Abstract
Understanding the fine-structure molecular architecture of bacterial epidemics has been a long-sought goal of infectious disease research. We used short-read-length DNA sequencing coupled with mass spectroscopy analysis of SNPs to study the molecular pathogenomics of three successive epidemics of invasive infections involving 344 serotype M3 group A Streptococcus in Ontario, Canada. Sequencing the genome of 95 strains from the three epidemics, coupled with analysis of 280 biallelic SNPs in all 344 strains, revealed an unexpectedly complex population structure composed of a dynamic mixture of distinct clonally related complexes. We discovered that each epidemic is dominated by micro- and macrobursts of multiple emergent clones, some with distinct strain genotype–patient phenotype relationships. On average, strains were differentiated from one another by only 49 SNPs and 11 insertion-deletion events (indels) in the core genome. Ten percent of SNPs are strain specific; that is, each strain has a unique genome sequence. We identified nonrandom temporal–spatial patterns of strain distribution within and between the epidemic peaks. The extensive full-genome data permitted us to identify genes with significantly increased rates of nonsynonymous (amino acid-altering) nucleotide polymorphisms, thereby providing clues about selective forces operative in the host. Comparative expression microarray analysis revealed that closely related strains differentiated by seemingly modest genetic changes can have significantly divergent transcriptomes. We conclude that enhanced understanding of bacterial epidemics requires a deep-sequencing, geographically centric, comparative pathogenomics strategy.
Keywords: Streptococcus pyogenes, evolution, invasive disease, phylogeography, population genetics
Bacterial molecular population genetics and evolution research have long been hobbled by the lack of comprehensive genome-wide polymorphic markers. This limitation has resulted in imprecisely defined genetic relationships between strains. The recent advent of massively parallel DNA sequencing techniques (“NextGen” sequencing) now permits full-genome sequences to be generated rapidly from large samples of bacterial strains (1–4). NextGen sequencing opens the door to addressing longstanding but heretofore economically intractable questions in all areas of biomedical research—for example, the fine-structure molecular architecture of bacterial epidemics.
Group A Streptococcus (GAS), a Gram-positive bacterial pathogen, is an important cause of human morbidity and mortality worldwide (5). The organism is responsible for an estimated 600 million episodes of human infection each year globally and more than 10,000 cases of severe invasive disease annually in the United States. It has been known for more than a century that GAS has the capacity to cause epidemics characterized by rapid increase in disease frequency and severity. For example, Weech (6) described an epidemic of septic scarlet fever in Yunnanfu, China, that killed 50,000 people, fully 25% of the population of the province. One unusual feature of GAS epidemics caused by strains of some M protein serotypes is a periodicity of infection peaks recurring at 4- to 7-year intervals (7, 8). The molecular genetic events contributing to the genesis and cyclic recurrence of GAS epidemics are essentially unknown. For example, the nature and extent of genetic differentiation of strains comprising individual or recurrent epidemics is not known.
We used serotype M3 strains causing invasive infections in Ontario as a model for understanding the molecular genetic basis of bacterial epidemics for several reasons (9, 10). First, a comprehensive, population-based sample of strains causing invasive infections since 1992 is available, giving us the ability to define, rather than infer, many epidemiologic characteristics (11–13). Second, the GAS genome is relatively small (∼1.9 Mb), so analysis of large sets of strains is technically feasible (14, 15). Third, clinical and geographic data available for these strains provide important additional parameters that can contribute to understanding the features/dynamics of these epidemics.
Here, we report the results of studies designed to probe the molecular contours of three consecutive epidemics of invasive disease caused by serotype M3 GAS strains. We provide a genome-wide portrait of successive bacterial epidemics and present evidence that in the environment of invasive infections positive selection contributes to variation in certain GAS genes.
Results and Discussion
Genome Sequencing and Polymorphism Discovery.
This study was based on 344 serotype M3 GAS strains (Table S1) recovered between January 1992 and December 2007 in a prospective population-based surveillance study of invasive GAS infections conducted in Ontario, Canada. Whole-genome short-read-length (36 nt) single-end reads were generated for 87 of the 344 invasive strains, one-fourth of the invasive isolates studied. Polymorphisms such as biallelic SNPs and insertions/deletions (indels) were identified relative to the MGAS315 core genome (that is, the ∼1,670-kbp portion lacking mobile genetic elements that is largely unique in sequence and conserved in gene content relative to other sequenced GAS serotypes) using Variant Ascertainment Algorithm software (16). Cumulatively, 21,060 biallelic SNPs and 2,962 short indels were identified among the 87 strains sequenced, but the vast majority of these polymorphisms (18,740) mapped erroneously to repetitive sequences within MGAS315 prophages. These falsely identified polymorphisms were excluded from the analysis of genetic relationships among the strains studied.
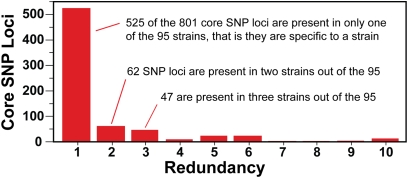
We found 5,243 polymorphisms (SNPs and indels) that mapped to the core genome of strain MGAS315, ranging from 29 to 133 polymorphisms per strain. The SNPs identified by short-read sequencing were combined with SNPs previously found in eight additional invasive M3 strains from the epidemics identified by microarray hybridization comparative genomic sequencing (9, 17). In the aggregate, 4,269 SNPs occurring at 801 polymorphic loci (Table S2) distributed across the core genome were identified among these combined 95 invasive strains. On average, the core genome of each strain differed from the reference MGAS315 strain by only 49 SNPs and 11 indels. Two-thirds of the GAS SNP loci (525, 65.5%) were identified in only 1 of the 95 strains studied (Fig. 1 and Fig. S1). However, in contrast to the population as a whole, for any single strain 10% of the SNPs were strain specific. Thus, although the 95 serotype M3 invasive strains are very closely related genetically, each strain has a unique genome sequence.
Fig. 1.
Distribution of SNPs among the sequenced serotype M3 genomes. As a population, 65% of SNP loci (525/801) and 10% of the SNPs (525/5,243) are strain specific. The vast majority of SNP loci are present in only one or just a few strains. Individually, each strain has on average ∼6 unique SNPs (range, 0–47 SNPs) and ∼41 informative SNPs (range, 10–70 SNPs) in the core genome relative to the others.
Conversely, about one-third of the SNP loci (276; 34.5%) were present in two or more of the 95 strains and therefore were phylogenetically informative. Predicted coding sequence accounts for 87% of the MGAS315 genome, and a relatively similar portion of the core SNPs (631; 79%) occur in coding sequences. Nonsynonymous (amino acid-altering) SNPs account for 69% (432), and synonymous SNPs account for 31% (199) of the core coding SNPs, values that approximate the expected distributions for randomly occurring mutations.
We found 496 insertions at 87 loci and 469 deletions at 106 loci (965 indels total) in the core chromosome among the 87 strains for which short-read-length sequence data were generated. Insertions ranged in size from 1 to 21 bp and deletions from 1 to 117 bp. The bulk of the indels (815, 85%) at most of the loci (151; 80%) were a single-nucleotide event occurring in a homopolymeric nucleotide tract (for example the gain or loss of a T nucleotide from a run of five consecutive Ts). About two-thirds of the indel loci (134; 69%) were strain specific, and the other one-third (59; 31%) were present in more than one of the genomes sequenced. However, in contrast to the SNPs, the distribution of the indel loci among coding sequence (73; 39%) and intergenic sequence (115; 61%) does not reflect the proportion of these sequences within the genome. That is, we found that the majority of indel loci were present in the intergenic regions rather than in the more abundant coding sequences.
Inferred Premature Stop Codons.
In principle, each nucleotide change may alter the phenotype of the strain containing it. Polymorphisms that create or remove stop codons are especially likely to alter strain phenotype because they result in loss of protein expression or in expression of an altered form of a protein. Among the 95 sequenced strains, we identified 21 nonsense SNP loci (3.3% of the 632 core coding SNP loci; Table S3) and 53 indel loci (72.6% of the 73 core coding indel loci; Table S4) predicted to terminate 61 gene products prematurely. These polymorphisms reduce the length of the inferred products on average by 50% (range, 0–98%). Although indels are underrepresented among coding sequences and constitute only 19% (193 of 995) of the total polymorphic loci identified, they account for the majority (72%; 53 of 74) of the loci inferred to cause premature translational termination. Importantly, consistent with the likelihood of phenotypic consequences, among the 61 genes encoding inferred prematurely terminated proteins are known GAS virulence factors including covRS, a two-component pleiotropic virulence regulator; mtsR, a metal uptake and metabolism regulator; speB, a secreted cysteine protease; and hasB, a key hyaluronic acid capsule synthesis gene.
Population Genetic Structure Assessments.
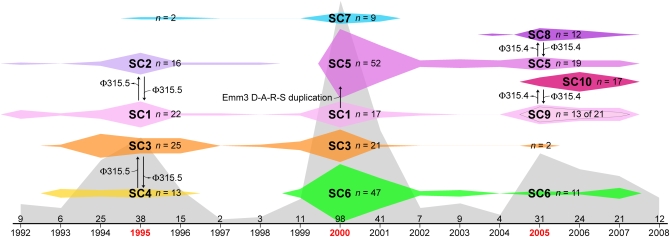
We assessed the molecular population genetic structure of the strain sample at three different levels. First, for the complete invasive strain sample, we modeled the genetic structure (Fig. 2) using a limited set of informative polymorphic loci distributed around the genome. Strains were parsed into 1 of 10 major subclones (SCs) based on the combination of emm3 allele, prophage content, and a 15-locus SNP haplotype as previously described (10). Although the model is based on only a small aggregate portion of the GAS genome, a critical finding was that the population is neither genetically homogeneous nor static but is composed of distinct subclones that emerge or are lost with each epidemic. Based on this limited genetic information, we hypothesized how the subclones probably are related to each other (Fig. 2). For example, SC-1 strains in the first epidemic wave probably are progenitors of SC-5 strains emerging in the second wave, which in turn give rise to the SC-8 strains in the third wave. We stress that Fig. 2 is only an estimate of the true population genetic structure, and therefore the conclusions that can be drawn from these estimated relationships are modest.
Fig. 2.
Model summarizing changes in M3 subclones over time. The frequency distribution of all strains in the three epidemics is shown in gray, with three peaks of infection centered around 1995, 2000, and 2005. Ten major subclones (SC-1 to SC-10) were identified among the 344 strains collected from 1992 through 2007 based on a 15-SNP haplotype, prophage content, and emm3 allele (as given in Table S1). SC-9 is inset within SC-1 in the third peak to indicate that at this level of genetic interrogation they are nearly identical, differing by a single synonymous polymorphism in emm3, and therefore produce M proteins that have the same amino terminal sequence. The widths of the colored SC symbols show the temporal distribution of the SCs, and the heights are proportional to the annual abundance. Arrows between SCs indicate estimated relationships and give differences found in the loci assessed. The total number of isolates per year is given above the time line at the bottom.
We next assessed the population genetic structure for 95 strains using a comprehensive set of 801 SNPs (Table S2) identified by whole-genome sequencing relative to the serotype M3 strain MGAS315 core genome. Strains differed from their nearest neighbor on average by 14 SNPs (range, 1–63 SNPs), and from the other strains by 54 SNPs on average (range, 1–105 SNPs). Consistent with our hypothesis of overall genetic relationships shown in Fig. 2, strains of the same subclone differ on average by only 28 SNPs (averages ranged from 14 to 45 SNPs among the 10 subclones).
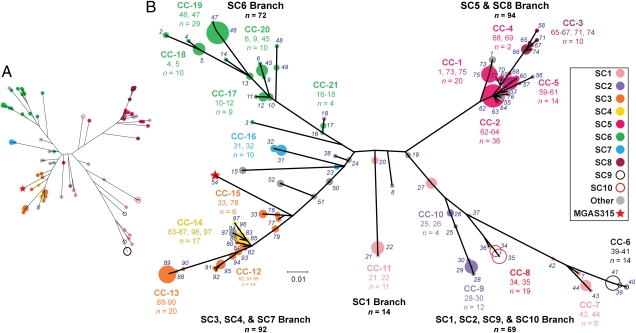
To test our hypothesis (Fig. 2) of genetic relationships among the 95 invasive strains, we analyzed strains using the entire sample of 276 informative SNPs. A neighbor-joining tree (Fig. 3A) fully supports the subclone model of overall genetic relationships. Similarly, all neighbor-joining trees generated using all SNPs (n = 801), intergenic SNPs (n = 170), nonsynonymous SNPs (n = 432), synonymous SNPs (n = 199), or indels (n = 192) gave a similar topology (Fig. S2). That is, each tree has four major branches, an SC5-SC8 branch, an SC1-SC9-SC10 branch, an SC3-SC4 branch, and an SC6 branch.
Fig. 3.
Genetic relationships among serotype M3 strains. (A) Neighbor-joining tree for 95 sequenced strains based on 276 informative SNPs. Nodes of the tree are color coded by subclone as shown in the legend. (B) Neighbor-joining tree for 344 invasive strains (plus reference strain MGAS315, red star, haplotype 54) based on 280 sequence-validated biallelic SNPs. The 97 haplotypes (numbers in italics) defined by the concatenated SNP sequences are illustrated on the tree as color-coded circles that have areas proportional to the number of strains represented. Clonal complexes (CCs) of related haplotypes (CC-1 to CC-21) are indicated, with grouped haplotypes, and the number of strains encompassed is given for each complex. Haplotypes and clonal complexes are color coded by subclone as shown in the legend. The topologies of the trees in A and B are analogous; common to both are four major branches similar in composition, separation, and length.
Last, we assessed the population structure for all 344 invasive strains and reference genome MGAS315 by analysis of 280 biallelic SNPs sequence-validated using a high-throughput mass-spectrometry method (344 strains at 280 loci = 96,320 base calls) (18, 19). These 280 SNPs were a random subset of the larger sample of 801 SNPs identified by whole-genome sequencing and included synonymous, nonsynonymous, and intergenic SNPs. By this process we determined a nucleotide for 95,949 (99.6%) of the SNP assessments performed. The topology of the resulting tree is consistent with the genetic relationships derived using all informative SNPs (Fig. 3B).
Genetic Polymorphisms with Respect to Time.
At the level of genetic resolution provided by our subclone model (Fig. 2), we were unable to resolve adequately several important aspects of the epidemics. For example, we could not determine whether the presence of SC-1 strains in each of the epidemics was caused by the reemergence of genetically identical strains from evolutionarily quiescent reservoirs, or, alternatively, if genotypically distinct strains emerged with each new peak. The availability of full-genome sequence data for strains from the three epidemics gave us the opportunity to resolve this matter. Thus, we assessed the nature and extent of genetic variation among invasive serotype M3 strains over time. Because of the considerable genetic complexity in the population revealed by the full-genome data, we chose to analyze the data by the single-locus-variant method of eBURST (20). This strategy permitted us to reduce the complexity of the population structure by grouping strains into complexes of clonally related haplotypes. Twenty-one clonal complexes accounted for 64 (67%) of the haplotypes and 275 (80%) of the strains (Fig. S3). Importantly, we found that SC-1, -2, -3, -5, and -6 are not genetically homogeneous; instead, each can be resolved into multiple clonal complexes (Fig. 3B). Inspection of the temporal distribution of the clonal complexes found that nearly all strains comprising a clonal complex are restricted to a single epidemic. Thus, the comparative genome sequence data unambiguously show that each epidemic is composed of strains genetically distinct from strains present in the preceding wave rather than reemerging genetically identical organisms.
To estimate a rate of SNP accretion, we determined the average number of SNPs that accumulated from one epidemic to the next (i.e., over ∼5 years) between sequenced strains of progenitor–descendant clonal complex pairs (Fig. 3B). The tree structure suggests that the evolutionary path between strains of these clonal complexes is direct, leading to a conservative estimate of accumulated SNPs. We estimate a rate of accumulation of 1.7 net core SNPs (95% confidence interval: 0.2–3.2) per strain per year in this strain sample (Table S5).
Identification of Genes Potentially Under Diversifying Selection.
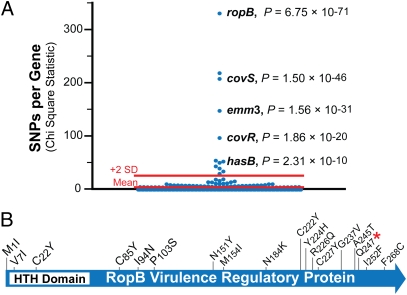
We next used the genome sequence data to test the hypothesis that some genes have a molecular signal of evolving under diversifying selection. To test this hypothesis, we assessed SNPs in coding sequences of the core genome for nonrandom distribution. Twenty-two of the total of 462 variant genes had a statistically significant overabundance of SNPs (Table S6), including genes encoding several well-characterized GAS virulence factors (e.g., emm3, covR, and covS). Notably ropB (also designated rgg), a gene that encodes a transcriptional regulator (21), had the highest rate of nucleotide sequence diversification (Fig. 4A). RopB controls expression of the speB gene that encodes an extracellular cysteine protease virulence factor (22). Surprisingly, all 12 SNPs in ropB were nonsynonymous—that is, were mutations producing amino acid replacements in RopB. These findings suggested that alleles of ropB resulting in RopB variants are under positive (Darwinian) selection. We further investigated the frequency and nature of polymorphism in ropB by conventional sequencing of the gene in another 70 invasive strains of the study set. We identified six more SNPs in ropB, including five nonsynonymous mutations and one nonsense mutation that produce a stop codon that would terminate RopB prematurely. Thus for all the invasive isolates examined, all ropB mutations would produce sequence changes in RopB (Fig. 4B). The data lead us to conclude that selection in the host shapes variation at this locus.
Fig. 4.
Identification of GAS genes with significantly increased nucleotide diversity. (A) Illustrated is the distribution of χ2 statistic determined for observed versus expected numbers of SNPs for all 1,549 core genes. Indicated are known virulence genes with nucleotide diversity significantly exceeding mean chance expectation (P ≤ 0.05; χ2 adjusted for multiple testing). (B) Schematic of RopB showing locations of inferred amino acid changes. HTH, predicted DNA-binding helix-turn-helix motif.
Temporal–Spatial Subclone Distribution (Phylogeography).
Our current understanding of the temporal–spatial characteristics of bacterial epidemics lacks precision and thus is inadequate. Many factors contribute to this knowledge deficit, including lack of analysis of comprehensive population-based samples and the relatively large genome sizes of bacteria (compared with viruses such as influenza). Understanding the geographic pattern and rate of strain dissemination has relevance to public health, vaccine development, and pathogenesis. Current thinking holds that most bacterial epidemics are short in time frame and clonal in nature and therefore are fairly homogenous in virulence characteristics. However, these ideas may be incorrect, because most studies have been performed by assessing strain variation in pulsed-field gel electrophoresis pattern or multilocus sequence type, techniques that index very small portions of the genome and are not useful for fine-structure differentiation of closely related strains (23). For example, 90% of emm3 strains in our study have the same multilocus sequence type.
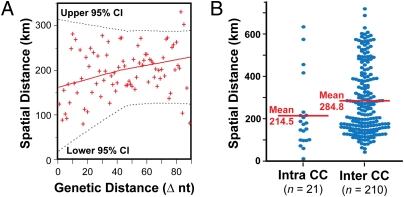
We hypothesized that spatial structure exists in clone distribution because insufficient time has elapsed in the human-to-human transmission chain for clonally related descendants to become distributed at random throughout the province. To test this idea, we assessed the relationship between genetic distance based on full-genome data and physical distance based on patient postal code information. Consistent with the hypothesis, among the 95 sequenced strains, genetic distance strongly correlated with geographic distance (P = 0.0005; Spearman’s test) (Fig. 5A). Moreover, expanding the analysis to the 275 strains encompassed by the 21 clonal complexes, we found the average geographic distance between strains within a clonal complex (214.5 km; range, 13–635 km) was significantly less than that between strains of different clonal complexes (284.8 km; range, 44–720 km) (P = 0.025; Mann–Whitney test) (Fig. 5B). Thus, the correlation between genetic relatedness and spatial proximity persists across larger groups of strains. Illustrative of this spatial structure, many clonal complexes were confined to one or a small number of geographic regions of the province (Fig. S4).
Fig. 5.
Phylogeographic structure. (A) Plotted for the sequenced genomes are the mean spatial distances between invasive infection cases versus the number of core nucleotide differences. Shown is a locally weighted least-squares fit (LOWESS; n = 33) of the data. In general, geographic distance between the cases increased with genetic distance between strains (P = 0.0005; Spearman’s test). (B) The distribution of mean spatial distances separating invasive infection cases, calculated pairwise for all strains within a clonal complex (intra CC), and similarly for all strains that are of different clonal complexes (inter CC). In general, the geographic distance separating strains of the same CC is significantly less (∼70 km, on average) than that separating strains of different CCs (P = 0.025; Mann-Whitney test).
Comparative Expression Microarray Analysis.
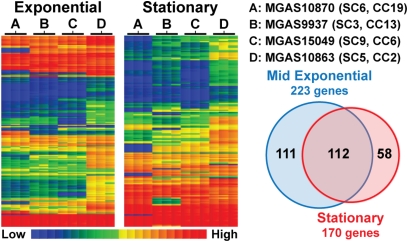
Considering the level of genetic diversity and complexity revealed by our population genomic analysis, we began to investigate corresponding phenotypic diversity. As an initial assessment we examined strain genotype–patient phenotype (invasive infection type) associations for each of the 21 clonal complexes. One-third of the clonal complexes (7/21) had significantly nonrandom infection-type associations (P ≤ 0.05; Fisher’s exact test; Table S7). Nonrandom associations between strain genotype and patient phenotype led us to hypothesize that variation in gene expression between strains contributed to these associations. Thus, we tested the hypothesis that genetically distinct serotype M3 lineages within this population differ in global gene expression. The four strains studied are genetically representative of strains of the four major phylogenetic branches (Fig. 3). Importantly, each strain lacks polymorphisms in major regulatory genes such as covRS and mga, known to influence the expression of large numbers of virulence genes. A custom Affymetrix GeneChip was used to compare the transcriptome of four strains at midexponential and stationary phases of growth. Growth in vitro was virtually identical for all four strains (Fig. S5). Consistent with our hypothesis, the analysis revealed considerable differences in the global transcript patterns of these four strains (Fig. 6). At midexponential phase, 223 genes had significant variation (P ≤ 0.05; ANOVA) in transcripts among the four strains, and for the stationary phase 170 transcripts were significantly different (Fig. 6). Cumulatively for both growth phases, 281 genes (∼15% of the GAS genome) differed significantly in expression for one or more of the strains tested. However, in individual pairwise comparisons between any two strains at a single growth phase, transcripts were significantly different on average for only 36 genes (1.9% of the genome). Thus, we found that the modest genetic differences between these strains of different clonal lineages (both SC and clonal complex) resulted in considerable differences in transcriptome. Although expression-array analysis was conducted for only a small number of strains, the findings are consistent with association between the population genetic structure and clinical manifestation that is supported by correspondences in transcriptional profiles.
Fig. 6.
Comparative transcriptome analysis. The four strains analyzed were each genetically representative of four numerically dominant subclones, as labeled for each lane. Microarray analysis was performed in triplicate on samples harvested at midexponential and stationary phases of growth. The heat map illustrates genes significantly (P ≤ 0.05; ANOVA) changed in expression at each time point. In total, 281 genes had altered transcripts, with 112 genes showing changes at both time points.
Concluding Comment.
We used deep-genome sequencing data integrated with epidemiologic information to resolve the molecular key features of three successive epidemics caused by the human bacterial pathogen GAS. The analysis was made possible by a unique population-based strain sample recovered over a period of 16 years. This comparative pathogenomic analysis delineated the fine-structure molecular population genetics of these epidemics and defined relationships among the 344 invasive strains responsible for the three epidemics. The resulting evolutionary genetic framework permitted us to identify overall patterns of strain genotype–patient phenotype relationships, to assess phylogeographic features of the epidemics, and rationally to choose representative strains for comparative transcriptome analysis. Of significant note, the extensive data set revealed information about GAS biology. Especially intriguing is the very strong signal of positive selection we observed for several genes, for example ropB that encodes a regulatory protein implicated in virulence.
Our work will facilitate subsequent study and enhanced understanding of the genomic relationships between serotype M3 strains causing pharyngitis and invasive infections. For example, are the strains recovered from patients with invasive infections a nonrandom genetic subset of pharyngitis strains? Similarly, is the rate of accretion of genetic variation in the pharyngitis strains essentially identical to invasive strains? These and other questions bearing on the evolutionary events contributing to bacterial clone emergence, strain differentiation, and epidemics are now readily amendable to analysis at the full-chromosomal level in large samples using the comparative molecular pathogenomics strategies employed herein.
Materials and Methods
For further details, see SI Materials and Methods.
Bacterial Strain Samples.
The 344 invasive serotype M3 strains (Table S1) that are the basis of this investigation were recovered between January 1992 and December 2007 from a prospective population-based surveillance study of invasive GAS infections conducted in Ontario, Canada (population = 12.2 million in 2006 census). This study has been described in several publications (11–13). The sequence of serotype M3 strain MGAS315, recovered in the late 1980s from a patient with invasive disease, was used as a reference for genome analyses (NCBI reference sequence NC004070) (14).
Subclone Assignment.
PCR amplification, gel electrophoresis, and conventional Sanger DNA sequencing were used to determine emm3 allele, prophage content, and an SNP haplotype, using 15 informative biallelic loci distributed around the genome, for all 344 invasive strains. Primers, reagents, and reaction conditions used were as previously described (10).
Genome Sequencing and Polymorphism Discovery.
Genome sequence data were generated for 87 invasive isolates using the Illumina Genome Analyzer System according to the manufacturer’s protocols. Resultant sequencing reads were mapped, and polymorphisms (SNPs and indels) were called relative to the strain MGAS315 reference genome using Variant Ascertainment Algorithm software (16). We included genome data for eight additional strains from the Ontario invasive sample previously obtained by comparative microarray hybridization-based sequencing (17), resulting in genome sequence data for 95 strains. Sequence data were deposited in the NCBI Short Read Archive (SRA) in the study “Streptococcus pyogenes Pathogenomics SRP000775” under accession numbers SRX004870–SRX004952 and SRX005690.
Mass Spectroscopy SNP Analysis.
One-third (n = 280 of 801; 35%) of the biallelic core SNPs identified by full-genome sequencing were analyzed in all 344 invasive emm3 strains studied and in reference strain MGAS315. This random subset of SNP loci was distributed throughout the core genome and reflected the distribution of unique versus informative and coding versus intergenic SNPs of the whole set. Polymorphic loci were assessed by a mass-spectrometry–based method (18, 19) using the iPLEXTM Gold assay and MassARRAY system according to the manufacturer’s instructions (Sequenom). Primers for multiplex PCR amplification and SNP interrogation by single-base extension reactions were designed using Assay Designer software (Sequenom) (Table S8).
Phylogenetic Reconstruction and Clonal Complex Assignment.
Concatenated SNP and indel loci nucleotide sequences were aligned using ClustalX (24). Phylogenetic relationships based on the aligned sequences were inferred using the neighbor-joining method implemented in SplitsTree v. 4 (25). Schematics of the resultant trees were made using DendroScope (26). Haplotypes were enumerated, and genetic distances were calculated using molecular evolutionary genetics analysis software v. 4.0 (27). Complexes of clonally related strains (Fig. S3) were inferred systematically by the single-locus-variant-grouping method of eBURST (20).
Statistical Analyses.
As an indice of positive selection, genes having more SNPs than expected for a random distribution were identified by χ2 test (Table S6). Probabilities were stringently and liberally corrected for multiple testing, using the Bonferroni and Benjamini–Hochberg methods, respectively. As an indice of potential bacterial genetic content influencing clinical disease manifestation, the association between strain genotype and infection phenotype, was assessed for all 21 of the clonal complexes using Fisher’s exact test (Table S7). As an indice of phylogeographic structure, the correlation between genetic distance based on nucleotide differences and spatial distance based on postal code was assessed among the 95 sequenced strains using the nonparametric two-tailed Spearman’s test. Similarly case-to-case geographic distance, intra- and interclonal complex, was assessed using the nonparametric two-tailed Mann–Whitney test. Various other distribution statistics (means, standard deviations, confidence intervals, and others) were calculated using Prism (www.graphpad.com).
Comparative Expression Microarray Analysis.
Global transcriptional analysis was conducted for four strains representative of four phylogenetically divergent emm3 clones using a custom GeneChip (Affymetrix) by methods described previously (28). GAS cells were cultured at 37 °C in 5% CO2 atmosphere in Todd Hewitt medium with 0.2% wt/vol yeast extract. Culture growth was assessed by measuring optical density at 600 nm and by quantitative plating CFU enumeration (Fig S5). Genes significantly differently expressed under the conditions compared were determined by ANOVA with probabilities corrected for multiple testing by the method of Bonferroni. Heat maps were generated with CHIPST2C software (www.chipST2c.org).
Supplementary Material
Acknowledgments
We are indebted to Dr. M. Giovanni, National Institutes of Allergy and Infectious Diseases, National Institutes of Health, for support; S. Pong-Porter, C. Cantu, and A. Ayeras for assistance with strains; P. Sumby for help with microarray analysis; the Broad Institute Genome Sequencing Platform team, K. Stockbauer, and F. R. DeLeo for critical reading of the manuscript, and members of the University of Houston Bioinformatics Laboratory for computational assistance. This project was supported in part with funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract HHSN272200900007C, and a Collaborative Scholar Award from The Methodist Hospital Research Institute (to J.M.M. and Y.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911295107/DCSupplemental.
References
- 1.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLean D, Jones JD, Studholme DJ. Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat Rev Microbiol. 2009;7:287–296. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- 3.Orsi RH, et al. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics. 2008;9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 5.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 6.Weech AA. Scarlet fever in China. Some impressions concerning the value of serum treatment in a malignant form of the disease. N Engl J Med. 1931;204:968–974. [Google Scholar]
- 7.Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980-1990 and their association with disease. J Med Microbiol. 1993;39:165–178. doi: 10.1099/00222615-39-3-165. [DOI] [PubMed] [Google Scholar]
- 8.Köhler W, Gerlach D, Knöll H. Streptococcal outbreaks and erythrogenic toxin type A. Zentralbl Bakteriol Mikrobiol Hyg [A] 1987;266:104–115. doi: 10.1016/s0176-6724(87)80024-x. [DOI] [PubMed] [Google Scholar]
- 9.Beres SB, et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beres SB, et al. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc Natl Acad Sci USA. 2004;101:11833–11838. doi: 10.1073/pnas.0404163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies HD, et al. Ontario Group A Streptococcal Study Group. Invasive group A streptococcal infections in Ontario, Canada. N Engl J Med. 1996;335:547–554. doi: 10.1056/NEJM199608223350803. [DOI] [PubMed] [Google Scholar]
- 12.Kaul R, McGeer A, Low DE, Green K, Schwartz B. Population-based surveillance for group A streptococcal necrotizing fasciitis: Clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Ontario Group A Streptococcal Study. Am J Med. 1997;103:18–24. doi: 10.1016/s0002-9343(97)00160-5. [DOI] [PubMed] [Google Scholar]
- 13.Sharkawy A, et al. Ontario Group A Streptococcal Study Group. Severe group A streptococcal soft-tissue infections in Ontario: 1992–1996. Clin Infect Dis. 2002;34:454–460. doi: 10.1086/338466. [DOI] [PubMed] [Google Scholar]
- 14.Beres SB, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: Phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferretti JJ, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nusbaum C, et al. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat Methods. 2009;6:67–69. doi: 10.1038/nmeth.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albert TJ, et al. Mutation discovery in bacterial genomes: Metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- 18.Honisch C, et al. Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc Natl Acad Sci USA. 2007;104:10649–10654. doi: 10.1073/pnas.0704152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honisch C, Cullinan A. Applications of nucleic acid analysis by MALDI-TOF mass spectrometry in clinical microbiology. European Infectious Disease. 2009;3:82–85. [Google Scholar]
- 20.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukomski S, et al. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foxman B, Zhang L, Koopman JS, Manning SD, Marrs CF. Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol Perspect Innov. 2005;2:10. doi: 10.1186/1742-5573-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 26.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Shelburne SA, 3rd, et al. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.