Abstract
The type 4 melanocortin receptor MC4R, a key relay in leptin signaling, links central energy control to peripheral reserve status. MC4R activation in different brain areas reduces food intake and increases energy expenditure. Mice lacking Mc4r are obese. Mc4r is expressed by hypothalamic paraventricular Thyrotropin-releasing hormone (TRH) neurons and increases energy usage through activation of Trh and production of the thyroid hormone tri-iodothyronine (T3). These facts led us to test the hypothesis that energy homeostasis should require negative feedback by T3 on Mc4r expression. Quantitative PCR and in situ hybridization showed hyperthyroidism reduces Mc4r mRNA levels in the paraventricular nucleus. Comparative in silico analysis of Mc4r regulatory regions revealed two evolutionarily conserved potential negative thyroid hormone-response elements (nTREs). In vivo ChIP assays on mouse hypothalamus demonstrated association of thyroid hormone receptors (TRs) with a region spanning one nTRE. Further, in vivo gene reporter assays revealed dose-dependent T3 repression of transcription from the Mc4r promoter in mouse hypothalamus, in parallel with T3-dependent Trh repression. Mutagenesis of the nTREs in the Mc4r promoter demonstrated direct regulation by T3, consolidating the ChIP results. In vivo shRNA knockdown, TR over-expression approaches and use of mutant mice lacking specific TRs showed that both TRα and TRβ contribute to Mc4r regulation. T3 repression of Mc4r transcription ensures that the energy-saving effects of T3 feedback on Trh are not overridden by MC4R activation of Trh. Thus parallel repression by T3 on hypothalamic Mc4r and Trh contributes to energy homeostasis.
Keywords: energy homeostasis, gene transcription, leptin pathway, ligand-dependent repression, thyrotropin-releasing hormone
Reversing the increasing incidence of obesity requires knowledge of physiological regulations controlling energy homeostasis. Understanding how central genes involved in endocrine and metabolic axes are regulated is crucial to these problems.
Thyroid hormones (THs) regulate metabolism and appetite (1, 2). THs, particularly the biologically active form tri-iodothyronine, T3, stimulate the basal metabolic rate. Hyperthyroidism leads to increased catabolism and weight loss; hypothyroidism causes weight gain (3). TH levels are kept within physiological ranges through hypothalamo-pituitary neuroendocrine feedback loops. Increased T3 represses transcription of hypothalamic Thyrotropin-releasing hormone, Trh (4), the master regulator of the hypothalamo-pituitary-thyroid (HPT) axis (5). In turn, decreased T3 output reduces metabolism and energy usage (6).
Hypothalamic TRH neurons integrate numerous metabolic, endocrine, and neuronal signals (7). T3-responsive TRH neurons in the paraventricular nucleus (PVN) express all the functional TH nuclear receptors (TRs) and a key membrane receptor involved in energy homeostasis, namely the type 4 melanocortin receptor (MC4R). In situ hybridization studies show that nearly all TRH neurons in the caudal-medial parvocellular PVN express MC4R (8).
MC4R, a membrane α-melanocyte stimulating hormone (αMSH) receptor, is integral to central leptin/melanocortin signaling (9). Leptin, a major satiety hormone, regulates energy homeostasis through food intake, energy partition, and thermogenesis (9). Centrally, leptin signaling is relayed mainly through the hypothalamic melanocortin system and MC4R-expressing neurons. Mc4r expression is widespread in the brain (10, 11), with high levels in the hypothalamus and brainstem, areas involved in energy homeostasis (12). The hypothalamus governs metabolism through complex neuroendocrine regulations. The brainstem also integrates metabolic signaling, notably regulating thermogenesis via the autonomic nervous system (13).
In the PVN, leptin stimulates TRH production by coordinating pathways that culminate in MC4R activation and increased intracellular cAMP levels. A cAMP response binding (CREB)-responsive element in the Trh promoter (8) drives Trh transcription, stimulating T3 production and energy expenditure. Further, hypothalamic T3 stimulates orexigenic pathways (14, 15) and dampens anorexigenic signals (16, 17). Despite these roles, no studies have addressed how brain TH levels contribute to central metabolic control through Mc4r expression.
Because MC4R activation stimulates the HPT axis, we examined whether homeostasis involved negative feedback regulation by T3 on Mc4r expression. We found that thyroid status modulated endogenous Mc4r mRNA levels in brain areas relevant to metabolism. In silico examination of human and mouse Mc4r proximal regulatory regions showed two conserved potential negative thyroid hormone responsive element (nTREs). To assess if TRs were associated with nTREs, we used TR antibodies for ChIP on regulatory regions from Mc4r and Trh. Differential patterns of TR binding to hypothalamic Mc4r and Trh nTREs were found as a function of thyroid status. Three different in vivo gene transfer (iGT) approaches in wild-type and TR-mutant mice were used to analyze further T3 regulation of Mc4r transcription and to define the roles of different TRs. iGT was used first for reporter studies with control and mutated Mc4r promoter constructs in wild-type and TRα°/° and TRβ−/− mice (18, 19), then to introduce shRNA against TR isoforms, and finally, to overexpress TRs. The results confirmed that T3 exerts direct regulation on Mc4r and that different TR isoforms have distinct transcriptional regulatory roles.
The data provide insight into ligand-dependent repression mechanisms and reveal an integrative function of the hypothalamus in metabolic homeostasis through coordinated, T3-dependent feedback on hypothalamic Mc4r gene, together with that of Trh.
Results
Thyroid Status Regulates Endogenous Mc4r Gene Expression.
MC4R activates PVN Trh transcription (8). Thus we hypothesized that, as for Trh in the PVN, Mc4r could undergo feedback by T3. To test this possibility, hypo- and hyperthyroidism were induced in 2-month-old mice by 6-n-propyl-2-thiouracil (PTU) and thyroxine (T4) treatment, respectively (Fig. S1A). The physiological consequences of modified thyroid status during this late growth phase were confirmed by the differential weight gain of treated mice (Fig. S1B). Thyroid effects on endogenous Mc4r and Trh mRNA expression were assessed by quantitative PCR (qPCR) and normalized with Gapdh mRNA (SI Materials and Methods). Levels were analyzed in the PVN, where Mc4r is expressed in hypophysiotropic TRH neurons (8), and in the arcuate nucleus (ARC) and the brainstem, areas also involved in metabolism. The frontal cortex served as a control because it expresses Mc4r but is not linked to energy homeostasis.
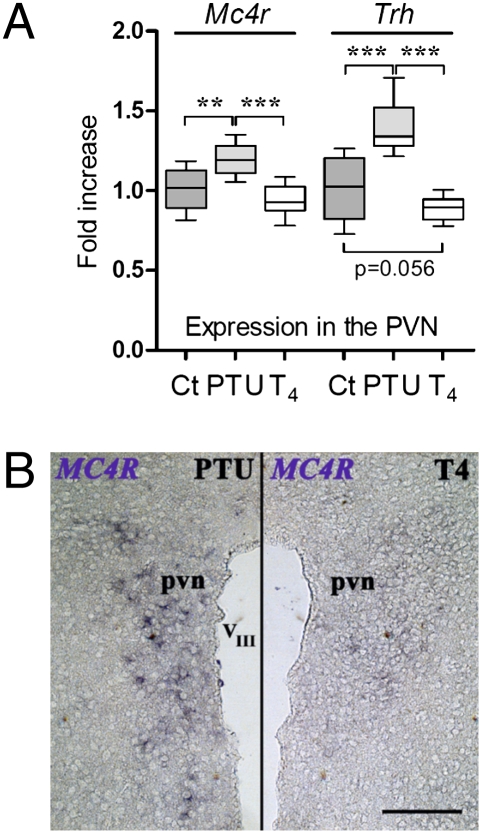
In the PVN, hypothyroidism significantly increased both Mc4r (P < 0.01) and Trh (P < 0.001) mRNA levels versus levels in euthyroid controls (Fig. 1A). However, hyperthyroidism modified neither Trh (P = 0.07) nor Mc4r expression in the PVN within the time frame used (13 days). In situ hybridization confirmed that PTU-induced hypothyroidism strongly increased Mc4r mRNA expression in the PVN compared with T4-treated animals (Fig. 1B). In the ARC, hypothyroidism increased Mc4r (P < 0.05) but, as expected (20), did not modify Trh (Fig. S2A). Conversely, hyperthyroidism decreased Mc4r expression in the ARC (P < 0.05), whereas, somewhat surprisingly, it increased Trh levels (P < 0.05) (Fig. S2A). As predicted (17), thyroid status modified Neuropeptide Y (Npy) and Pro-opiomelanocortin (Pomc) expression in the ARC (Fig. S2D). In the brainstem, hypothyroidism slightly, but significantly, reduced Mc4r mRNA, and hyperthyroidism induced a dramatic, almost 10-fold, decrease (P < 0.001) (Fig. S2B). In the cortex, Mc4r was not altered (Fig. S2C). As expected (21), neither in brainstem nor in cortex was Trh transcription affected by thyroid status (Fig. S2 B and C).
Fig. 1.
Thyroid status alters Mc4r expression in the hypothalamic PVN. (A) Real-time PCR quantification of Mc4r and Trh mRNA from PVN of adult male mice treated for 13 days by PTU (hypothyroid) or T4 (hyperthyroid), or euthyroid controls (Ct). Gene expression was normalized with Gapdh. Shown are pooled results of two independent experiments (n = 7 or 8 per group). Nonparametric ANOVA followed by permutation test was used to assess statistical significance. **, P < 0.01; ***, P < 0.001. (B) In situ hybridization shows a decrease in Mc4r mRNA expression in the PVN following T4 treatment, compared with PTU-treated mice. pvn, paraventricular nuclei; VIII, third ventricle. (Scale bar, 1 mm.)
T3-Dependent Mc4r and Trh Repression Requires Gene-Specific TR/DNA Interactions.
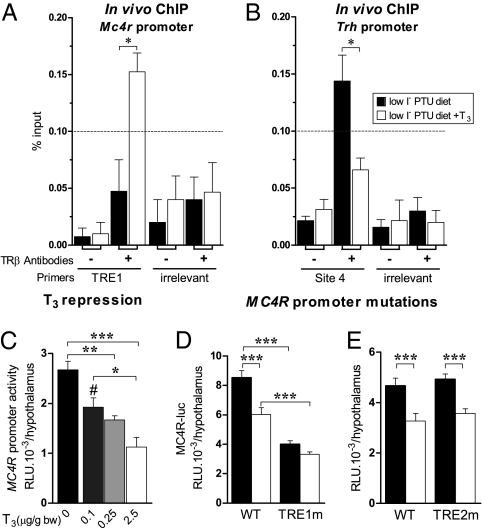
To determine if TH-induced regulatory responses involve direct interactions of TR, ChIP assays were used. Human and mouse Mc4r proximal regulatory regions share two putative monomeric TRE half-sites (Fig. S3A): TRE1 and TRE2. TRE1, at −92 bp before the transcription start site (TSS) in hMC4R, is conserved in mouse, rat, pig, and human (22). TRE2, at +374 bp after the TSS, resembles the nTRE conserved in mouse and human Trh (23).
TR binding was analyzed on the most conserved nTREs identified in Mc4r (TRE1) and Trh (site 4) regulatory regions. ChIP using anti-TRβ antibodies (Upstate Biotechnologies) was carried out on hypothalami from hypothyroid newborn mice. In these conditions, TRβ was absent from the Mc4r TRE1 (Fig. 2A), whereas TRβ was present on the Trh TRE site 4 (Fig. 2B). Following T3 treatment, TRβ was recruited to the Mc4r TRE1 region (Fig. 2A), but it decreased significantly on the Trh regulatory region (Fig. 2B). In no case, with or without T3, was any binding found on the irrelevant control regions of Trh or Mc4r promoters (Fig. 2 A Right and B Right). A second set of anti-TRβ antibodies (provided by R. Denver, University of Michigan, Ann Arbor, MI) gave similar TR/DNA binding patterns for both genes (Fig. S4 A and B).
Fig. 2.
T3 directly regulates Mc4r expression. (A and B) PCR quantification of ChIP assays carried out on hypothalami from hypothyroid newborn mice treated with T3 (2.5 μg/g bw) 20 h before sacrifice. Samples were immunoprecipitated with TRβ-specific antibodies and amplified with either MC4R-TRE1 or TRH site 4 primers or their respective irrelevant control primers. For negative controls, samples were processed through immunoprecipitation without antibody (−, background). Results represent the occupancy of TRβ isoforms at the TRE1 site in the Mc4r promoter and at the TRE-site 4 in the Trh promoter. Data are presented as percentage of input (starting sonicated DNA used for ChIP). The threshold value for a positive signal was set at 0.1% of input (dashed line). Graphs represent means of four independent experiments. Student’s t test assessed difference between the groups. (A) TRβ isoforms are present at the TRE1 site in Mc4r promoter only after T3 treatment. (B) TRβ isoforms are present at the TRE site 4 in the Trh promoter in hypothyroid animals but are absent after T3 treatment. Negative controls included primers spanning irrelevant sequences in both gene sequences (right half of each graph). (C) Dose-dependent repression of Mc4r promoter activity by T3 in hypothyroid mice. (D) Mutation of the putative TRE1 half-site abrogates T3-independent Mc4r promoter activity and its T3-dependent repression. (E) Mutation of the TRE2 putative half site is without effect. One-day-old pups were transfected in the hypothalamus with 2 μL of a 250 ng/μL solution of PEI-complexed MC4R-f.luc and then were injected s.c. with T3 [2.5, 0.25, or 0.1 μg/g body weight (bw)] or vehicle (NaCl 0.9%). Luciferase was measured 24 h later. RLU, relative light units. Means ± SEM are shown. One-way ANOVA statistical analysis was followed by Bonferroni’s multiple comparisons to assess statistical differences. In T3 treatment at 2.5 μg/g bw, *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0.05 for control group vs. group treated with T3 at 0.1 μg/g bw.
Given the association of TRs with the nTREs shown by in vivo ChIP assays, the functionality of the putative nTREs in mediating T3 signaling was tested in a physiological context using iGT (24). Transcription from Mc4r reporter plasmids bearing the nTREs driving Firefly (f.) or Renilla (r.) luciferase (luc) expression (Fig. S2 E–G) was followed in hypothalami of euthyroid or hypothyroid pups. Hypothyroidism increased T3-independent MC4R-luc transcription 2-fold versus euthyroid mice (Fig. S2E). T3 treatment of hypothyroid pups reduced MC4R-luc transcription dose dependently (Fig. 2C), with repression reaching more than 65% at the maximal T3 dose used (P < 0.001) or 50% with 3,5,3-triiodothyroacetic acid, a T3 analogue (Fig. S2F). The T3 repression of MC4R-luc paralleled that of TRH-luc (Fig. S2G).
When either the MC4R-luc or TRH-luc was cotransfected with an appropriate control (cytomegalovirus-luciferase, CMV-luc), only the physiologically relevant promoters, and not the CMV promoter, were repressed (Fig. S5 A and B). Given the parallel regulation of Mc4r and Trh transcription, we used a cotransfection approach, mixing an Mc4r reporter construct coupled to the Renilla luciferase sequence (MC4R-r.luc) with TRH-f.luc. In this cotransfection paradigm, T3 repressed transcription by 30% and 52% for Mc4r and Trh reporter plasmids, respectively (Fig. S5C).
To test the roles of the putative Mc4r TREs, site-directed mutations of either or both of the Mc4r promoter nTREs was done, producing three constructs (Fig. S3). Mutation of TRE1 (TRE1m) decreased T3-independent expression (P < 0.001) and abolished T3-dependent repression of Mc4r transcription (Fig. 2D). In contrast, mutating TRE2 (TRE2m) had no effect (Fig. 2E). When both TREs were mutated (DBm), a similar profile to the TRE1m construct was observed (Fig. S4C). In all these experiments, the non-mutated Trh construct was cotransfected as a positive control for T3-dependent regulation (Fig. S4C).
TRs Exhibit Isoform-Specific Contributions to Mc4r Expression.
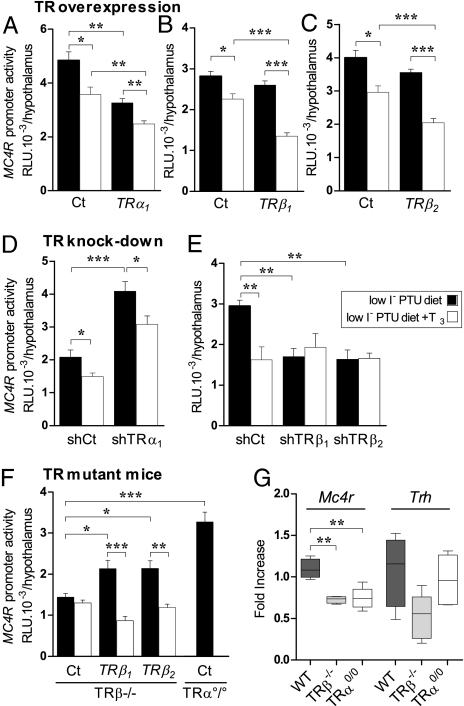
Given that site-directed mutagenesis of Mc4r promoter and ChIP analyses argued for direct regulation of Mc4r by T3 and TRs, the roles of TR isoforms were tested using expression vectors encoding TRα1 (Fig. 3A), TRβ1 (Fig. 3B), or TRβ2 (Fig. 3C). TRα1 reduced T3-independent transcription by 30% (P < 0.05). The TRβ isoforms did not modify T3-independent transcription. However, overexpression of TRβ1 or TRβ2 enhanced the repressive effects of T3 on Mc4r transcription. T3 repressed Mc4r-r.luc transcription by 20% (P < 0.05) in the presence of a control empty vector, whereas repression reached 48% with TRβ1 (P < 0.001) and 42% with TRβ2 (P < 0.001) (Fig. 3 B and C).
Fig. 3.
TRs display isoform-specific roles on Mc4r promoter. (A) TRα1 overexpression diminishes T3-independent Mc4r promoter activity. (B and C) TRβ1 or TRβ2 overexpression reinforces T3-dependent repression of Mc4r-promoter activity. Pups were transfected in the hypothalamic region of the brain with 2 μL of a 250 ng/μL solution of MC4R-f.luc with pSG5-TRα1, pSG5-TRβ1, pSG5-TRβ2, or empty pSG5 vector (25 ng/μL). (D) shTRα1 activates T3-independent MC4R-f.luc expression and abrogates T3-dependent repression. (E) shTRβ1 or shTRβ2 represses T3-independent Mc4r promoter activity and abrogates T3-induced repression. Hypothalamic transfections used MC4R-f.luc (250 ng/μL) and either pCMV-H1-shTRα1, pCMV-H1-shTRβ1, pCMV-H1-shTRβ2, or the empty vector pCMV-H1 (400 ng/pup). (F) TRα°/° and TRβ−/− animals were transfected with MC4R-f.luc with or without TRβ1- or TRβ2-expressing plasmid. T3 treatment, 2.5 μg/g bw. (G) Mc4r and Trh mRNA expression levels in adult TRβ (TRβ−/−) and TRα (TRα°/°) knockout mice. Real-time PCR quantification of Mc4r and Trh mRNA from the PVN of adult male wild-type (WT), TRβ−/−, or TRα°/° mice. Gene expression was normalized with Gapdh. Results are pooled from two independent experiments (n = 7 or 8 per group). One-way ANOVA statistical analysis was performed, followed by Bonferroni's multiple comparisons to assess statistical differences.
To examine TR-specific effects further, knockdown with plasmids containing the hybrid cytomegalovirus-polymerase III histone H1-RNA promoter (pCMV-H1) expressing shRNAs was applied. Because neither the pCMV-H1 empty vector nor the pCMV-H1 vectors coding for irrelevant shRNA sequences (shGFP) or a scrambled sequence (shSCR) affected TRH-, MC4R- or control CMV-driven transgenes (Figs. S6 and S7), pCMV-H1 was used as a control. Two shRNAs against each TR isoform (shTRs, see Table S2) were designed from published TRβ1 and TRβ2 siRNAs (25).
The shRNA method was validated against the well-documented (24, 26, 27) specificity of TRα1, TRβ1, and TRβ2 isoforms on hypothalamic Trh regulation (Fig. S6) and then was applied to the Mc4r promoter. ShTRα1 significantly (55%) increased T3-independent Mc4r transcription in hypothyroid newborn mice and maintained T3-dependent repression (Fig. 3D). Conversely, expression of shRNA against TRβ1 or TRβ2 significantly decreased T3-independent transcription (by 42% and 44%, respectively) (Fig. 3E) and abolished T3 repression. Knockdown of TRβ1 and TRβ2 thus confirmed the lack of differential effect between these two isoforms on Mc4r promoter activity seen with overexpression. These results were reproduced using another set of TRβ1 or TRβ2 shRNA (Fig. S7 B and C).
TR-specific contributions to Mc4r regulation also were investigated in mice lacking TRβ or TRα gene products (TRβ−/− or TRα°/° mice). In TRβ−/− mice, MC4R-luc was insensitive to T3 (Fig. 3F, Left). Overexpression of TRβ1 or TRβ2 restored T3-dependent Mc4r promoter repression and increased T3-independent Mc4r promoter expression (P < 0.001 in each case), confirming the roles of both TRβ isoforms seen with overexpression or shTR experiments. Moreover, T3-independent MC4R-luc expression was increased significantly in TRα°/° pups (P < 0.001) compared with TRβ−/− pups (Fig. 3F), confirming the repressive role of TRα in T3-independent Mc4r transcription. Analysis of endogenous Mc4r levels using qPCR on mutant mice showed that loss of either TR significantly modified Mc4r expression in the PVN (Fig. 3G) but not in the ARC (Fig. S7D).
Discussion
Metabolic homeostasis requires coordination of activity in specific hypothalamic nuclei, notably those governing food intake and energy expenditure. One key actor in these energy-regulating networks is the αMSH receptor MC4R (18). MC4R mutations in humans are associated with obesity and ≈5% rate of associated morbidity (28). Mice lacking the Mc4r gene are severely obese, and heterozygous mice show intermediate phenotypes, demonstrating a gene–dosage effect for Mc4r in energy balance (29). Another key player is T3, which determines metabolic rate (3). Here, using multiple in vivo approaches, we show that these two signaling pathways are linked centrally by T3 repression of Mc4r.
Thyroid Status Modifies Mc4r Expression in Brain Areas Relevant to Metabolic Regulation.
Mc4r is widely expressed in the brain, with high levels in brainstem and hypothalamus, notably the PVN where Mc4r is found in Trh neurons (10, 11). qPCR and in situ hybridization showed hypothyroidism to increase endogenous Mc4r expression in the PVN. This increase parallels the well-known rise in PVN Trh mRNA seen in hypothyroidism (21, 30). The increase in Mc4r mRNA was modest, but small changes in regulatory genes can significantly impact targets in terms of physiology, development, and evolution (31). Hypothyroidism also substantially raised, and hyperthyroidism repressed, Mc4r expression in the ARC. In the brainstem, also targeted by the melanocortin pathway, Mc4r again was repressed by hyperthyroidism, but to a much larger extent. As expected, neither in brainstem nor in cortex was Trh expression affected by thyroid status. Mc4r expression also was unchanged in the cortex. Thus thyroid status specifically alters Mc4r expression in brain areas relevant to metabolic regulation.
Evidence for Direct T3-Dependent Regulation of Mc4r Expression.
The putative nTRE, TRE1, identified in the Mc4r 5′ region, provides a molecular basis for T3 action on Mc4r expression. TRE1 mutation dramatically decreases Mc4r promoter activity and leads to loss of T3-dependent repression. TRE1, a nonclassical sequence conserved in mammals, is thought to bind monomeric nuclear receptors. TRE1 is situated in the core-promoter region and next to a CAAT box (22), suggesting a role for TRs in transcriptional machinery recruitment and direct transcriptional regulation of Mc4r by T3. This hypothesis is supported by the in vivo ChIP results showing TRβ recruitment on the putative TRE1 sequence of Mc4r promoter and on site 4 in the Trh promoter. Data from many in vitro studies on positively regulated genes suggest a model wherein TRs bind to TREs with or without ligand, T3 (32). In vivo, the TREs studied from the Mc4r and Trh promoters showed distinct TRβ recruitment patterns as a function of T3 presence or absence. Mc4r TRE1 recruited only low levels of TRβ in the absence of T3, and T3 induced a large increase in TRβ binding. In contrast, TRβ was found on Trh TRE site 4 without hormone, as previously shown (33), and T3 dissociated TRβ from Trh TRE site 4. These differences may contribute to the different sensitivities of the two genes to T3-dependent repression. Indeed, Mc4r TRE1 and Trh site 4 display very different sequences (Fig. S8 B and C), and the neighboring chromatin contexts differ for each TRE. The Mc4r TRE1 sequence could bind nuclear receptors as monomers (22), allowing TR to exert its own distinct transcriptional properties. The Trh site 4, on the contrary, preferentially binds TR/retinoid X receptor heterodimers (23). This TRE also overlaps with a canonical CREB-response element (8) that mediates rapid responses to cAMP/protein kinase A pathways. Thus Trh site 4 could recruit different sets of transcriptional coregulators and induce T3 response kinetics different from that of the Mc4r TRE1.
The data thus provide in vivo data on the molecular mechanisms of T3-dependent gene repression by TRβ in a physiological context. It should be borne in mind, however, that other TR response elements also could participate in the regulations studied, and that the contribution of TRα remains to be clarified.
Specific Contributions of TRα and TRβ Isoforms to Mc4r Regulation.
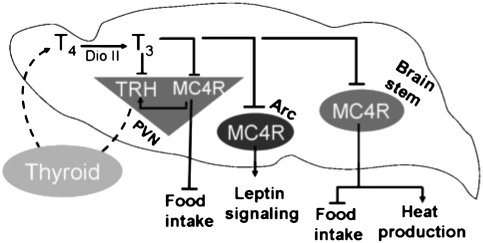
To analyze further the transcriptional regulations induced by TH, we used an iGT paradigm in the newborn mouse hypothalamus. With iGT, hypothalamic neurons are transfected with plasmid DNA complexed with polyethylenimine (PEI). PEI is one of the most exploited nonviral gene transfer agents and can be used in the same manner as its viral counterparts (e.g., adeno-associated virus or lentivirus). Previous results using this technique established that it provides region- and neuron-specific regulations (24). Somatic iGT transgenesis in pups confirmed that hypothyroidism upregulates transcription from the Mc4r promoter, whereas T3 represses hypothalamic MC4R-luc. These changes are synchronous with T3 effects on Trh transcription (Fig. 4).
Fig. 4.
Schema of T3 repression of Mc4r expression in brain. The physiological consequences of modified thyroid status on Trh and Mc4r gene regulation and metabolic output are played out through modulation of Mc4r expression in key brain areas affecting metabolism, in parallel with Trh repression. Coordinated T3 repression of Mc4r has energy-saving consequences. See Discussion for commentary.
Functional TRs (TRα1, TRβ1, and TRβ2) colocalize in many hypothalamic nuclei (5, 21). Each isoform exerts specific transcriptional activities on the Trh promoter (4, 24, 26, 27), differences which are caused by variations in N-terminal sequence (34). Overexpression and knockdown studies in newborn mice revealed TR isoform-specific and age-dependent effects on Mc4r transcription. For TRα, transient, hypothalamic knockdown of TRα activated Mc4r transcription in the newborn, but its long-term, generalized knockout resulted in repression in the adult. These stage-dependent differences could result from developmental compensatory effects in the mutant mouse. For TRβ, similar effects on T3-dependent regulation of Mc4r were seen in newborn and adult mice. Comparing the roles of each TRβ isoform in Mc4r and Trh transcription, that of TRβ1 appears common to Mc4r and Trh promoters, and that of TRβ2 seems to be gene specific. TRβ2 activates T3-independent Mc4r basal expression, but for Trh regulation, it mediates T3-dependent repression (26). As discussed earlier, the mechanisms underlying such gene-specific regulations might be related to differences in the TRE sequences seen in Mc4r and Trh promoters, leading to gene-specific recruitment of transcription factors.
Physiological Relevance of T3-Dependent Repression of Mc4r Expression.
The leptin/melanocortin pathway impinges on the HPT axis, adapting energy expenditure to metabolic reserves (8). Our results reveal a feedback loop targeting the melanocortin relay, with T3 repressing Mc4r expression not only via hypothalamic Trh but also in other key energy-related brain regions. TH-dependent repression of Mc4r will have multiple physiological consequences. A first short-term consequence is orexigenia and increased weight gain, as seen in the T4-treated mice (Fig. S1B). Indeed, hypothalamic T3-dependent repression of Mc4r expression should reduce the sensitivity of TRH neurons to leptin/melanocortin inputs. Moreover, increased T3 also would reduce hypothalamic sensitivity to Agouti-related protein (AgRP) signaling and other MC4R ligands. More than 50% of hypophysiotropic TRH neurons receive AgRP fibers, and αMSH fibers represent 38% of the innervation to PVN TRH neurons. Thus T3-dependent repression of Mc4r would dampen brain global responsiveness to melanocortin anorectic signaling while stimulating orexigenic pathways through NPY signaling. Again, our findings are consistent with this prediction. T3 modulated the leptin-sensitive neuropeptides Pomc and Npy in the ARC, downregulating the Pomc precursor for αMSH and increasing Npy mRNA, in line with published data (15, 17). In contrast, because hypothyroidism increases Mc4r expression in the PVN and ARC, low T3 would sensitize TRH neurons and their ARC counterparts to MC4R ligands (35), enhancing central metabolic responses (Fig. S8A). Future work should address whether the effects of TH on Mc4r expression are uniform across different hypothalamic cell populations or are limited to given neuronal categories. For instance, corticotropin-releasing hormone neurons, as well as TRH neurons, express MC4R (36). Thus T3 regulation of Mc4r could affect other neuroendocrine circuits. The largest repressive effect of T3 on Mc4r expression was in brainstem, which controls thermogenesis and meal size (37–39). Here, T3 repression of Mc4r could modulate autonomic outputs governing peripheral energy homeostasis and food intake.
The fact that Mc4r is regulated by thyroid status in different brain areas having distinct effects on metabolism has major physiological significance. Overall increased MC4R signaling will decrease food intake, increase thermogenesis, and restrict meal size. Each of these effects will reduce energy reserves and will, in parallel, be amplified by high T3. To avoid excess catabolism, T3 feeds back on Trh (thereby reducing T3 production), providing a first line of defense against energy dissipation. However, if this process occurs without putting a brake on MC4R signaling, the energy-saving effects of T3 feedback on Trh would be neutralized or overridden: MC4R stimulates Trh transcription through cAMP and CREB (8), potentially overriding T3 feedback on Trh. Thus, coordination of leptin/melanocortin signaling and HPT axis pathways is achieved by a double T3-dependent feedback loop, reinforcing maintenance of metabolic homeostasis (Fig. 4 and Fig. S8A).
During evolution, the selection of a syn-repression control by T3 on these two central genes could have provided a selective advantage through optimal regulation of energy conservation and homeostasis. The presence of the nTRE we identified in all mammals examined (mouse, rat, human, and pig) is a strong argument for the significance of this endocrine regulation in homeotherms.
In conclusion, this study demonstrates that thyroid status regulates Mc4r expression in major brain areas relevant to metabolic homeostasis. T3 directly mediates the regulation of Mc4r expression. Taken together, the data reinforce the concept that T3 has a major role in integrating endocrine and metabolic signaling at the central level.
Materials and Methods
Animals.
Animal care and experimentation were in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee, Veterinary Services Direction, Paris, France. Swiss wild-type mice were from Janvier. Mice lacking all TRβ or all TRα gene products [TRβ−/− (19) or TRα°/° (18)] were provided by J. Samarut (Ecole Normale Supérieure, Lyon, France). For investigations of hypothyroidism, 8-week-old male mice were given iodine-deficient food containing 0.15% PTU (Harlan) and drinking water with 0.5 g/L PTU (Sigma-Aldrich) for 13 days before they were killed. To induce fetal and neonatal hypothyroidism, dams were given the iodine-deficient PTU diet from day 14 of gestation through lactation. For investigations of hyperthyroidism, adult mice were treated with 1.2 μg/mL of T4 in drinking water for 13 days before they were killed.
Assessment of T4 Levels.
Total T4 blood concentrations were quantified using ELISA kits (AbCys, Paris, France).
In Situ Hybridization.
In situ hybridization protocol was adapted from (40) (see SI Materials and Methods).
Plasmids.
TRH-f.luc and TRα1, TRβ1, and TRβ2 plasmids were as described (27). MC4R-f.luc plasmid, provided by H. Krude (Charity Medical University, Berlin, Germany), contains −653 to +448 bp of the MC4R promoter cloned upstream of the Firefly luciferase-coding sequence of the pGL3 basic Luciferase reporter vector (Promega, Charbonniéres les Bains, France). MC4R-Renilla luciferase (MC4R-r.luc) was designed in the laboratory (see SI Materials and Methods).
Two putative TREs in the hMC4R promoter region were mutated using the QuikChange II XL Site-directed Mutagenesis kit (Stratagene, La Jolla, CA) per the manufacturer’s protocol. Primers for mutagenesis (Table S1) were from Invitrogen. Three MC4R-r.luc transgenes were generated and sequenced: TRE1m, TRE2m, and DBm (mutations in both TRE1 and TRE2).
The plasmid encoding shRNA against TRα1 (shTRα1) has been published (41). The two sets of shRNA against TRβ isoforms (25), shGFP and shSCR, bear the same backbone as shTRα1. Design (Table S2 and S3) and cloning are described in SI Materials and Methods.
In Vivo Transfection and Luciferase Assays.
DNA/PEI complexes and iGT were adapted from (24). For single and cotransfection iGT protocols and T3 treatments see SI Materials and Methods.
In Vivo Chromatin Immunoprecipitation.
Pups were treated with T3 or vehicle and killed 20 h later. Dorsal hypothalamic regions including the PVN were dissected. Samples were fixed in 1% formaldehyde solution and sonicated. Control and T3-treated samples were used for ChIP with a mix of anti-TRβ1 and anti-TRβ2 antibodies (Millipore) or without antibody (negative control). Precipitated DNA fragments were purified. Primers spanning the TRE sequences in Mc4r and Trh promoter were used in qPCR to measure enrichment of DNA samples. Negative controls comprised primers spanning irrelevant sequences in both gene sequences. The detailed ChIP protocol is provided in SI Materials and Methods.
qPCR.
qPCR procedures used for Mc4r, Trh, Pomc, and Npy mRNA detection are described in SI Materials and Methods. Primers for specific TR mRNA detection were as described (21).
Statistical Analysis.
qPCR results: boxes represent the fifth to ninety-fifth percentiles around the median with whiskers for minimum and maximum values. Statistical analysis for qPCR data compared the means of fold increase values ± SD, using nonparametric ANOVA, followed by a permutation test (Cytel Studio software, Cambridge, MA) to compare the control and treated groups. Independent experiments (5 ≤ n ≤ 8) were repeated three times, providing similar results and data were pooled.
iGT results are presented as means ± SEM per group. ANOVA was used to compare groups followed by multiple-comparisons Bonferroni’s post-test. Each experiment was carried out with n ≥ 10 and was repeated at least three times providing the same results. In all cases, typical experiments are shown, differences were considered significant at P < 0.05 with *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Supplementary Material
Acknowledgments
We thank J. Elmquist (Southwestern University) for providing the plasmid for mouse Mc4r probes; Z. Hassani, P. Bilesimo, L. Sachs (UMR 7221, Paris), and C. Fekete (Hungarian Academy of Science, Budapest) for advice on experimental techniques; and G. Levi and G. Morvan (UMR 7221, Paris) for insightful comments. S. Sosinsky and P. Durand provided excellent animal care. This work was supported by European Union PIONEER and CRESCENDO contracts. S.D. received fellowships from Nestlé Nutrition and the Fondation pour la Recherche Médicale. A.L-J received a PhD grant from PolyPlus Transfection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905190107/DCSupplemental.
References
- 1.Barker SB. Mechanism of action of the thyroid hormone. Physiol Rev. 1951;31:205–243. doi: 10.1152/physrev.1951.31.3.205. [DOI] [PubMed] [Google Scholar]
- 2.Blair T, Forbes JM. Changes in voluntary food intake, body-weight and metabolic rate with thyroxine treatment in sheep. Proc Nutr Soc. 1974;33:78A. [PubMed] [Google Scholar]
- 3.Silva JE. Thyroid hormone control of thermogenesis and energy balance. Thyroid. 1995;5:481–492. doi: 10.1089/thy.1995.5.481. [DOI] [PubMed] [Google Scholar]
- 4.Lezoualc’h F, et al. Assignment of the beta-thyroid hormone receptor to 3,5,3′-triiodothyronine-dependent inhibition of transcription from the thyrotropin-releasing hormone promoter in chick hypothalamic neurons. Mol Endocrinol. 1992;6:1797–1804. doi: 10.1210/mend.6.11.1480171. [DOI] [PubMed] [Google Scholar]
- 5.Lechan RM, Fekete C. Feedback regulation of thyrotropin-releasing hormone (TRH): Mechanisms for the non-thyroidal illness syndrome. J Endocrinol Invest. 2004;27(6 Suppl):105–119. [PubMed] [Google Scholar]
- 6.Krotkiewski M. Thyroid hormones in the pathogenesis and treatment of obesity. Eur J Pharmacol. 2002;440:85–98. doi: 10.1016/s0014-2999(02)01420-6. [DOI] [PubMed] [Google Scholar]
- 7.Hollenberg AN. The role of the thyrotropin-releasing hormone (TRH) neuron as a metabolic sensor. Thyroid. 2008;18:131–139. doi: 10.1089/thy.2007.0251. [DOI] [PubMed] [Google Scholar]
- 8.Harris M, et al. Transcriptional regulation of the thyrotropin-releasing hormone gene by leptin and melanocortin signaling. J Clin Invest. 2001;107:111–120. doi: 10.1172/JCI10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wikberg JE, Mutulis F. Targeting melanocortin receptors: An approach to treat weight disorders and sexual dysfunction. Nat Rev Drug Discov. 2008;7:307–323. doi: 10.1038/nrd2331. [DOI] [PubMed] [Google Scholar]
- 10.Mountjoy KG, Mortrud MT, Low MJ, Simerly RB, Cone RD. Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Mol Endocrinol. 1994;8:1298–1308. doi: 10.1210/mend.8.10.7854347. [DOI] [PubMed] [Google Scholar]
- 11.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 12.Garza JC, Kim CS, Liu J, Zhang W, Lu XY. Adeno-associated virus-mediated knockdown of melanocortin-4 receptor in the paraventricular nucleus of the hypothalamus promotes high-fat diet-induced hyperphagia and obesity. J Endocrinol. 2008;197:471–482. doi: 10.1677/JOE-08-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Horvath TL. Neuronal control of energy homeostasis. FEBS Lett. 2008;582:132–141. doi: 10.1016/j.febslet.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong WM, et al. Triiodothyronine stimulates food intake via the hypothalamic ventromedial nucleus independent of changes in energy expenditure. Endocrinology. 2004;145:5252–5258. doi: 10.1210/en.2004-0545. [DOI] [PubMed] [Google Scholar]
- 15.Coppola A, et al. A central thermogenic-like mechanism in feeding regulation: An interplay between arcuate nucleus T3 and UCP2. Cell Metab. 2007;5:21–33. doi: 10.1016/j.cmet.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perello M, et al. Thyroid hormones selectively regulate the posttranslational processing of prothyrotropin-releasing hormone in the paraventricular nucleus of the hypothalamus. Endocrinology. 2006;147:2705–2716. doi: 10.1210/en.2005-1609. [DOI] [PubMed] [Google Scholar]
- 17.Ishii S, et al. Hypothalamic neuropeptide Y/Y1 receptor pathway activated by a reduction in circulating leptin, but not by an increase in circulating ghrelin, contributes to hyperphagia associated with triiodothyronine-induced thyrotoxicosis. Neuroendocrinology. 2003;78:321–330. doi: 10.1159/000074885. [DOI] [PubMed] [Google Scholar]
- 18.Gauthier K, et al. Genetic analysis reveals different functions for the products of the thyroid hormone receptor alpha locus. Mol Cell Biol. 2001;21:4748–4760. doi: 10.1128/MCB.21.14.4748-4760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauthier K, et al. Different functions for the thyroid hormone receptors TRalpha and TRbeta in the control of thyroid hormone production and post-natal development. EMBO J. 1999;18:623–631. doi: 10.1093/emboj/18.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyess EM, et al. Triiodothyronine exerts direct cell-specific regulation of thyrotropin-releasing hormone gene expression in the hypothalamic paraventricular nucleus. Endocrinology. 1988;123:2291–2297. doi: 10.1210/endo-123-5-2291. [DOI] [PubMed] [Google Scholar]
- 21.Clerget-Froidevaux MS, Seugnet I, Demeneix BA. Thyroid status co-regulates thyroid hormone receptor and co-modulator genes specifically in the hypothalamus. FEBS Lett. 2004;569:341–345. doi: 10.1016/j.febslet.2004.05.076. [DOI] [PubMed] [Google Scholar]
- 22.Lubrano-Berthelier C, et al. The human MC4R promoter: Characterization and role in obesity. Diabetes. 2003;52:2996–3000. doi: 10.2337/diabetes.52.12.2996. [DOI] [PubMed] [Google Scholar]
- 23.Hollenberg AN, et al. The human thyrotropin-releasing hormone gene is regulated by thyroid hormone through two distinct classes of negative thyroid hormone response elements. Mol Endocrinol. 1995;9:540–550. doi: 10.1210/mend.9.5.7565802. [DOI] [PubMed] [Google Scholar]
- 24.Guissouma H, Ghorbel MT, Seugnet I, Ouatas T, Demeneix BA. Physiological regulation of hypothalamic TRH transcription in vivo is T3 receptor isoform specific. FASEB J. 1998;12:1755–1764. doi: 10.1096/fasebj.12.15.1755. [DOI] [PubMed] [Google Scholar]
- 25.Guissouma H, Froidevaux MS, Hassani Z, Demeneix BA. In vivo siRNA delivery to the mouse hypothalamus confirms distinct roles of TR beta isoforms in regulating TRH transcription. Neurosci Lett. 2006;406:240–243. doi: 10.1016/j.neulet.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 26.Abel ED, Ahima RS, Boers ME, Elmquist JK, Wondisford FE. Critical role for thyroid hormone receptor beta2 in the regulation of paraventricular thyrotropin-releasing hormone neurons. J Clin Invest. 2001;107:1017–1023. doi: 10.1172/JCI10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupré SM, et al. Both thyroid hormone receptor (TR)beta 1 and TR beta 2 isoforms contribute to the regulation of hypothalamic thyrotropin-releasing hormone. Endocrinology. 2004;145:2337–2345. doi: 10.1210/en.2003-1209. [DOI] [PubMed] [Google Scholar]
- 28.Vaisse C, et al. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huszar D, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 30.Koller KJ, Wolff RS, Warden MK, Zoeller RT. Thyroid hormones regulate levels of thyrotropin-releasing-hormone mRNA in the paraventricular nucleus. Proc Natl Acad Sci USA. 1987;84:7329–7333. doi: 10.1073/pnas.84.20.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 32.Shibusawa N, Hollenberg AN, Wondisford FE. Thyroid hormone receptor DNA binding is required for both positive and negative gene regulation. J Biol Chem. 2003;278:732–738. doi: 10.1074/jbc.M207264200. [DOI] [PubMed] [Google Scholar]
- 33.Froidevaux MS, et al. The co-chaperone XAP2 is required for activation of hypothalamic thyrotropin-releasing hormone transcription in vivo. EMBO Rep. 2006;7:1035–1039. doi: 10.1038/sj.embor.7400778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guissouma H, et al. Feedback on hypothalamic TRH transcription is dependent on thyroid hormone receptor N terminus. Mol Endocrinol. 2002;16:1652–1666. doi: 10.1210/mend.16.7.0868. [DOI] [PubMed] [Google Scholar]
- 35.Smith MA, et al. Melanocortins and agouti-related protein modulate the excitability of two arcuate nucleus neuron populations by alteration of resting potassium conductances. J Physiol. 2007;578:425–438. doi: 10.1113/jphysiol.2006.119479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu XY, Barsh GS, Akil H, Watson SJ. Interaction between alpha-melanocyte-stimulating hormone and corticotropin-releasing hormone in the regulation of feeding and hypothalamo-pituitary-adrenal responses. J Neurosci. 2003;23:7863–7872. doi: 10.1523/JNEUROSCI.23-21-07863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan W, Morrison SF, Cao WH, Yu P. Thermogenesis activated by central melanocortin signaling is dependent on neurons in the rostral raphe pallidus (rRPa) area. Brain Res. 2007;1179:61–69. doi: 10.1016/j.brainres.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Skibicka KP, Grill HJ. Energetic responses are triggered by caudal brainstem melanocortin receptor stimulation and mediated by local sympathetic effector circuits. Endocrinology. 2008;149:3605–3616. doi: 10.1210/en.2007-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng H, Patterson LM, Phifer CB, Berthoud HR. Brain stem melanocortinergic modulation of meal size and identification of hypothalamic POMC projections. Am J Physiol Regul Integr Comp Physiol. 2005;289:R247–R258. doi: 10.1152/ajpregu.00869.2004. [DOI] [PubMed] [Google Scholar]
- 40.Becker N, Seugnet I, Guissouma H, Dupre SM, Demeneix BA. Nuclear corepressor and silencing mediator of retinoic and thyroid hormone receptors corepressor expression is incompatible with T(3)-dependent TRH regulation. Endocrinology. 2001;142:5321–5331. doi: 10.1210/endo.142.12.8531. [DOI] [PubMed] [Google Scholar]
- 41.Hassani Z, et al. A hybrid CMV-H1 construct improves efficiency of PEI-delivered shRNA in the mouse brain. Nucleic Acids Res. 2007;35:e65. doi: 10.1093/nar/gkm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.