Abstract
microRNAs (miRNAs) play key roles in modulating a variety of cellular processes through repression of mRNA targets. In a screen for miRNAs regulated by myocardin-related transcription factor-A (MRTF-A), a coactivator of serum response factor (SRF), we discovered a muscle-enriched miRNA, miR-486, controlled by an alternative promoter within intron 40 of the Ankyrin-1 gene. Transcription of miR-486 is directly controlled by SRF and MRTF-A, as well as by MyoD. Among the most strongly predicted targets of miR-486 are phosphatase and tensin homolog (PTEN) and Foxo1a, which negatively affect phosphoinositide-3-kinase (PI3K)/Akt signaling. Accordingly, PTEN and Foxo1a protein levels are reduced by miR-486 overexpression, which, in turn, enhances PI3K/Akt signaling. Similarly, we show that MRTF-A promotes PI3K/Akt signaling by up-regulating miR-486 expression. Conversely, inhibition of miR-486 expression enhances the expression of PTEN and Foxo1a and dampens signaling through the PI3K/Akt-signaling pathway. Our findings implicate miR-486 as a downstream mediator of the actions of SRF/MRTF-A and MyoD in muscle cells and as a potential modulator of PI3K/Akt signaling.
Keywords: Akt signaling, microRNA, muscle growth, myocardin related transcription factor-A, cardiomyocyte
microRNAs (miRNAs) play key roles in a broad range of biological processes, and in many cases have been shown to modulate intracellular signaling pathways involved in development and disease (1). miRNAs inhibit translation or promote mRNA degradation by annealing to complementary sequences in mRNA 3′ untranslated regions (UTRs). Individual miRNAs typically target dozens of mRNAs, often encoding proteins with related functions. Thus, although their inhibitory effects on individual mRNAs are generally modest, their combined effects on multiple mRNAs can evoke strong biological responses. miRNAs play especially powerful roles in muscle cells, where they have been implicated in cell fate specification, proliferation, differentiation, and stress responsiveness (2–4).
Members of the myocardin family of transcription factors control growth and development of cardiac, skeletal, and vascular smooth muscle cells, as well as their responses to physiological and pathological signaling (5–8). Myocardin and myocardin-related transcription factors (MRTFs) are recruited to downstream target genes through interaction with serum response factor (SRF). Recently, we discovered that MRTFs directly activate the transcription of a cardiovascular-specific miRNA cluster encoding miR-143 and miR-145 (9). These miRNAs, in turn, regulate multiple modulators of actin dynamics and SRF function, thereby influencing smooth muscle growth and responsiveness to mechanical stimuli.
Here we show that MRTF-A also regulates the expression of miR-486, a muscle-enriched miRNA encoded by intron 40 of the Ankyrin-1 (Ank-1) gene. Among the targets of miR-486 are the mRNAs encoding PTEN and Foxo1a, which serve as negative components of phosphoinositide-3-kinase (PI3K)/Akt signaling. Through gain- and loss-of-function experiments, we show that miR-486 modulates PI3K/Akt signaling by directly targeting phosphatase and tensin homolog (PTEN), an inhibitor of PI3K phosphorylation, thereby promoting the phosphorylation of Akt and the activity of downstream components of the pathway. Consistent with a role for MRTF-A in the regulation of this signaling pathway via miR-486, MRTF-A enhances PI3K/Akt signaling in cardiomyocytes, an effect that is blunted by inhibition of miR-486 expression. Our findings reveal a role for miR-486 as an intermediary between MRTF-A and the PI3K/Akt-signaling pathway and suggest the potential involvement of miR-486 in regulating muscle growth and homeostasis.
Results
Regulation of miR-486 by MRTF-A.
In an effort to identify miRNAs that mediate the actions of MRTF-A and its partner SRF in muscle cells, we performed microarray analysis to compare the miRNA expression profiles of neonatal rat cardiomyocytes (CMCs) infected with adenoviruses expressing MRTF-A or β-galactosidase as a control (Fig. S1). Among several miRNAs that were up-regulated by MRTF-A (9), miR-486 showed the most dramatic increase in expression in MRTF-A–transduced CMCs. The induction of miR-486 by MRTF-A was confirmed by Northern blot and real-time RT–PCR (Fig. 1 A and B).
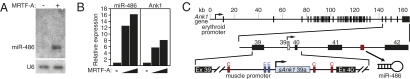
Fig. 1.
miR-486 and host gene, sAnk1, are induced by MRTF-A. (A) Cardiac myocytes (CMCs) were infected with adenovirus directing expression of β-gal (−) or MRTF-A (+). miR-486 was detected by Northern blot. (B) The response of miR-486 and sAnk1 to increasing multiplicity of infection (MOI) of adenovirus-mediated MRTF-A expression in CMCs was examined by real-time RT–PCR. Expression levels are relative to that in β-gal-infected control CMCs. (C) miR-486 is encoded by the final intron of the Ank1 gene (Top), which is regulated by an alternative promoter containing two E-boxes. The first intron contains two CArG boxes. (Middle) The genomic organization of the Ank1 gene and miR-486 (red box). (Bottom) The genomic organization of the muscle-enriched sAnk1 isoform, including the putative regulatory elements.
miR-486 is a unique miRNA with no known family members and is conserved in mammals, but does not exist in birds, fish, amphibians, or lower metazoans. miR-486 is transcribed from an intron of the Ank1 gene (Fig. 1C and Fig. S2), which encodes an ankyrin-repeat protein that links the cytoskeleton to the plasma membrane. Ank1 is expressed specifically in erythroid cells under control of an erythroid-specific promoter (Fig. 1C and ref. 10). The final three exons (exons 40–42) of the Ank1 gene, preceded by an alternative exon (exon 39a), code for a muscle-specific Ank1 protein, referred to as small Ank1 (sAnk1), which connects the sarcomere to the sarcoplasmic reticulum (11).
The expression of the sAnk1 transcript is regulated by an alternative promoter immediately upstream of exon 39a of the Ank1 gene, which contains two conserved E-boxes that confer responsiveness to MyoD (Fig. 1C) (12, 13). Genomic sequence upstream and within the first intron of the sAnk1 gene also contains putative binding sites for SRF [CC(A/T)6GG], referred to as CArG boxes, which may mediate responsiveness to MRTF-A (Fig. 1C and Fig. S3). Like miR-486, sAnk1 was also induced in CMCs by MRTF-A (Fig. 1B). The coregulation of miR-486 and sAnk1 suggests that miR-486 is generated by the processing of Ank1 intronic RNA.
Expression of miR-486 and sAnk1 in Cardiac and Skeletal Muscle.
We examined the expression of miR-486 and sAnk1 by Northern blot and RT–PCR, respectively, to compare their tissue distribution. miR-486 is enriched in cardiac and skeletal muscle (Fig. 2A and Fig. S4), as previously reported for sAnk1 (12). The tissue distribution of sAnk1 transcripts recapitulated the cardiac and skeletal muscle expression of miR-486 (Fig. 2A).
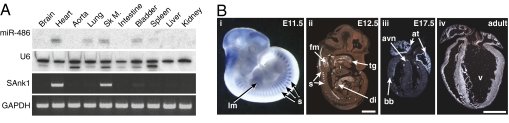
Fig. 2.
Muscle-specific expression of miR-486 and sAnk1. (A) Northern blot of miR-486 in adult mouse tissues. RT–PCR demonstrates the adult expression of the miR-486 host gene sAnk1. Controls for RNA input in the Northern blot and RT–PCR are U6 and GAPDH, respectively. (B) In situ hybridization demonstrates embryonic and adult expression of the sAnk1 gene. (i) sAnk1 is observed specifically in the developing somites and limb muscles of a whole-mount E11.5 embryo; head staining is nonspecific. Radioactive-section in situ hybridization demonstrates sAnk1 expression at E12.5 (ii) and in the embryonic and adult heart (iii and iv). (ii) sAnk1 is robustly expressed in the somites, tongue, facial muscles, and diaphragm. Expression within the heart is localized to the atria, AV node, and bundle branches at E17.5 (iii). sAnk1 expression is observed throughout all cardiac chambers in the adult heart (iv). (Scale bar: 1 mm.) at, atrium; avn, atrioventricular node; bb, bundle branches; di, diaphragm; fm, facial muscles; lm, limb muscle; s, somites; tg, tongue; v, ventricle.
By in situ hybridization with a probe designed against the sAnk1-specific exon 39a, sAnk1 transcripts were initially detected specifically in the somites at embryonic day (E)11.5 (Fig. 2B). By E12.5, sAnk1 expression was observed throughout the skeletal muscle of the tongue, diaphragm, and the myotome layer of the somites, but not in the heart (Fig. 2B). We first observed cardiac expression of sAnk1 at E17.5, specifically in the atrial chambers, the atrio-ventricular (AV) node, and the bundle branches in the trabecular layer of the ventricles (Fig. 2B). Atrial and AV node expression became more robust by postnatal day 1 (P1), and ventricular expression became detectable by P5 (Fig. S5). The adult heart displayed sAnk1 expression throughout all cardiac chambers (Fig. 2B). We conclude that miR-486 is a skeletal muscle and cardiac-enriched miRNA, which is coexpressed with sAnk1.
Regulation of sAnk1/miR-486 by MRTF-A.
We examined noncoding DNA upstream and within the first intron of the sAnk1 gene for the ability to direct MRTF-A–dependent transcription. A luciferase reporter linked to 1080 bp of genomic DNA upstream of alternate exon 39a displayed strong responsiveness to MyoD in transfected COS cells (Fig. 3A). Mutation of two E-boxes in this genomic fragment completely abolished MyoD directed promoter activity (Fig. 3A). In contrast, MRTF-A failed to activate this reporter (Fig. 3A), despite the presence of a putative CArG box, suggesting that cis-regulatory elements responsible for MRTF-A responsiveness are located elsewhere.
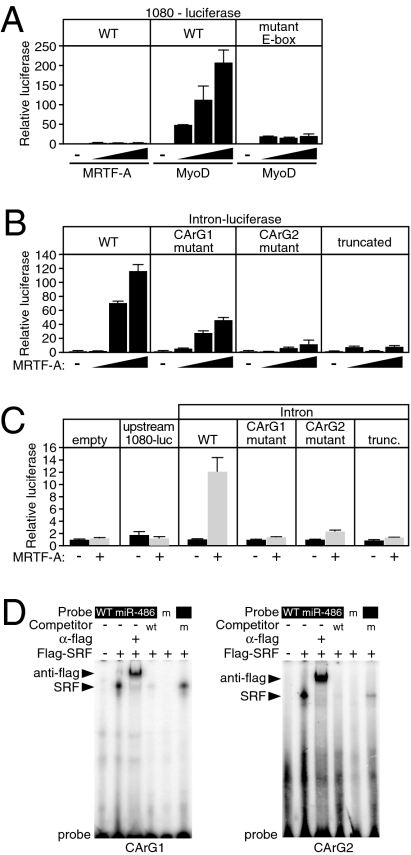
Fig. 3.
Identification of miR-486/sAnk1 regulatory sequences in vitro. (A) Luciferase assay demonstrating the response of 1080 bp of upstream DNA (WT) or of the DNA fragment containing a mutation of the E-boxes to increasing amounts of MRTF-A and MyoD. A total of 50, 100, and 200 ng of expression plasmids were cotransfected with the luciferase reporter construct in COS cells. Error bars represent SD. (B) Response of the 649 bp of sAnk1 intron 39a to MRTF-A examined by luciferase reporter assay in COS cells. A total of 50, 100, and 200 ng of expression plasmid were transfected with full-length intron–luciferase (WT), a luciferase reporter construct with a mutation in the proximal CArG1, a mutation in the distal CArG2, or a truncation that consists of the 5′ most 300 bp. Error bars represent the SD. (C) Response of the empty luciferase vector or the 1,080-bp upstream luciferase or various intron–luciferase reporters to adenovirus-mediated MRTF-A expression in CMCs. Error bars represent SD. (D) Gel electromobility shift assay demonstrating the binding of flag epitope-tagged SRF overexpressing COS cell lysate to a radiolabeled oligonucleotide probe consisting of the sequences of the CArG1 or CArG2 of sAnk1 intron 39a. Flag antibody results in a supershift, and wild-type unlabeled competitor abolishes the binding of CArG probe. Mutant probe does not bind SRF, nor does mutant unlabeled competitor abolish WT probe binding to SRF. m, mutant CArG.
Both mouse and human sAnk1 contain CArG-like sequences in intron 39a (Fig. 1C and Fig. S3). Like the endogenous gene, the sAnk1 intron-luciferase reporter was responsive to MRTF-A in COS cells (Fig. 3B). Mutation of CArG1 reduced responsiveness to MRTF-A, and a CArG2 mutation nearly abolished transcriptional activity (Fig. 3B). A 3′ truncation of the intron to ∼300 bp, which contains only CArG1, also resulted in a pronounced loss of reporter activity. The intronic reporter was also responsive to adenoviral-expressed MRTF-A in CMCs, whereas constructs containing a 3′ truncation of the intron or a mutation of either CArG1 or CArG2 were not induced by MRTF-A (Fig. 3C). The upstream reporter was not responsive to MRTF-A, further suggesting that the upstream CArG is not functional (Fig. 3C).
In gel mobility shift assays, SRF bound efficiently and specifically to both intronic CArG sequences, and unlabeled CArG oligonucleotide effectively competed for SRF binding, whereas a mutant CArG oligonucleotide did not compete for binding (Fig. 3D). We conclude that the first intron of the sAnk1 gene contains two functional CArG boxes that are required to direct responsiveness to SRF/MRTF-A in transient transfection assays.
Regulation of sAnk1/miR-486 Transcription in Cardiac and Skeletal Muscle.
To identify cis-regulatory elements responsible for muscle-specific expression in vivo, we generated transgenic mice harboring a lacZ reporter controlled by upstream and intronic genomic regions of sAnk1/miR-486. Consistent with the responsiveness of the 1080-bp upstream region to MyoD in vitro, this genomic region directed lacZ activity in muscle cells within developing somites by E11.5 and throughout skeletal muscles at E13.5 and into adulthood (Fig. 4 A and C). However, this regulatory region was inactive in the heart at all embryonic and adult time points examined. The full-length intron did not direct expression in heart or skeletal muscle, but the ∼300-bp 3′ truncated intron, which contains only the proximal CArG1, directed robust expression in adult skeletal muscle and in the atria and the AV canal of the heart, partially recapitulating endogenous sAnk1/miR-486 expression (Fig. 4 B and C and Fig. S6). We never observed embryonic lacZ reporter expression with intronic sequences, nor did we identify sequences directing ventricle expression of miR-486. Mutation of CArG1 within the truncated intron completely abolished expression of the lacZ reporter in adult muscle (Fig. 4C), further suggesting that SRF and MRTF-A contribute to the postnatal muscle-specific expression of miR-486.
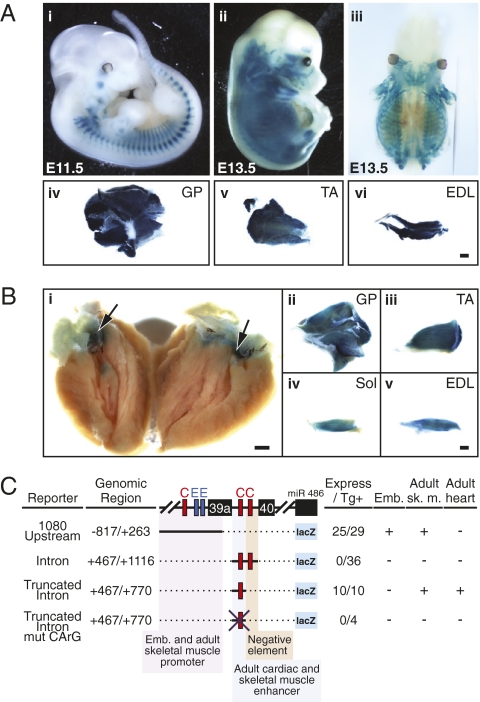
Fig. 4.
Identification of miR-486/sAnk1 regulatory sequences in vivo. (A) Embryonic and adult transgenic mice harboring a lacZ transgene controlled by 1080 bp upstream of the sAnk1 gene exhibit skeletal-muscle–specific β-gal activity. Reporter expression is observed (i) in somites and facial muscles at E11.5 and (ii) in the entire developing musculature at E13.5. (iii) Dorsal view of a cleared E13.5 embryo demonstrating internal muscle staining and absence of smooth muscle or cardiac staining. (iv–vi) lacZ expression detected in whole-mount (iv) GP, (v) TA, and (vi) EDL muscles of adult transgenic mice. (Scale bar: 1 mm.) (B) Adult transgenic mouse harboring a lacZ transgene controlled by the 5′-most 300 bp of intron 39a of the sAnk1 gene. Expression of lacZ is observed in the (i) atria and AV canal of the heart and in the (ii–v) GP, TA, Sol, and EDL of adult mice. (i) Arrows mark AV canal. (Scale bar: 1 mm.) EDL, extensor digitorum longus; GP, gastrocnemieus plantaris; Sol, soleus; TA, tibialis anterior. (C) Summary of the transgenic constructs used to examine the regulation of sAnk1/miR-486. Number of transgenic embryos displaying muscle-specific expression per total number of transgenic embryos is shown in right columns. Embryonic and adult expression was assessed in F0 transgenics and stable transgenic lines. The 1,080-bp upstream reporter containing the two E-boxes directed embryonic and adult skeletal muscle expression, but was insufficient for cardiac expression. The full-length intron reporter, containing two CArG boxes, was insufficient to direct expression in transgenic mice; however, a truncated intron consisting of the 5′ most 300 bp and a single CArG box directed adult skeletal muscle and heart expression. Mutation of the single CArG box within the truncated intron abolished lacZ expression in the skeletal muscle and heart.
Targeting of the PI3K/Akt-Signaling Pathway by miR-486.
To begin to decipher the functions of miR-486, we searched for evolutionarily conserved, predicted mRNA targets of this miRNA. Multiple positive and negative components of the PI3K/Akt-signaling pathway, including PTEN, Foxo1a, PI3KRα (p85α), and insulin-like growth factor 1 (IGF1) are among the strongest predicted targets of miR-486 using the TargetScan5.0 and miRanda algorithms (Table S1). Activation of the PI3K/Akt-signaling pathway by IGF1 results in the phosphorylation of Akt and the promotion of muscle growth and survival via the inactivation of glycogen synthase3β (GSK3β) and Foxo1 (14, 15). PTEN is a PI3K-specific phosphatase that inactivates the Akt-signaling pathway, functioning as a potent inhibitor of growth and survival signaling (16).
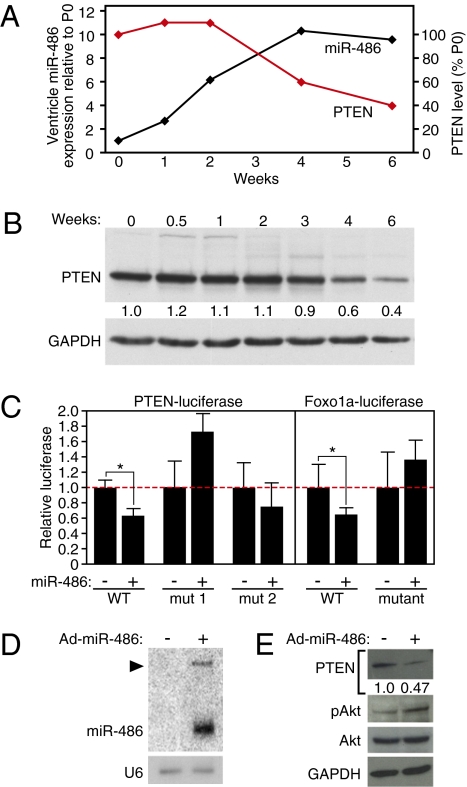
Examination of the expression levels of miR-486 and the protein levels of components of the PI3K/Akt-signaling pathway at various postnatal time points revealed an inverse correlation between expression of miR-486 and PTEN in the maturing heart (Fig. 5A). miR-486 was elevated ≈10-fold in the ventricles between birth and 4 weeks of age, whereas PTEN protein levels decreased ∼40% over the same time period (Fig. 5 A and B). These findings suggested an intriguing physiological relationship between miR-486 and PTEN concomitant with a time of dramatic postnatal cardiac growth, potentially implicating miR-486 in a molecular pathway controlling muscle growth. In line with the observed correlation of miR-486 expression and muscle growth, the expression of miR-486 and its host gene, sAnk1, was decreased in atrophied skeletal muscle following surgical denervation (Fig. S7).
Fig. 5.
miR-486 is predicted to target PTEN and Foxo1a. (A) Normalized expression level of miR-486 in the ventricles detected by real-time RT-PCR at various postnatal ages and represented relative to postnatal day 0 (PO) in a line graph. Normalized levels of PTEN protein are plotted as a percentage of the level at P0. (B) The level of endogenous PTEN protein from ventricular lysates of indicated postnatal ages was detected by Western blot. PTEN band intensity, normalized to GAPDH and relative to P0, is shown below image of Western blot. (C) miR-486 targets the 3′ UTRs of PTEN and Foxo1a mRNAs. The 3′ UTR of the PTEN and Foxo1a mRNAs was linked to the pMIR-REPORT luciferase plasmid (Ambion) and examined in COS cells transiently transfected with CMV6-miR-486 or CMV6-β-gal as a negative control. “mut” refers to mutation of the predicted miR-486 seed region. *P value < 0.05. Error bars represent the SD. (D) Adenoviral-mediated overexpression of miR-486 in CMCs demonstrated by Northern blot. Arrowhead marks the precursor miRNA. U6 was detected as the control for RNA input. (E) The endogenous protein levels of PTEN, phospho-Akt, and total Akt were detected by Western blot in the control or miR-486–transduced CMCs. GAPDH was detected as the control for protein loading.
Because miR-486 is expressed in a manner more consistent with promotion of muscle growth or inhibition of muscle atrophy and is correlated with decreased PTEN protein levels, we examined the potential of miR-486 to directly target the 3′ UTRs of PTEN and Foxo1a, two critical negative components of the Akt-signaling cascade. The 3′ UTRs of PTEN and Foxo1a mRNAs each contain thermodynamically stable predicted binding sites for miR-486 (Table S1 and Fig. S8). Luciferase reporter constructs linked to the 3′ UTRs of either the PTEN or Foxo1a mRNAs were repressed by cotransfection of miR-486 expression plasmid (Fig. 5C). Mutation of the miR-486 binding sites abrogated the repression of the luciferase reporter construct (Fig. 5C).
To test more directly the potential involvement of miR-486 in the process of Akt signaling, we asked whether overexpression of miR-486 in CMCs was sufficient to activate Akt signaling. Adenoviral-mediated overexpression of miR-486 resulted in ≈10-fold higher levels of miR-486 than in control-infected CMCs (Fig. 5D). Concomitant with elevated miR-486 levels, we observed a reduction of PTEN protein levels and phosphorylation of Akt at Ser-473 (Fig. 5E), a site responsible for Akt activation. These results implicate miR-486 in the activation of PI3K/Akt signaling by inhibiting PTEN.
MRTF-A Activates PI3K/Akt Signaling via miR-486 Induction.
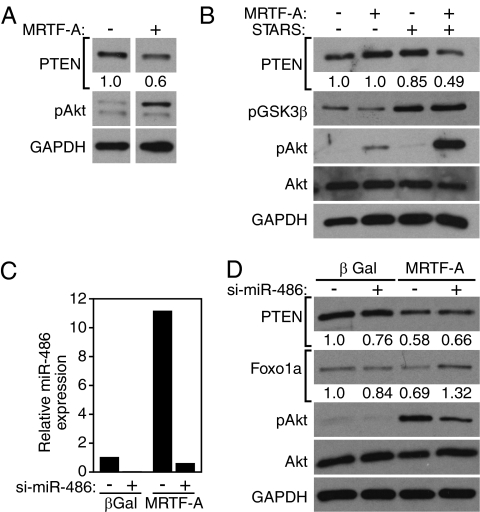
The ability of MRTF-A to up-regulate miR-486, which, in turn, inhibits PTEN, a negative regulator of the PI3K/Akt pathway, suggested that MRTF-A might stimulate Akt signaling. Consistent with this hypothesis, PTEN expression was down-regulated and phospho-Akt was enriched in CMCs in response to adenoviral-mediated MRTF-A overexpression (Fig. 6A). The actin-binding protein STARS enhances MRTF-A activity by promoting actin polymerization (7, 8), thereby releasing MRTF-A from G-actin and allowing its nuclear translocation. Low levels of MRTF-A–expressing adenovirus only modestly induced Akt signaling whereas STARS did not induce Akt signaling. However, MRTF-A and STARS synergized to induce robust activation of the Akt-signaling pathway, as demonstrated by reduction of PTEN protein levels and phosphorylation of Akt and GSK3β (Fig. 6B). We note, however, that induction of phospho-Akt by MRTF-A was greater than by overexpression of miR-486, suggesting that MRTF-A may act through additional mechanisms to control this pathway.
Fig. 6.
Activation of PI3K/Akt signaling by MRTF-A and miR-486. (A) The endogenous protein levels of PTEN and phosphorylated Akt were detected by Western blot from CMCs transduced with 10 MOI of control or MRTF-A–expressing adenovirus. The intensity of the band for PTEN, normalized to GAPDH and relative to control infection, is depicted below the image. (B) The endogenous protein levels of PTEN, phosphorylated GSK3β, phosphorylated Akt, total Akt, and GAPDH were detected by Western blot from CMCs transduced with 1 MOI of MRTF-A and/or 10 MOI of STARS-expressing adenovirus. The intensity of the band for PTEN, normalized to GAPDH and relative to control infection, is depicted below the image. (C) siRNA-mediated knockdown of miR-486 in CMCs demonstrated by real-time RT-PCR and normalized to that of U6 expression. si-miR-486 abolishes the induction of miR-486 expression in response to MRTF-A. (D) The endogenous protein levels of PTEN, Foxo1a, phospho-Akt, total Akt, and GAPDH were detected by Western blot in si-miR-486 treated CMCs. (Upper) Transduction of CMCs with 10 MOI of control or MRTF-A–expressing adenovirus. si-miR-486 or control transfection is indicated at the top by “+” or “−,” respectively. Intensity of the band for PTEN or Foxo1a, normalized to GAPDH and relative to the control lane, is shown below the image.
To determine whether miR-486 is required for the induction of Akt signaling by MRTF-A, we used 2′ O-Me–modified antisense oligonucleotides to knock down miR-486 expression in CMCs overexpressing MRTF-A. Whereas MRTF-A induced miR-486 expression ∼10-fold, si-miR-486 reduced the level of miR-486 to background levels (Fig. 6C). Western blot for endogenous PTEN and Foxo1a demonstrated reduced protein levels upon overexpression of MRTF-A, whereas knockdown of miR-486 attenuated the decrease of both PTEN and Foxo1a protein levels (Fig. 6D). Phospho-Akt showed a corresponding increase in the presence of MRTF-A, whereas Akt phosphorylation was reduced upon knockdown of miR-486 (Fig. 6D). These results demonstrate that MRTF-A activates the PI3K/Akt-signaling pathway at least partially via the induction of miR-486 and concomitant reduction of PTEN protein levels (Fig. 7).
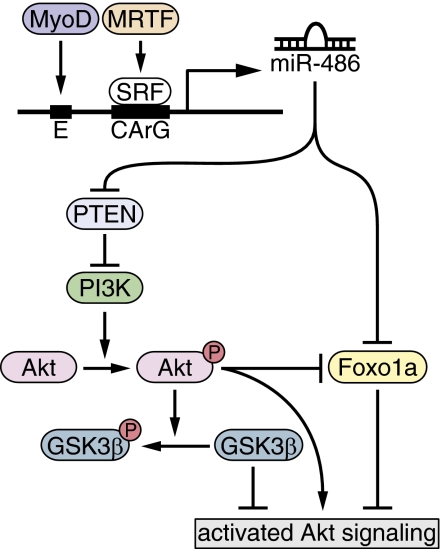
Fig. 7.
Proposed model of miR-486 regulation and function in muscle cells. miR-486 transcription is activated by MyoD and MRTF-A. miR-486 directly represses the translation of PTEN and Foxo1a, two crucial negative regulators of PI3K/Akt signaling, resulting in the phosphorylation of Akt and the activation of the pathway. Activated Akt results in the phosphorylation of GSK3β and the inhibition of Foxo1a activity.
Discussion
In this study, we identify a unique muscle-enriched miRNA, miR-486, that enhances PI3K/Akt signaling by repressing expression of PTEN and Foxo1a, which function as negative components of this pathway. The regulation of miR-486 by MRTF-A also reveals a previously unknown role of MRTF-A in the modulation of PI3K/Akt signaling.
Regulation of miR-486 by MRTF-A.
miR-486 is generated by processing intronic RNA from the sAnk1 gene, which is directly regulated by SRF/MRTF-A and MyoD. It is intriguing that, although MRTF-A is present in the heart during embryogenesis, miR-486 is not significantly expressed in the ventricle until postnatal day 5. It is possible that the expression of miR-486 is positively or negatively influenced by additional muscle regulatory factors. Indeed, although the full-length intronic enhancer directs MRTF-A responsiveness in vitro, only a truncated construct directs muscle expression in transgenic mice. These results suggest the presence of a negative regulatory element in the 3′ end of the intronic enhancer that may be responsible for temporal or tissue-specific expression of miR-486.
miR-486 joins a collection of muscle-restricted miRNAs with roles in muscle growth, development, and signal responsiveness, many of which are also regulated by SRF together with members of the myocardin family (9, 17–19). miR-1 promotes cardiac gene activation in stem cells and inhibits myoblast proliferation, and miR-133 regulates multiple aspects of muscle growth and maturation at least in part through repression of SRF (20, 21, 22). SRF also regulates the expression of miR-143 and -145, which are expressed specifically in cardiac and smooth muscle cells and mediate multiple steps in actin signaling and SRF function (9, 18). The integration of miRNAs into the regulatory circuits controlled by SRF and its coactivators provides signal responsiveness to miRNA-regulated networks and also serves to stabilize and modulate SRF-signaling pathways. It remains to be determined if other transcriptional circuits are as reliant on miRNA regulation as SRF-dependent circuits.
Modulation of PI3K/Akt Signaling by MRTF-A and miR-486.
The ability of miR-486 to target the mRNAs encoding PTEN and Foxo1a point to this miRNA as a potential regulator of PI3K/Akt signaling in muscle cells. The PI3K/Akt-signaling pathway is integral to the process of postnatal muscle growth and physiological hypertrophy (23–25). IGF1 and insulin stimulate hypertrophic growth and glucose uptake via the PI3K-dependent phosphorylation and activation of Akt. Activated Akt results in the phosphorylation-dependent activation of the progrowth protein mTOR, concomitant with a phosphorylation-dependent inhibition of Foxo and GSK3β, negative regulators of protein synthesis and muscle growth (14, 15, 23, 26). PTEN inhibits PI3K phosphorylation and blocks the subsequent initiation of phospho-Akt–dependent hypertrophic and survival signaling. PTEN deletion in the heart results in exaggerated PI3K/Akt signaling, promoting physiological hypertrophy and preventing maladaptive ventricular remodeling (16).
Although a role for MRTF-A in regulating the physiological hypertrophy of cardiac or skeletal muscle has not yet been reported, MRTF-A expression is increased during exercise-induced hypertrophy (27) and reduced in aged muscles, which display significant atrophy (28). Moreover, expression of a dominant negative MRTF mutant protein in skeletal muscle causes muscle hypoplasia (29). In the heart, pathological stimuli up-regulate the expression of STARS, triggering nuclear accumulation of MRTF-A and hypertrophic growth (7, 8).
Our study identifies miR-486 as a potential link between MRTF-A and the PI3K/Akt-signaling pathway. However, knockdown of miR-486 does not completely block the phosphorylation of Akt, suggesting that MRTF-A may exhibit miR-486–independent mechanisms of Akt activation. miR-486 may also influence both positive and negative aspects of the Akt-signaling pathway, perhaps acting as a buffer to minimize dramatic changes in activity. Indeed, miR-486 is predicted to target positive regulators of the Akt-signaling pathway, including IGF1 and p85α. Negative regulation of IGF1 and p85α by miR-486 would be predicted to counteract the positive influence on the PI3K/Akt-signaling pathway via inhibition of PTEN and Foxo1a; however, it is currently unclear whether miR-486 regulates these predicted targets. Finally, miR-486 is not limited to the regulation of components of the Akt-signaling pathway because predicted targets include myriad mRNAs involved in a wide range of biological processes.
The expression of miR-486 is altered during the processes of muscle growth and atrophy in a manner consistent with the regulation of muscle hypertrophy. miR-486 levels increase in the heart postnatally, concomitant with the switch from proliferative to hypertrophic growth (30). Increased miR-486 levels correlate with a decrease in postnatal PTEN levels in the heart, potentially promoting the hypertrophic phase of cardiac growth. Conversely, miR-486 levels decrease in atrophied muscle following denervation (Fig. S7). The miR-486 target and Akt pathway component, Foxo1a, induces atrophy-associated gene expression in adult skeletal muscle (15, 26). By impinging on the PI3K/Akt pathway and Foxo1a levels, miR-486 may affect the postnatal switch between proliferative and hypertrophic cardiac growth and may influence the balance of skeletal muscle growth versus atrophy.
Numerous other miRNAs have been shown to act as positive and negative regulators of the PI3K/Akt-signaling pathway. The miR-29 family activates p53, presumably by directly inhibiting the PI3K regulatory subunit, p85α (31). Likewise, miR-126 regulates angiogenesis at least partially via repression of the PI3K regulatory subunit, p85β (32). Multiple miRNAs, including miR-21, -26a, -216a, and -217, also appear to positively influence PI3K/Akt signaling by targeting PTEN for inhibition (33–36). It is possible that considerable overlap or redundancy exists between miRNAs that maintain the PI3K/Akt pathway in a state of equilibrium under normal conditions.
Previous studies have shown that miR-486 is down-regulated in Duchenne’s muscular dystrophy (37), and our results demonstrate that miR-486 is down-regulated during denervation-induced muscle atrophy, suggesting a potential role for miR-486 in muscle atrophy. Pharmacological interventions designed to elevate miR-486 levels during disease states may therefore impede the progression of muscle wasting.
Materials and Methods
RNA Isolation and miRNA Microarray Analysis.
RNA was purified from CMCs using TRIzol reagent (Invitrogen), and microRNA microarray analysis was performed using the mammalian microRNA probeset (LC Sciences). Details are described in SI Materials and Methods.
Northern Blot.
Ten micrograms of total RNA was loaded onto a denaturing 20% polyacrylamide gel and probed with an antisense STARFIRE probe (IDT) as described in SI Materials and Methods.
In Situ Hybridization.
Whole-mount and radioactive section in situ hybridization was performed, using a probe designed against exon 39a of sAnk1. Details are in SI Materials and Methods.
lacZ Reporter Assay.
Transgenic mice harboring various fragments of miR-486 regulatory DNA fused to a hsp68-lacZ reporter construct were generated and assayed for β-galactosidase activity as described in SI Materials and Methods.
Surgical Procedures.
All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center. Denervation was performed on anesthetized 6- to 8-week-old mice by cutting the left sciatic nerve at the mid-thigh region.
Cell Culture and Luciferase Assay.
COS cells were transfected with various expression plasmids and miR-486–luciferase reporter constructs using Fugene 6.0 (Roche) according to the manufacturer’s protocol as described in SI Materials and Methods. Primary rat cardiomyocytes were prepared and infected with adenovirus or transfected with siRNA (IDT) as described in SI Materials and Methods.
Electrophoretic Mobility Shift Assay.
In vitro binding analysis was performed using conditions and oligonucleotides as described in SI Materials and Methods.
Plasmid Construction.
A miR-486 genomic fragment, various putative enhancers of miR-486, and 3′ UTRs of putative miR-486 targets were amplified using LA Taq (TAKARA) and cloned into pCRII-TOPO. Site-directed mutagenesis was carried out using the Quickchange II kit (Stratagene). Details and primer sequences are described in SI Materials and Methods.
Western Blot.
Antibodies directed against PTEN, Akt, Akt-phospho-Ser473, GSK3β-phospho-Ser9 (Cell Signaling Technologies), and Foxo1a (Abcam) were used to determine protein level by Western blot. GAPDH (Calbiochem) was detected as loading control.
Supplementary Material
Acknowledgments
We thank Nadia Rosenthal and Gianluigi Condorelli for insightful comments, Andrew Williams for critical reading of the manuscript, Jose Cabrera for graphics, Jennifer Brown for editorial assistance, and John Shelton for technical help. Work in the laboratory of E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Center for Clinical Cardiovascular Research, The Robert A. Welch Foundation, the Fondation Leducq’s Transatlantic Network of Excellence in Cardiovascular Research Program, and the American Heart Association-Jon Holden DeHaan Foundation. E.M.S. was supported by a National Institutes of Health postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest. E.N.O. is a cofounder of miRagen Therapeutics and holds equity in the company.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000300107/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condorelli G, Dimmeler S. MicroRNAs: Components of an integrated system controlling cardiac development, physiology, and disease pathogenesis. Cardiovasc Res. 2008;79:551–552. doi: 10.1093/cvr/cvn189. [DOI] [PubMed] [Google Scholar]
- 5.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 6.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 7.Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuwahara K, et al. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J Clin Invest. 2007;117:1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin M, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher PG, Romana M, Tse WT, Lux SE, Forget BG. The human ankyrin-1 gene is selectively transcribed in erythroid cell lines despite the presence of a housekeeping-like promoter. Blood. 2000;96:1136–1143. [PubMed] [Google Scholar]
- 11.Porter NC, et al. Association of small ankyrin 1 with the sarcoplasmic reticulum. Mol Membr Biol. 2005;22:421–432. doi: 10.1080/09687860500244262. [DOI] [PubMed] [Google Scholar]
- 12.Gallagher PG, Forget BG. An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene. J Biol Chem. 1998;273:1339–1348. doi: 10.1074/jbc.273.3.1339. [DOI] [PubMed] [Google Scholar]
- 13.Birkenmeier CS, Sharp JJ, Gifford EJ, Deveau SA, Barker JE. An alternative first exon in the distal end of the erythroid ankyrin gene leads to production of a small isoform containing an NH2-terminal membrane anchor. Genomics. 1998;50:79–88. doi: 10.1006/geno.1998.5305. [DOI] [PubMed] [Google Scholar]
- 14.Antos CL, et al. Activated glycogen synthase-3 beta suppresses cardiac hypertrophy in vivo. Proc Natl Acad Sci USA. 2002;99:907–912. doi: 10.1073/pnas.231619298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 16.Crackower MA, et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110:737–749. doi: 10.1016/s0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 17.Niu Z, et al. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci USA. 2008;105:17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 20.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 22.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 24.Musarò A, et al. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- 25.Lai KM, et al. Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol Cell Biol. 2004;24:9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamon S, Wallace MA, Léger B, Russell AP. Regulation of STARS and its downstream targets suggest a novel pathway involved in human skeletal muscle hypertrophy and atrophy. J Physiol. 2009;587:1795–1803. doi: 10.1113/jphysiol.2009.168674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakuma K, Akiho M, Nakashima H, Akima H, Yasuhara M. Age-related reductions in expression of serum response factor and myocardin-related transcription factor A in mouse skeletal muscles. Biochim Biophys Acta. 2008;1782:453–461. doi: 10.1016/j.bbadis.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Li S, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson EN, Schneider MD. Sizing up the heart: Development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 31.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 32.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji R, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 34.Roy S, et al. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res. 2009;82:21–29. doi: 10.1093/cvr/cvp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenberg I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.