletion, and 17q gain, the breaks associated with these specific aberrations were omitted in comparisons. For practical reasons, all notations of gain and loss were considered in relation to a nominal tumor karyotype. For example, some of the cases in the numerical-only group are in the triploid range; thus, our principle when presenting these data is that in a triploid tumor, the presence of three chromosomes represents “normality,” the presence of two copies of a chromosome constitutes loss, and the presence of four chromosomes constitutes gain. Plots were constructed using SPSS 16.0 for Windows (SPSS Inc.). Student’s two-sided t test was used to compare groups.
The relative distribution of genomic breaks was compared among MYCN-amplified, 11q-deleted, and 17q-gained tumors using the χ2 test, ignoring data from 11q, 2p, and 17q.
Expression Analysis.
cDNA preparation and expression analysis was performed essentially as described previously (28). The cDNA was verified to not contain genomic DNA before real-time RT-PCR analysis using TaqMan gene expression assays (Applied Biosystems). Reactions were run in duplicate, and quantification was performed by the standard curve method. All samples were normalized by dividing the concentration of the H2AFX gene by the concentration of the endogenous control GUSB in the same cDNA sample. The logarithms of the expression levels in the groups were compared using Student’s two-sided t test. Fold change between groups was calculated using geometric means of the relative expression levels.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Cancer Society, the Children’s Cancer Foundation, the Nilsson-Ehle Foundation, the Assar Gabrielsson Foundation, the Wilhelm and Martina Lundgren Research Foundation, and the Sahlgrenska University Hospital Foundation. H.C. received a fellowship from the Swedish Knowledge Foundation through the Industrial PhD program in Medical Bioinformatics at the Strategy and Development Office at Karolinska Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910684107/DCSupplemental.
References
- 1.Spitz R, et al. Favorable outcome of triploid neuroblastomas: A contribution to the special oncogenesis of neuroblastoma. Cancer Genet Cytogenet. 2006;167:51–56. doi: 10.1016/j.cancergencyto.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Spitz R, et al. Oligonucleotide array–based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer. 2006;45:1130–1142. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 4.Schwab M, Westermann F, Hero B, Berthold F. Neuroblastoma: Biology and molecular and chromosomal pathology. Lancet Oncol. 2003;4:472–480. doi: 10.1016/s1470-2045(03)01166-5. [DOI] [PubMed] [Google Scholar]
- 5.Cohn SL, et al. INRG Task Force. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaruffi P, et al. Identification and characterization of DNA imbalances in neuroblastoma by high-resolution oligonucleotide array comparative genomic hybridization. Cancer Genet Cytogenet. 2007;177:20–29. doi: 10.1016/j.cancergencyto.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Chen QR, Bilke S, Khan J. High-resolution cDNA microarray–based comparative genomic hybridization analysis in neuroblastoma. Cancer Lett. 2005;228:71–81. doi: 10.1016/j.canlet.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 8.Chen QR, et al. cDNA array–CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma. BMC Genomics. 2004;5:70. doi: 10.1186/1471-2164-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Preter K, et al. Combined subtractive cDNA cloning and array CGH: an efficient approach for identification of overexpressed genes in DNA amplicons. BMC Genomics. 2004;5:11. doi: 10.1186/1471-2164-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michels E, et al. Array CGH–based classification of neuroblastoma into genomic subgroups. Genes Chromosomes Cancer. 2007;46:1098–1108. doi: 10.1002/gcc.20496. [DOI] [PubMed] [Google Scholar]
- 11.Michels E, et al. Genome-wide measurement of DNA copy number changes in neuroblastoma: Dissecting amplicons and mapping losses, gains and breakpoints. Cytogenet Genome Res. 2006;115:273–282. doi: 10.1159/000095924. [DOI] [PubMed] [Google Scholar]
- 12.Mosse YP, et al. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer. 2007;46:936–949. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]
- 13.Selzer RR, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 14.Tomioka N, et al. Novel risk stratification of patients with neuroblastoma by genomic signature, which is independent of molecular signature. Oncogene. 2008;27:441–449. doi: 10.1038/sj.onc.1210661. [DOI] [PubMed] [Google Scholar]
- 15.Janoueix-Lerosey I, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 16.Carén H, et al. High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: Four cases of homozygous deletions of the CDKN2A gene. BMC Genomics. 2008;9:353. doi: 10.1186/1471-2164-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwab M, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 18.Guo C, et al. Allelic deletion at 11q23 is common in MYCN single-copy neuroblastomas. Oncogene. 1999;18:4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- 19.Schleiermacher G, et al. Société Française des Cancers de l’Enfant (SFCE) Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br J Cancer. 2007;97:238–246. doi: 10.1038/sj.bjc.6603820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeda O, et al. There may be two tumor-suppressor genes on chromosome arm 1p closely associated with biologically distinct subtypes of neuroblastoma. Genes Chromosomes Cancer. 1994;10:30–39. doi: 10.1002/gcc.2870100106. [DOI] [PubMed] [Google Scholar]
- 21.Caron H, et al. Evidence for two tumour suppressor loci on chromosomal bands 1p35-36 involved in neuroblastoma: One probably imprinted, another associated with N-myc amplification. Hum Mol Genet. 1995;4:535–539. doi: 10.1093/hmg/4.4.535. [DOI] [PubMed] [Google Scholar]
- 22.Bassing CH, et al. Histone H2AX: A dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 23.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodeur GM, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 25.Monclair T, et al. INRG Task Force. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nannya Y, et al. A robust algorithm for copy number detection using high-density oligonucleotide single-nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto G, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carén H, Fransson S, Ejeskär K, Kogner P, Martinsson T. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer. 2007;97:1416–1424. doi: 10.1038/sj.bjc.6604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.