Abstract
For the promise of human induced pluripotent stem cells (iPSCs) to be realized, it is necessary to ask if and how efficiently they may be differentiated to functional cells of various lineages. Here, we have directly compared the neural-differentiation capacity of human iPSCs and embryonic stem cells (ESCs). We have shown that human iPSCs use the same transcriptional network to generate neuroepithelia and functionally appropriate neuronal types over the same developmental time course as hESCs in response to the same set of morphogens; however, they do it with significantly reduced efficiency and increased variability. These results were consistent across iPSC lines and independent of the set of reprogramming transgenes used to derive iPSCs as well as the presence or absence of reprogramming transgenes in iPSCs. These findings, which show a need for improving differentiation potency of iPSCs, suggest the possibility of employing human iPSCs in pathological studies, therapeutic screening, and autologous cell transplantation.
Keywords: neural development, regeneration, reprogramming, transplantation, motor neurons
Human embryonic stem cells (hESCs) can efficiently differentiate into functional neurons and glia with a mechanism akin to in vivo development (1–6). However, the optimism surrounding hESCs has been hampered by ethical concerns as well as the potential immune rejection on transplantation because of the nature of allograft. Induced pluripotent stem cells (iPSCs), which have been shown to be truly pluripotent in mouse studies (7–9), have also been generated from human somatic cells, such as skin cells, by a set of core pluripotent transcription factors (10–12). These human iPSCs would overcome the drawbacks of using hESCs and allow for the direct examination of diseased cells for pathological studies and drug screening; this has recently been performed on iPSCs from patients of amyotrophic lateral sclerosis (13), spinal muscular atrophy (14), and familial dysautonomia (15). Indeed, based on morphology or gene-expression profiles, all of the iPSC lines generated independently in various laboratories show remarkable similarity to hESCs (10–12). Most recently, human iPSCs free of transgenes have been generated (16–18) and are expected to grant human iPSCs true pluripotency unfettered by exogenous gene expression.
Regardless of the aims and methods of reprogramming iPSCs, fundamental questions that must be addressed are whether or not human iPSCs, like hESCs, can efficiently differentiate into target cell lineages and whether or not those iPSC-derived cells are functional. Human iPSCs are capable of generating hematopoietic and neural cells, at least from the reported lines (19, 20). However, whether or not human iPSCs differentiate after an intrinsic developmental program is not known. In addition, iPSCs used in these studies were generated with integrating viral methods. Thus, it is unclear if human iPSCs free of integrated transgenes will show improved differentiation.
Here, we compared the neural-differentiation potential of human iPSCs, including those with or without integrating transgenes, with hESCs. We found that human iPSCs differentiate to neuroepithelial (NE) cells and functional neurons or glia after the same time course and through the same transcriptional networks as hESCs do but with increased variability, which is independent of transgene integration. This indicates that although human iPSCs are invaluable for future regenerative medicine and disease studies, techniques other than simple removal of reprogramming transgenes are needed to improve the differentiation efficiency and potency for iPSCs.
Results
Human iPSCs Convert to NE Cells After the Same Differentiation Program as hESCs.
To test whether or not human iPSCs, like hESCs, can differentiate to NE cells, we compared 12 human iPSC lines, established through lentiviral (11, 21), retroviral, or nonintegrating episomal methods (16), and 5 hESC lines (22–24) (Table S1). The human iPSCs exhibited typical characteristics of hESCs (i.e., large nucleus and multiple, prominent nucleoli; growing as round colonies and expressing pluripotency markers such as OCT4) (Fig. 1 A and B and Fig. S1 A–D). After separation from the feeder fibroblasts, both the iPSCs and hESCs formed spherical aggregates known as embryoid bodies (EBs), which were indistinguishable across lines by gross morphological analysis (Fig. S1 E–H).
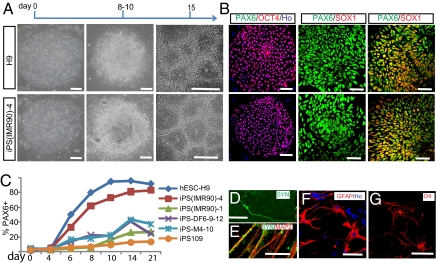
Fig. 1.
Human iPSCs and hESCs follow the same temporal course of neural differentiation. (A) Phase contrast images show that hESCs and iPSCs grew as individual colonies, differentiated to columnar epithelial cells at days 8–10, and formed neural tube-like rosettes at day 15. (B) Both iPSCs and hESCs were positive for OCT4 at day 0, for PAX6 but not SOX1 at days 8–10, and for both PAX6 and SOX1 at day 15. (C) FACS analyses indicate that differentiating cells from H9 hESCs, iPS(IMR90)-1 and 4, iPS-M4-10, iPS-DF6-9–12, and iPS109 began to generate PAX6-expressing cells at days 6–8, and this reached a plateau at day 14 but with different efficiency. Shown are curves of the average from three replicates for each cell line. (D and E) By 12 weeks in culture, many MAP2+ neurons also expressed synapsin; higher magnification indicated a punctuate staining pattern on the cell bodies and neurites. (F) GFAP+ astrocytes were present in differentiated cultures at 12 weeks. (G) O4+ ramified oligodendrocytes were observed in cultures after 16 weeks. Except when noted elsewhere, images of iPSCs are presented with iPS(IMR90)-4 in this and the following figures. (Scale bar, 50 μm.)
The iPSCs aggregates (EBs) were plated at day 7 for NE differentiation (25, 26). Columnar NE cells appeared at day 10 and formed typical neural tube-like rosettes at day 15 (Fig. 1A). The rosettes were morphologically indistinguishable from those differentiated from hESCs (Fig. 1A), although iPSC-derived individual colonies either had homogeneous neural rosettes or no neural cells. Immunostaining indicated that at day 10, the columnar cells in rosettes uniformly expressed PAX6 but not SOX1 (Fig. 1B). By day 15, most of these cells became positive for both PAX6 and SOX1 (Fig. 1B). This same temporal progression, as observed in hESC (2, 25), occurred in all human iPSC lines regardless of how iPSCs were reprogrammed (Fig. 1 A and C), indicating that the two types of stem cells follow the same neural-differentiation program.
Human iPSCs Sequentially Differentiate into Functional Neurons and Glia in Vitro.
hESC-derived NEs differentiate into neurons in the first month and into astrocytes or oligodendrocytes after 2–3 months (2, 6, 27). To determine whether or not human iPSC-derived NEs behave similarly, we enriched the NEs from four representative iPSC lines (iPS(IMR90)-1 and 4, M4-10, and DF6-9–12) and differentiated the neural precursors on a laminin substrate at day 25. iPSC-derived neurons, labeled with βIII-tubulin, extended long processes by day 42 (Fig. S2A). These neurons gradually expressed synapsin; by 12 weeks, this was distributed in neurites in a punctuate pattern (Fig. 1 D and E), an indication of synaptic maturation that has been observed previously in hESC-derived neurons (28). Whole-cell patch clamping further showed that iPSC-derived neurons attained the capacity to fire trains-of-action potentials by 7–8 weeks (Fig. S2C), the same time point for neurons differentiated from hESCs (27).
In cultures for glial differentiation, S-100β+ astrocyte progenitors were first observed after 4 weeks of differentiation (Fig. S2B). At around 3 months, glial fibrillary acidic protein (GFAP) expressing astrocytes appeared and exhibited a satellite-like morphology in the presence of serum (Fig. 1F and Fig. S2 D and E). Oligodendrocytes labeled with O4 began to appear after 4 months of differentiation (Fig. 1G), similar to the time course of oligodendroglial differentiation from hESCs (6). Together, these results indicate that the order and timing of neurogenesis and gliogenesis in human iPSC differentiation resemble those in hESCs and normal brain development.
Human iPSCs Differentiate into NE with Increased Variability Between Lines.
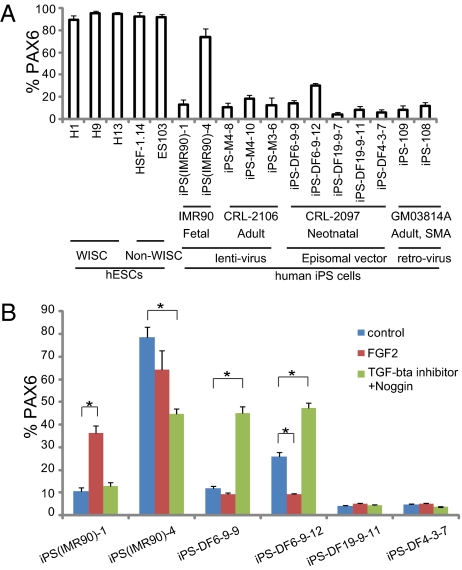
Various hESC lines differentiate into NE at a similar efficiency under our chemically defined adherent colony culture (29), thus minimizing the propensities that may manifest if using alternative protocols (30, 31). FACS analysis indicated that all hESC lines, generated from different laboratories (22–24), yielded a similar high proportion (90–97%) of PAX6-expressing NE cells at day 15 (Fig. 2A). In contrast, iPSCs reprogrammed with lentiviral methods (11) exhibited lower and variable neural differentiation (15–79%) (Fig. 2A), regardless of the origins of fibroblasts from which the iPSCs were derived (neonatal lung vs. adult forearm skin). Even among those lines generated from the same fibroblasts, the neural-differentiation efficiency varied significantly as well [15% in iPS(IMR90)-1 vs. 79% in iPS(IMR90)-4]. Similarly, the two retrovirally induced iPSC lines also showed less efficient neural differentiation (∼15% PAX6+ cells) (Fig. 2A). Thus, integrating viral methods or reprogramming genes alone is unlikely to account for the observed variability and reduced efficiency of neural differentiation from human iPSCs.
Fig. 2.
Variation of iPSC neural differentiation. (A) At day 15, FACS analyses revealed a similar proportion of PAX6-expressing cells from five hESC lines but variable percentages for the iPSC lines. (B) The human iPSC lines responded to FGF2 or dual SMAD inhibition variably. Data are shown as mean ± SEM (n = 3). Asterisk denotes P < 0.001.
Residual transgene expression in iPSCs generated using integrating viral approaches may affect pluripotency and differentiation (10, 11). Hence, nonintegrating strategies may overcome this problem. Somewhat surprisingly, iPSCs generated using the nonintegrating episomal vectors (16) exhibited similarly low and variable neural-differentiation rates (Fig. 2A). This low and variable neural differentiation was not caused by an altered developmental course, because the neural-differentiation process of these iPSCs followed the same time line (Fig. 1C).
Neural Differentiation of Some but Not All iPSC Lines Can Be Improved by Regulating FGF and Bone Morphogenetic Protein (BMP) Signaling.
Efficient NE differentiation in our protocol is largely because of endogenous FGF signaling and/or inhibition of bone morphogenetic protein (BMP) signaling (32), and it was recently reported that SMAD (a family of proteins that are homologous to the gene products of the Drosophila gene ‘mothers against decapentaplegic’ (Mad) and the C. elegans gene sma, and that modulate the activity of transforming growth factor-beta ligands) inhibition by two inhibitors together can significantly increase neural differentiation of hESCs and two iPSC lines (33). To test if FGF2 or SMAD inhibition could improve their neural differentiation, we selected six lines from both transgenic and transgene-free human iPSCs. FGF2 significantly increased the proportion of PAX6+ cells in only one iPSC line, (IRM90)-1 (Fig. 2B). Application of two SMAD antagonists improved NE differentiation of some iPSC lines (iPS-DF6-9–9 and iPS-DF6-9-12) but not others (Fig. 2B). Two iPSC lines (iPS-DF19-9–11 and iPS-DF4-3-7) responded to neither FGF2 nor SMAD inhibition and exhibited consistently poor neural differentiation (Fig. 2B). Together, these data indicate that lower neural-differentiation efficiency of human iPSC lines is caused by the variability in response to neural inducers.
Enriched iPSC-Derived NE Cells Can Be Efficiently Patterned to Regional Neurons.
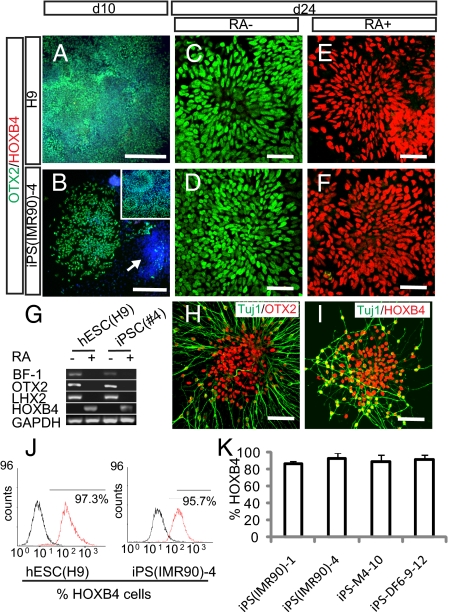
As in development, NE cells differentiated from hESCs in our chemically defined system initially expressed an anterior transcription factor OTX2 but not homeobox protein HOXB4 (Fig. 3A) (2, 25). Similarly, human iPSC-derived NE uniformly expressed OTX2 but not HOXB4 (Fig. 3B), particularly after enrichment by manually selecting typical neural tube-like rosettes or scratching off nonepithelial colonies (29) (Fig. S3). This expression pattern was retained for at least 4 weeks of differentiation in the absence of morphogens. RT-PCR confirmed that these cells expressed anterior transcription factors BF1 and OTX2 but not HOXB4 (Fig. 3G) or the mid/hindbrain transcription factors EN1 and GBX2. Thus, human iPSCs, like hESCs, differentiate to NE cells with a default anterior phenotype.
Fig. 3.
iPSC-derived neuroepithelial cells can be patterned to regional progenitors and differentiated to neurons and glia. (A and B) Human iPSC- and ESC-derived neuroepithelial cells were positive for OTX2 and negative for HOXB4 at day 10. Some cell clusters (Hoechst labeled; arrows) in the iPSC cultures were negative for OTX2. Most of the OTX2+ cluster formed typical rosettes 4 days later as shown in B Inset. (C–F) By day 24, cultures treated with RA were now positive for HOXB4 but negative for OTX2, whereas those without RA treatment remained OTX2 positive. (G) RT-PCR showed expression of BF-1, OTX2, and LHX2 but not HOXB4 in neuroepithelial cultures without RA treatment and with HOXB4 expression in RA-treated neuroepithelia at day 24. (H) Further differentiation shows that without RA, iPSC-derived βIII-tubulin+ neurons were positive for OTX2 but were labeled for HOXB4 in the presence of RA (I). (J) FACS showed that the majority of hESC- and iPSC-derived neuroepithelia was positive for HOXB4 in response to RA treatment. (K) For the representative iPS cell lines tested, more than 90% of the neural epithelial cells could be caudalized to HOXB4-expressing progenitors.
To determine if the iPSC-derived NE cells can be patterned in a similar manner as hESC-derived cells, NE cells from four iPSC lines [iPS-M4-10, iPS-DF6-9–12, iPS(IMR90)-1, and iPS(IMR90)-4] were treated with 0.1 μM retinoic acid (RA) from day 10. Similar to cells derived from hESCs (Fig. 3 C and E), immunostaining indicated that iPSC-derived NE cells cultured without RA retained OTX2 expression until day 24 (Fig. 3D), whereas cells treated with RA became HOXB4+ (Fig. 3F). FACS confirmed that when patterning with RA, 90% of the iPSC-derived and 97% of hESC-generated neural progenitors expressed HOXB4 (Fig. 3 J and K). Therefore, iPSC-derived NE, like those derived from hESCs (2, 25), can be effectively patterned to caudal progenitors in response to the same factors at the same concentration.
The patterned progenitors were then differentiated to neurons according to previously established methods (2, 25, 28). Neurons with extensive neurites were readily observed in all cultures. The Tuj1+ neurons generated from the anterior progenitors expressed OTX2, whereas those from RA-patterned progenitors were positive for HOXB4 at 4 weeks of differentiation (Fig. 3 H and I). Thus, using methods established for hESC-derived neurons, human iPSCs can be specified to neurons that bear regional identities of the central nervous system.
Human iPSCs Differentiate into Functional Neuronal Subtypes.
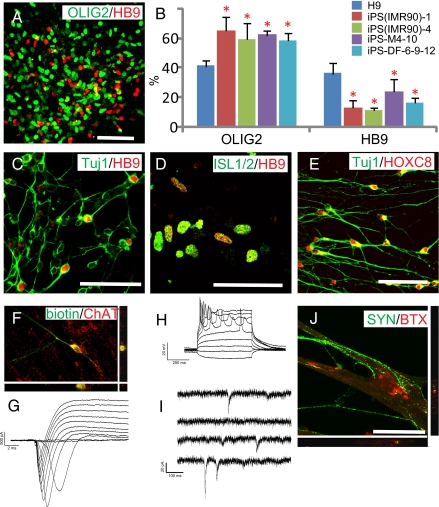
One of the ultimate goals of using human iPSCs is to generate functional target cells for medical screening and therapeutic applications. In the present study, we used the well-established motoneuron differentiation system (2, 29) to address whether or not human iPSCs can differentiate into functionally specialized neurons. Treatment of NE cells from four iPSC lines and H9 hESCs with RA and sonic hedgehog (SHH) for 2 weeks resulted in an efficient induction of motoneuron progenitors that were identified by OLIG2 expression at day 28 (Fig. 4A), which further generated motoneurons with complex processes and coexpression of the motor neuron transcription factors HB9 and ISL1/2 at the fifth and sixth week (Fig. 4 A, C, and D). Furthermore, the majority of the βIII-tubulin+ neurons expressed HOXC8 (Fig. 4E), indicating that iPSC-derived motoneurons attain an appropriate rostro-caudal identity similar to hESC-derived cells.
Fig. 4.
iPSCs differentiate to functional motor neurons. (A) iPSC-derived neural cells expressed OLIG2 and HB9 at day 35. (B) FACS analysis of HB9+ motor neurons differentiated from hESCs and iPSCs. Data are shown as mean ± SEM (n = 3). Asterisk denotes P < 0.01 by Dunnett's test with H9 as a reference. (C) HB9 was expressed on differentiation to TuJ1+ neuronal cells. (D) iPSC-derived HB9+ cells were positive for ISL1/2 shown by a single confocal section. (E) Confocal imaging shows HOXC8 staining in the nuclei of iPSC-derived Tuj1+ neurons. (F) Cells targeted for physiological analysis were labeled with neurobiotin and stained for ChAT+ to verify their motor neuron (MN) identity. (G) I-Clamp traces revealed multiple APs generated during 500-ms current injections of various amplitudes. (H) Expanded view of step-induced currents revealed large, rapidly inactivating inward currents followed by sustained outward currents. (I) Example traces of spontaneous postsynaptic currents detected in iPSC-derived MNs. (J) Confocal imaging showed that BTX was localized to the area where synapsin+ neurites contacted the myotube. (Scale bar, 50 μm.)
Interestingly, the proportion of OLIG2+ progenitors was higher in iPSC-derived (60–70%) than in hESC-derived progenies (41 ± 4%), but the population of HB9+ motor neurons was only about 10–23%, in contrast to 30–50% achieved from hESC differentiation (Fig. 4B). Altering the concentration and duration of RA or SHH did not increase the proportion of HB9+ motoneurons.
Whole-cell patch clamp recording indicated that human iPSC-generated ChAT+ spinal motoneurons (Fig. 4F) expressed large inward currents and outward currents by 8 weeks (Fig. 4G). A single action potential (AP) could be generated in all neurons tested, whereas trains of APs could be generated in a portion of these cells (3 of 11) after at least 8 weeks (Fig. 4H). In older cultures (>8 weeks), spontaneous synaptic currents were also observed, most of which had fast rise times indicative of excitatory postsynaptic currents (EPSCs) (Fig. 4I); however, some displayed slower kinetics typical of inhibitory postsynaptic current (IPSCs). These results indicate that human iPSC-generated motoneurons are electrophysiologically active and may form functional synapses with surrounding neurons, and the functional maturation is similar to that seen in hESC-differentiated neurons (28).
Cocultures of iPSC-derived motor neurons with C2C12 muscle-like cells for 2 weeks resulted in the formation of myotube-like structures and connections of neurites with the myotubes. Contractions of myotubes were readily observed, particularly in areas of robust neurite innervation. Aggregated Bungarotoxin (BTX) staining, which labels postsynaptic acetylcholine receptors, appeared in close apposition to the presynaptic marker synapsin that was present on iPSC-derived motor neurons, which was confirmed by confocal analysis (Fig. 4J). In myocytes with no neurites contacts, the BTX staining was diffusive. We conclude that iPSC-differentiated spinal motoneurons are indeed functional.
Discussion
Our present study shows that human iPSCs differentiate to the neural lineage according to the same temporal program as that of hESCs. Similarly, the iPSC-derived NEs, in response to a same set of extracellular molecules, differentiate to regional progenitors, which further produce functional neurons. However, neural differentiation of human iPSCs was variable and less efficient. Unexpectedly, transgene-free iPSCs did not show improved neural differentiation to a level comparable with that of hESCs. Although these findings raise hopes of applying human iPSCs in modeling diseases and potential autologous cell transplantation, they also suggest that the differentiation of individual human iPSC lines are variable and unpredictable, which does not seem to relate to the presence or absence of transgenes.
Human iPSCs, by definition, should behave like hESCs in their self-renewal and lineage differentiation. This was revealed in the present study through a direct comparison of human iPSC and hESCs on their neural differentiation. The temporal course and gene-expression pattern during NE specification was nearly identical between the two types of stem cells, regardless of if the iPSCs were from adult or neonatal cells, if the iPSCs were generated by transfection with 3 or 4 genes (M3-6 vs. other lines in the category reprogrammed with lentivirus), or if the iPSCs were established with or without transgene integration (transgenic vs. episomal lines). Even though the iPSCs originated from different tissue sources such as foreskin or lung fibroblast, the iPSC-derived NEs all uniformly bear a phenotype of anterior brain regions, which is virtually the same as that for in hESCs differentiation. Furthermore, human iPSC-derived NEs responded to morphogens in the same manner as did hESC-derived NEs, and the sequential neuron–glial differentiation is similar to that revealed in hESC differentiation (6). Together, these findings provide strong evidence that human iPSCs, like hESCs, differentiate into the neural lineage after an intrinsic developmental program.
A noticeable difference is the lower and variable NE and neuronal differentiation efficiencies among the human iPSC lines. The lower efficiency does not seem to stem from technical considerations, because the iPSCs and hESCs were cultured and differentiated under the same conditions and were initially morphologically and immunologically indistinguishable from one another. Also, the reduced efficiency is not caused by the differentiation protocol, because addition of FGF2 or inhibition of SMAD, which are known to enhance the neural differentiation of pluripotent stem cells (32, 33), did not consistently improve the iPSC differentiation. During neuronal differentiation, we also noticed that human iPSCs tended to generate more OLIG2+ ventral progenitors but relatively less HB9+ postmitotic motoneurons (Fig. 4B). We have previously shown that expression of OLIG2 is strictly dependent on SHH, (2, 6) and sustained OLIG2 expression prevents the generation of HB9+ motoneurons (34). However, increasing RA to promote neurogenesis or reducing SHH to accelerate cell-cycle exit did not increase the motor neuron population, suggesting that progenies of human iPSCs may respond to extracellular signals differently from those of hESCs or the body at certain stages of differentiation. These phenomena again suggest that individual human iPSC lines might be epigenetically unique and predisposed to generate cells of a particular sublineage.
Cell differentiation is preprogrammed and coordinated by down-regulation of pluripotent genes and concomitant up-regulation of stage-specific lineage genes. During hESC differentiation to NE cells, NANOG and OCT4 are down-regulated before Pax6 becomes highly expressed (25). Initially, we attributed the lower and variable differentiation of iPSCs to transgene expression, because iPSCs generated using lentiviruses or retroviruses showed residual transgene expression (10, 11) that we believed inhibited the iPSCs from efficiently differentiating. Thus, we expected that human iPSCs free of transgene integration may overcome this drawback. However, our present study clearly shows that removal of transgenes during iPSC generation does not improve the low efficiency and variability of neural differentiation.
Our findings that neural differentiation of human iPS cells follows the intrinsic developmental program and that the neurons are functional suggest that the human iPSCs are suitable for applications that simply require generating certain cell types. For pathological and related studies, such as modeling neural developmental disorders, the variability and low efficiency of differentiation needs to be taken into consideration. The fact that mouse iPSCs are capable of germline transmission and tetraploid complementation (7, 8) shows that iPSCs hold the pluripotent differentiation potential at least in the blastocyst and ovary/testis environment. In this regard, modifying the culture environment for establishing and maintaining human iPSCs may overcome the low and variable neural-differentiation efficiency, particularly in light of the different culture systems that have been employed for derivation of iPSCs and hESCs (23, 24). The finding that neural-differentiation efficiency of some but not all lines can be improved by applying extra neural inducers suggests that the lower neural-differentiation efficiency cannot be explained by single mechanisms, and different iPSC colonies are likely different during derivation. From the application standpoint, additional criteria besides the pluripotency assays (e.g., differentiation to target cell types) may help select more uniform iPSCs.
Methods
Neural Differentiation from Human ESCs and iPSCs.
Human ESCs (H1, H9, H13, HSF-1.14, and ES103, passage 25–40) and human iPSC lines [iPS(IMR90)-1, iPS(IMR90)-4, M4-8, M4-10, M3-6, DF4-3-7, DF6-9-9, DF6-9-12, DF19-9-7, DF19-9-11, iPS-108, and iPS-109] (11, 16, 22–24) (SI Text and Table S1) were maintained and differentiated according to our previously established methods (24–26) and acclimated to the same culture condition for several passages before differentiation. Partially differentiated colonies were manually removed (29) before differentiation analysis. After separation from feeder cells and culture in suspension for 7 days, aggregates of human iPSCs or hESCs were differentiated to primitive NEs in an adherent culture in the neural medium consisting of DMEM/F12, N2 supplement, and nonessential amino acid, as detailed (26, 29). Neural tube-like rosettes at day 15 of differentiation were then detached mechanically and cultured in suspension in the same medium. FGF2 or Noggin were added to cultures for the first 15 days, SB43152 was added from day 0–5 according to published protocols (26, 33).
Neuron and Glial Differentiation.
Primitive NE cultures were treated with or without RA (100 nM) from day 10 and SHH (100 ng/mL) from day 14. On day 25, neural progenitors were differentiated on a laminin substrate in the differentiation medium consisting of neurobasal medium, N2 supplement, and cAMP (1 μM). For motoneuron differentiation, the patterned progenitors were adhered to laminin substrate and cultured in the presence of a mixture of BDNF, glial cell-derived neurotrophic factor (GDNF), and IGF1 (10 ng/mL) (2, 29). For glia differentiation, progenitors were expanded in suspension for another 2 months in a medium consisting of DMEM/F12, N1 supplement (Sigma; 100 ng/mL), and cAMP (1 μM), and for oligodendorcytes, T3 (60 ng/mL), platelet-derived growth factor-AA (PDGF-AA), insulin-like growth factor 1 (IGF1), and neurotrophin 3 (NT3), all at 10 ng/mL (6), were added. The progenitors were then adhered to plastic (for astrocytes) or ornithine substrate (for oligodendrocytes) and cultured for 7 days before immunocytochemical analysis. For coculture, C2C12 myoblasts from the American Type Culture Collection (ATCC) were differentiated for 2 days in DMEM containing 2% FBS. hESC- or human iPSC-derived motoneuron clusters were then plated onto the myocyte cultures, and the medium was changed to that for motoneuron differentiation as described (2).
Immunocytochemistry and Microscopy.
Immunofluorescence on coverslip cultures was described previously (2, 6), and primary antibodies were listed in Table S2. Acetylcholine receptors on differentiated C2C12 cells were labeled with Alexa Fluor 594 conjugated α-bungarotoxin (BTX, Molecular Probes Inc., Eugene, OR; 1:500) at 20 °C for 30 min (2). Images were obtained with a Nikon TE600 fluorescent scope with a SPOT camera (Diagnostic Instruments) or a Nikon C1 laser-scanning confocal microscope (2, 25).
Quantification and Statistics.
Randomly selected region of interest (ROI) from images of biological replicates were subjected to cell counting with a plug-in of ImageJ. Statistical analyses were performed using t test or multiple comparisons (Dennett) in R environment (R Development Core Team).
RNA Extraction and PCR.
RNA was extracted using the TRIzol reagent, and RT-PCRs were performed in a two-step way as described (25). Primers are listed in Table S3.
FACS.
The whole cultures (adherent from day 7 to 15 and suspension cultures before day 7 and after day 15) were trypsinized and stained for FACS. FACS were performed with a Becton Dickinson FACSCaliber and analyzed with CellQuest Pro (BD Biosciences) as described (25).
Electrophysiological Recording.
hESCs- or iPSCs-derived telencephalic neurons and motoneurons were recorded with patch clamp as described (28). One percent neurobiotin was added to the intracellular solution. Current clamp and voltage clamp recordings were performed using a MultiClamp 700B amplifier (Axon Instruments). Signals were filtered at 4 kHz and sampled at 10 kHz using a Digidata 1322A analog–digital converter (Axon Instruments). Data were acquired and analyzed using pClamp software v.9 (Axon Instruments). Access resistance was typically 8−15 MΩ and was typically compensated by 50−80%, but it was omitted for miniature postsynaptic current analysis.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Neurological Diseases and Stroke (Grants NS045926, NS057778, and NS064578) and the ALS Association; this study was partly supported by a core grant to the Waisman Center from the National Institute of Child Health and Human Development (P30 HD03352).
Footnotes
Conflict of interest statement: J.A.T. is a founder, stock owner, consultant, and board member of Cellular Dynamics International (CDI). He also serves as a scientific advisor to and has financial interests in Tactics II Stem Cell Ventures.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910012107/DCSupplemental.
References
- 1.Perrier AL, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li XJ, et al. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 3.Roy NS, et al. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12:1259–1268. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 6.Hu BY, Du ZW, Li XJ, Ayala M, Zhang SC. Human oligodendrocytes from embryonic stem cells: Conserved SHH signaling networks and divergent FGF effects. Development. 2009;136:1443–1452. doi: 10.1242/dev.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao XY, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 8.Boland MJ, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 9.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 10.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 14.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karumbayaram S, et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi KD, et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abeyta MJ, et al. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- 23.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 24.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 25.Pankratz MT, et al. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 27.Yan Y, et al. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson MA, Weick JP, Pearce RA, Zhang SC. Functional neural development from human embryonic stem cells: Accelerated synaptic activity via astrocyte coculture. J Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu BY, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, et al. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci USA. 2007;104:13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaVaute TM, et al. Regulation of neural specification from human embryonic stem cells by BMP and FGF. Stem Cells. 2009;27:1741–1749. doi: 10.1002/stem.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du ZW, Hu BY, Ayala M, Sauer B, Zhang SC. Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells. 2009;27:1032–1041. doi: 10.1002/stem.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.