Abstract
Autophagy is a catabolic process by which cells remove long-lived proteins and damaged organelles for recycling. Viral infections may also induce autophagic response. Here we show that hepatitis B virus (HBV), a pathogen that chronically infects ≈350 million people globally, can enhance autophagic response in cell cultures, mouse liver, and during natural infection. This enhancement of the autophagic response is not coupled by an increase of autophagic protein degradation and is dependent on the viral X protein, which binds to and enhances the enzymatic activity of phosphatidylinositol 3-kinase class III, an enzyme critical for the initiation of autophagy. Further analysis indicates that autophagy enhances HBV DNA replication, with minimal involvement of late autophagic vacuoles in this process. Our studies thus demonstrate that a DNA virus can use autophagy to enhance its own replication and indicate the possibility of targeting the autophagic pathway for the treatment of HBV patients.
Keywords: autophagy, hepatitis B virus DNA replication, hepatitis B virus X protein, PI3KC3
Autophagy is a catabolic process by which long-lived proteins and damaged organelles are sequestered in the cytoplasm and removed for recycling. It is important for maintaining cellular homeostasis. During autophagy, membrane crescents appear in the cytoplasm. These membranes will eventually form a double-membrane structure known as autophagosomes, which will mature by fusing with lysosomes to form autolysosomes. The contents of autophagosomes will subsequently be degraded by lysosomal enzymes. Autophagy has also been implicated in innate and adaptive immune responses to the infection of microbial pathogens (1, 2). A number of viruses have been shown to induce autophagy, either completely or partially, and often with either a destructive or beneficial result to themselves. For examples, several single-stranded RNA viruses such as poliovirus, coronavirus, dengue virus, and hepatitis C virus all seem to induce the accumulation of autophagic vacuoles and use these membrane vesicles to benefit their replication (3–6). In contrast, other viruses such as herpes simplex virus-1 (HSV-1), cytomegalovirus (CMV), and Kaposi’s sarcoma herpes virus (KSHV) have evolved mechanisms to suppress autophagy and, in the case of HSV-1, for its own survival (2).
Hepatitis B virus (HBV) belongs to the Hepadnavirus family. This virus has a 3.2-kb circular and partially double-stranded DNA genome that contains four genes named S, C, P, and X genes. The S gene codes for the surface antigens (i.e., envelope proteins), the C gene codes for the core protein and a related protein termed precore protein, the P gene codes for the viral DNA polymerase, and the X gene codes for a multifunctional regulatory protein. After its synthesis, the core protein packages its own mRNA, which is also known as the pregenomic RNA (pgRNA), to form the core particle. The pgRNA will be converted to the DNA genome in the core particle by the viral DNA polymerase, which is also a reverse transcriptase. The core particle will then interact with the surface antigens on the endoplasmic reticulum (ER) for the formation of the virion. The mature virion is then released from the infected cells through a process that is poorly understood (for a review, see ref. 7).
In this report, we have investigated whether and how HBV may induce autophagy. Our results demonstrate that HBV can enhance the autophagic response without increasing autophagic protein degradation. This induction of autophagic response enhances viral DNA replication with only a slight effect on viral RNA transcription and requires the HBV X protein (HBx), which binds to phosphatidylinositol 3-kinase class III (PI3KC3) to enhance its activity. Our studies thus delineated the molecular pathway by which HBV induces autophagy.
Results
Induction of Autophagic Response by HBV.
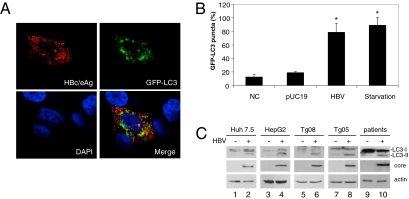
We have previously established from Huh7.5 human hepatoma cells a stable cell line that expresses the fusion protein of green fluorescence protein (GFP) and LC3 (5). This GFP–LC3 fusion protein is diffusely localized in the cytosol. However, it is localized to autophagosomes during autophagy. To investigate whether HBV could induce autophagy, we transfected HBV genomic DNA into Huh7.5–GFP–LC3 cells. As shown in Fig. 1A, the GFP–LC3 signal is weak in the absence of HBV, but it is bright and punctate in the presence of HBV, consistent with its localization to autophagosomes. When the cells were counted, ≈80% of cells transfected by the HBV DNA, as evidenced by the positive HBV core/e antigen staining, and 90% of nutrient-starved cells were positive for GFP–LC3 puncta (Fig. 1B). In contrast, ≈15% of control Huh7.5–GFP–LC3 cells and 20% of cells transfected by the pUC19 control plasmid DNA contained GFP–LC3 puncta. These results suggested that HBV, similar to nutrient starvation, could induce autophagosomes.
Fig. 1.
Induction of autophagic vacuoles by HBV. (A) Confocal microscopy of Huh7.5–GFP–LC3 cells transfected by the 1.3mer HBV genomic DNA. Cells were stained with the antibody that recognized both HBV core protein and e antigen (HBc/eAg; red) 48 h after DNA transfection. Note that the GFP–LC3 puncta (green) are apparent only in the HBV-positive cell, and a significant overlap of the signals of HBV c/e antigens and GFP puncta is also visible when the images are merged. DAPI (blue) stains for the nuclei. (B) Percentage of cells with GFP–LC3 puncta in nontransfected cells (NC), pUC19-transfected cells, 1.3mer HBV DNA-transfected cells, and nutrient-starved cells. GFP-positive cells were defined as cells that display bright punctate staining. Approximately 50 cells were counted and the experiment was repeated at least three times. Asterisks indicate statistical significance (P < 0.01). (C) Western blot analysis of LC3. Lanes 1 and 2 were Huh7.5 cells transfected with pUC19 and the 1.3mer HBV DNA, respectively. Lane 3 was the naive HepG2 cells and lane 4 was a stable HepG2 cell line that contained replicating HBV DNA (24). Tg08 (lane 6) and Tg05 (lane 8) were two independent transgenic mouse lines that carried the 1.3mer HBV genome (8). Their HBV-negative control littermates were shown in lanes 5 and 7, respectively. Lanes 9 and 10 were liver tissues from HBV noninfected and infected patients, respectively. (Top) LC3 proteins. (Middle) HBV core protein. (Bottom) Actin control.
To further confirm that HBV could indeed induce autophagosomes, we performed Western blot analysis of LC3, which is a protein that is converted from the cytosolic form (LC3-I) to the lipidated, autophagosome-associated form (LC3-II) during autophagy. As shown in Fig. 1C, the predominant form of LC3 detected in Huh7.5 cells transfected with the pUC19 control DNA was LC3-I, whereas a significant amount of LC3-II was detected in Huh7.5 cells transfected with the HBV genomic DNA. The induction of LC3-II was not specific to Huh7.5 cells, as similar results were observed in its parental cell line Huh7 (see below), another liver-derived cell line HepG2 cells, and in the liver of transgenic mouse lines harboring low (Tg08) and high (Tg05) replication levels of the HBV DNA (8, 9). Importantly, this induction was also observed in the liver of an HBV-infected patient but not in the liver that contained metastasized colon cancer of a noninfected patient. The induction of autophagosomes by HBV was also observed in the liver of HBV transgenic mice when the tissue sections were examined by electron microscopy (Fig. S1 A and B). Together, these results indicated that HBV could induce autophagosomes in liver cells.
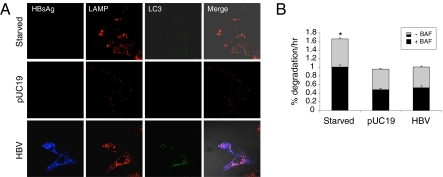
Autophagosomes mature by fusing with lysosomes to form autolysosomes. To investigate whether HBV could induce the formation of autolysosomes, we performed confocal microscopy on Huh7.5–GFP–LC3 cells that had been transfected with either pUC19 or the HBV genomic DNA. Cells were stained for HBV surface antigen (HBsAg) and lysosome-associated membrane protein 1 (LAMP1). Nutrient-starved cells were used as a positive control. As shown in Fig. 2A, an extensive colocalization of LAMP1 with GFP–LC3 puncta was detected in nutrient-starved cells (Top) (colocalization coefficient 0.76) and HBV-positive cells (Bottom) (colocalization coefficient 0.58), indicative of formation of autolysosomes. Few GFP–LC3 puncta could be detected in cells transfected with the control plasmid DNA pUC19 (Middle). To ensure that HBV could indeed induce the formation of these late autophagic vacuoles, we also performed electron microscopy using liver tissue sections prepared from HBV transgenic mice. As shown in Fig. S1C, late autophagic vacuoles could indeed be detected in mouse hepatocytes. These results indicated that, similar to nutrient starvation, HBV could enhance the autophagic flux.
Fig. 2.
Enhancement of autophagic flux by HBV without increasing autophagic protein degradation. (A) Confocal microscopy of HBV surface antigens (HBsAg) (blue), LAMP1 (red), and GFP–LC3 puncta (green) in Huh7.5 cells. (Top) Nutrient-starved cells. (Middle) pUC19-transfected cells. (Bottom) 1.3mer HBV DNA-transfected cells. (B) Long-lived protein degradation assay. Gray + black bar, overall protein degradation rate in the absence of BAF (−BAF); black bar, protein degradation rate in the presence of BAF (+BAF). The gray area of the bars represents the BAF-sensitive protein degradation rate. The results represent the average of three independent experiments. The asterisk indicates statistical significance (P < 0.05).
No Significant Induction of Autophagic Protein Degradation by HBV.
Autophagy removes long-lived proteins from cells. To determine whether the enhancement of autophagic flux by HBV also increases the autophagic protein degradation rate, we conducted the long-lived protein degradation assay. Cells transfected with either pUC19 or the HBV genomic DNA were metabolically labeled with 3H-leucine for 24 h and then chased for another 24 h to allow the removal of labeled, short-lived proteins. Cells were then rinsed and incubated with or without bafilomycin A1 (BAF), a vacuolar ATPase inhibitor that suppresses autophagic protein degradation (10). The culture supernatant and cell lysates were collected and the protein degradation rate was determined by measuring the release of TCA-soluble radiolabel. The overall protein degradation rates and those sensitive to BAF and hence mediated by autophagy are shown in Fig. 2B. Nutrient starvation, which induces autophagy, was included in the studies to serve as a positive control. As shown in Fig. 2B, nutrient starvation significantly increased both overall and BAF-sensitive protein degradation rates, when these degradation rates were compared with those of the control cells or cells transfected with pUC19. Interestingly, no significant increase of both overall and BAF-sensitive protein degradation was observed in HBV DNA transfected cells. These results indicated that HBV enhanced autophagic flux without increasing the autophagic protein degradation rate.
Enhancement of HBV DNA Replication by Autophagy.
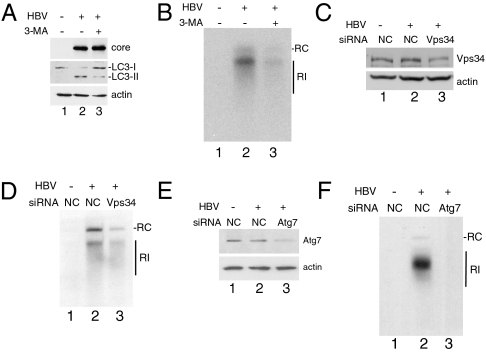
Autophagy may enhance or suppress viral replication, depending on the viruses. To understand the possible effect of autophagy on HBV replication, we treated cells with 3-methyladenine (3-MA), an inhibitor of PI3KC3, and examined its effect on HBV DNA replication. PI3KC3 is critical for initiating autophagy. As shown in Fig. 3A, the treatment of cells with 3-MA suppressed the lipidation of LC3 induced by HBV, in support of the role of PI3KC3 in HBV-induced autophagy. This treatment with 3-MA significantly reduced HBV DNA replication (Fig. 3B), suggesting a positive role of autophagy in HBV DNA replication. To ensure that this effect of 3-MA on HBV was not due to nonspecific effects of the drug, we also performed an siRNA knockdown experiment to suppress the expression of Vps34, the catalytic subunit of PI3KC3. As shown in Fig. 3C, the Vps34 siRNA, but not the control siRNA, reduced the expression level of Vps34. In agreement with the 3-MA result, this Vps34 siRNA also suppressed the HBV DNA replication (Fig. 3D). To further confirm that autophagy indeed plays a positive role in HBV DNA replication, we also used siRNA to suppress the expression of Atg7 (Fig. 3E), an enzyme that mediates the lipidation of LC3 and is critical for the formation of autophagosomes. Similar to the PI3KC3 results, the suppression of Atg7 expression also led to the suppression of HBV DNA replication (Fig. 3F). Thus, these results together indicate a positive effect of autophagy on HBV DNA replication.
Fig. 3.
Enhancement of HBV DNA replication by autophagy. Huh7.5 cells transfected with pUC19 (lane 1) or the 1.3mer HBV DNA (lanes 2 and 3) were used for the analysis. (A) Western blot analysis of the HBV core protein (Top), LC3 (Middle), and actin (Bottom). Cells transfected with the DNA for 24 h were further treated without (lanes 1 and 2) or with (lane 3) 10 mM 3-MA for 16 h. (B) Southern blot analysis of HBV DNA. Cells without or with 3-MA treatment as in A were lysed for the purification of replicated HBV DNA for the Southern blot analysis. RC, HBV relaxed circular DNA; RI, HBV replicative intermediates. (C) Western blot analysis of Vps34. DNA-transfected cells were further transfected with the negative control (NC), siRNA (lanes 1 and 2), or Vps34 siRNA (lane 3) for 48 h. (D) Southern blot analysis of replicated HBV DNA in cells treated with control or Vps34 siRNA. (E) Western blot analysis of Atg7 expressed in cells transfected with control or Atg7 siRNA. (F) Southern blot analysis of replicated HBV DNA in cells treated with control or Atg7 siRNA.
Mechanism of Autophagy-Enhanced HBV DNA Replication.
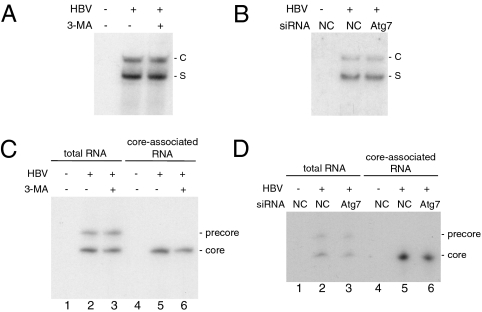
To understand how autophagy enhances HBV DNA replication, we performed Northern blot analysis on HBV RNAs. Huh7.5 cells transfected with the HBV DNA were incubated in the presence (+) or absence (−) of 3-MA. These cells were then lysed for RNA isolation, followed by Northern blot analysis. As shown in Fig. 4A, 3-MA had only a marginal effect on the HBV RNA level. This result is consistent with our Western blot analysis, which revealed no significant effect of 3-MA on the HBV core protein level (Fig. 3A), and with our ELISA analysis, which revealed only a marginal effect of 3-MA on the level of HBV surface antigens in the incubation media (Fig. S2). To confirm that autophagy indeed has no major effect on HBV RNAs, we also performed the Atg7 knockdown experiment. As shown in Fig. 4B, the suppression of Atg7 expression also had no significant effect on HBV RNAs.
Fig. 4.
Analysis of the autophagic effect on HBV RNAs. Huh7.5 cells were transfected with pUC19 or the 1.3mer HBV genome and treated with 3-MA or siRNA as described in the legend to Fig. 3. (A and B) Northern blot analysis of HBV RNAs. (C and D) Primer extension analysis of total HBV precore and core RNAs in cells (lanes 1–3) or the core RNA packaged in core particles (lanes 4–6). S, S gene transcripts; C, C gene transcripts.
To further understand how autophagy may enhance HBV DNA replication, we analyzed the effect of autophagy on the packaging of HBV core RNA (i.e., pgRNA). Huh7.5 cells transfected with the HBV DNA with (+) or without (−) 3-MA treatment for 16 h were lysed for the isolation of total cellular RNA and core particle-associated HBV RNA. These RNAs were then quantified by primer extension analysis. As shown in Fig. 4C, 3-MA had no significant effect on the HBV precore RNA level but it reduced slightly (∼20%) the core RNA level in cells and that packaged in core particles. To confirm this finding, we also analyzed HBV precore and core RNAs in cells that had been treated with either the control siRNA or the Atg7 siRNA. As shown in Fig. 4D, the Atg7 siRNA also reduced slightly the core RNA level in cells and that packaged in core particles. These results indicated that autophagy had a slight effect on the core RNA level, possibly on its transcription, and no apparent effect on the packaging of the core RNA, because the reduction of the packaged core RNA level correlated well with the reduction of the total core RNA level in cells. Furthermore, because 3-MA and the Atg7 siRNA reduced the HBV DNA level to an almost undetectable level (Fig. 3 B and F), these results also indicated that autophagy played an important role in HBV DNA replication at a step after the packaging of the core RNA.
No Major Effect of Autolysosomes on HBV DNA Replication.
The suppression of PI3KC3 activity and Atg7 expression significantly reduced HBV DNA replication, indicating that a step at or after the onset of autophagy is important for HBV DNA replication. To investigate the possible role of late autophagic vacuoles in HBV DNA replication, we treated cells with BAF, which suppresses the formation of autolysosomes. As shown in Fig. S3A, the treatment of control Huh7.5 cells and Huh7.5 cells that had been transfected with the HBV DNA resulted in the increase of the LC3-II level. This result indicated that BAF faithfully suppressed autolysosome formation, which is required for the removal of LC3-II. BAF reduced slightly the levels of HBV RNA transcripts (Fig. S3B) and ≈50% of the core RNA and the replicated HBV DNA (Fig. S3 C and D). This observation indicates that late autophagic events may enhance HBV RNA expression but not HBV DNA replication, because the degrees of the core RNA and the HBV DNA reduced by BAF were similar and thus the reduced HBV DNA replication might simply reflect the reduction of the total core RNA amount that is available for packaging.
Induction of Autophagy by HBV via HBx.
HBx is a regulatory protein. To investigate its possible role in HBV-induced autophagy, we transfected Huh7.5 cells with the wild-type HBV DNA genome and the HBV genome that is incapable of expressing only HBx (HBVX−). As shown in Fig. 5A, although the wild-type HBV DNA was able to induce the lipidation of LC3, the HBVX− mutant was not. The analysis of the liver of transgenic mice carrying either the wild-type HBV genome or the HBVX− genome revealed a similar result (Fig. 5B). These results indicate that HBx is important for HBV-induced autophagy. To further investigate the role of HBx in HBV-induced autophagy, we transfected an HA-tagged HBx-expression (HAX) plasmid and its control vector into Huh7.5 cells. As shown in Fig. 5C, HBx by itself was sufficient to induce the lipidation of LC3. The results shown in Fig. 5 A–C together indicate that HBV induces autophagy via the activities of its HBx protein.
Fig. 5.
Effects of HBx on autophagy and HBV DNA replication. (A) Huh7.5 cells were transfected with pUC19 (lane 1), 1.3mer HBV DNA (lane 2), or 1.3mer HBVX− DNA (lane 3) followed by Western blot analysis using anti-LC3, anti-HBV core, or anti-actin antibodies. (B) Western blot analysis of mouse liver lysates. Lane 1, control nontransgenic mouse; lane 2, Tg05 mouse that carried the wild-type 1.3mer HBV genome; and lane 3, Tg38 mouse that carried the X-negative 1.3mer HBVX− genome. (C) Huh7.5 cells were transfected with the control pECE1 vector (lane 1) or pECE1-HAX (lane 2) followed by Western blot analysis using anti-LC3 (Top), anti-HA (Middle), or anti-actin (Bottom) antibodies. (D) Huh7 cells were transfected with pUC19 (lane 1), 1.3mer HBV DNA (lanes 2 and 3), or p1.3mer HBVX− (lanes 4 and 5) and then treated without or with 3-MA for 16 h. Cells were then lysed for Southern blot analysis for HBV DNA. (E) The same experiment as shown in D was repeated using HepG2 cells.
To further investigate the role of HBx and autophagy in HBV DNA replication, we examined the effect of 3-MA, which suppresses autophagy, on the replication of wild-type HBV and HBVX− mutant. In agreement with the previous reports (8, 9, 11, 12), wild-type HBV and HBVX− mutant had similar replication efficiency in Huh7 cells (Fig. 5D, lanes 2 and 4), the parental cell line of Huh7.5 (13). However, 3-MA suppressed specifically the replication of the wild-type HBV DNA but not the replication of the HBVX− mutant (Fig. 5D). These results were consistent with the results shown in Fig. 3F and Fig. S4, which indicated that the suppression of Atg7 expression with siRNA could suppress the replication of the wild-type HBV DNA but not the HBVX− mutant. We also conducted a similar experiment using HepG2 cells. Again, in agreement with the previous reports (9, 14, 15), wild-type HBV had a significantly higher replication efficiency than the HBVX− mutant in this cell line (Fig. 5E, lanes 2 and 4). However, similar to Huh7 cells, 3-MA suppressed the replication of only the wild-type HBV but not the HBVX− mutant in HepG2 cells. These results indicated that, in both Huh7 and HepG2 cells, HBx could enhance HBV DNA replication via the induction of autophagy.
Activation of PI3KC3 by HBx.
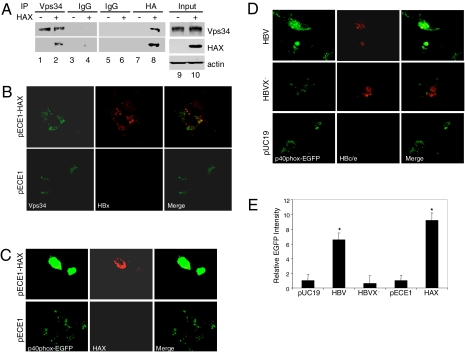
Our results indicate that HBV enhances the autophagic pathway via HBx. To determine how HBx may enhance autophagy, we examined whether HBx could interact with PI3KC3, an enzyme critical for HBV-induced autophagy (Fig. 2), by performing a coimmunoprecipitation experiment. Huh7.5 cells were transfected with the HAX expression plasmid or its control vector. These cells were then lysed and immunoprecipitated with the anti-Vps34 antibody or a control antibody, followed by Western blot analysis with an anti-HA antibody. As shown in Fig. 6A, HAX could be precipitated by the anti-Vps34 antibody but not by the control antibody. In a reciprocal experiment, the cell lysates were immunoprecipitated first with the anti-HA antibody followed by Western blot analysis using the anti-Vps34 antibody. As also shown in Fig. 6A, Vps34 could be precipitated by the anti-HA antibody in the presence of HAX but not in the absence of it. These coimmunoprecipitation results indicated that HBx and Vps34 could bind to each other. The ability of HBx to bind to Vps34 was also confirmed by confocal microscopy. Due to the inability of our anti-Vps34 to detect endogenous Vps34 in cells, we coexpressed Flag-tagged Vps34 and HAX in Huh7.5 cells. As shown in Fig. 6B, an extensive colocalization of Vps34 and HAX could be detected in cells. Thus, the confocal microscopy results were consistent with the coimmunoprecipitation results and in support of the physical interaction of these two proteins in cells.
Fig. 6.
Physical and functional interactions between HBx and PI3KC3. (A) Coimmunoprecipitation experiments. Huh7.5 cells were transfected with pECE1 (−) or pECE1-HAX (+). After 48 h, cells were lysed and immunoprecipitated with rabbit anti-Vps34 (lanes 1 and 2), control rabbit IgG (lanes 3 and 4), control mouse IgG (lanes 5 and 6), or the mouse anti-HA antibody (lanes 7 and 8). The immunoprecipitated samples were then analyzed by Western blot for Vps34 (Top) and HAX (Middle). Input, total cell lysates. (B) Colocalization of HAX and Flag-tagged PI3KC3 (Vps34). Huh7.5 cells were cotransfected with pFlag–PI3KC3 and pECE1–HAX (Upper) or pECE1 (Lower). Cells were then stained for Flag-tagged PI3KC3 (green) or HAX (red) and analyzed by confocal microscopy. (C) Induction of PI3P by HBx. Huh7.5 cells were cotransfected with p40PX–EGFP and pECE1–HAX (Upper) or its control vector pECE1 (Lower). After 48 h, cells were fixed, stained for HAX (red), and analyzed for the intensity of EGFP (green) by fluorescence microscopy. (D) Induction of PI3P by wild-type HBV but not the X-negative HBV. Huh7.5 cells were cotransfected with p40PX–EGFP and 1.3mer HBV DNA (Top), 1.3mer HBVX− mutant DNA (Middle), or pUC19 (Bottom). Cells were then fixed, stained for HBV core/e antigen, and analyzed by fluorescence microscopy. (E) Histogram of the relative intensities of EGFP shown in Fig. 6 C and D. The EGFP pixel densities of more than 50 cells were measured and that of pUC19-transfected cells was arbitrarily defined as one. The results represent the average of three independent experiments.
To investigate whether HBx and PI3KC3 could also functionally interact, we cotransfected Huh7.5 cells with the p40phox–EGFP expression plasmid and the HAX expression plasmid or its control vector. The p40phox–EGFP expression plasmid produces the fusion protein of p40phox and EGFP, and this fusion protein binds to phosphatidylinositol 3-phosphate (PI3P), a product of PI3KC3. As shown in Fig. 6C, the p40phox–EGFP signal was significantly brighter in cells transfected with the HAX expression plasmid than in cells transfected with the control pECE1 vector, indicating that HAX could enhance the production of PI3P. A similar enhancement of PI3P production was also observed in Huh7.5 cells transfected with the wild-type HBV genome but not with the HBVX− genome or the control vector pUC19 (Fig. 6D). Due to the lack of a good anti-HBx antibody, we were not able to test whether HBx expressed from the HBV genome could also physically interact with Vps34. Thus, this latter experiment using the HBV genome was of particular importance, as it indicated that HBx expressed from the HBV genome could also activate PI3KC3. The relative intensities of p40phox–EGFP, and hence the PI3P level, in various transfection experiments were measured and shown in Fig. 6E.
Discussion
A number of viruses have been shown to induce either partial or complete autophagic response with positive or negative effects on the virus. In this report, we demonstrate by confocal microscopy, electron microscopy and biochemical assays that HBV can enhance the autophagy in cell cultures and mouse liver as well as during natural infection (Figs. 1 and 2 and Fig. S1). Interestingly, in contrast to starvation-induced autophagy, this enhancement of autophagy by HBV does not lead to an increased autophagic protein degradation rate (Fig. 2). A recent report indicated that the mitochondria depolarization could induce recognition and sequestration of mitochondria by autophagosomes (16). Thus, it is possible that during nutrient starvation, cellular organelles and proteins are “marked” for autophagic recycling, whereas HBV does not induce such marking and hence does not increase the autophagic protein degradation rate.
By using the PI3KC3 inhibitor 3-MA and by suppressing the expression of Vps34, the catalytic subunit of PI3KC3, using siRNA, we demonstrate that, similar to nutrient starvation, PI3KC3 plays a critical role in HBV-induced autophagy (Fig. 3). This suppression of PI3KC3 activity or the suppression of expression of Atg7, an enzyme essential for the formation of autophagosomes, results in the suppression of HBV replication (Fig. 3). These results indicate that autophagy plays a positive role in HBV replication. This is unusual, as it is a unique example of a DNA virus that uses autophagy to enhance its replication. Our finding is reminiscent of two recent reports, which indicated that fasting could enhance HBV replication in the mouse liver (17, 18). Although in those two previous reports the enhancement of HBV replication was attributed to enhanced gluconeogenesis and the activation of peroxisome proliferator-activated receptor α (PPARα), it is conceivable that the enhanced HBV replication observed in those studies may also be due to the induction of autophagy by fasting.
The suppression of autophagy slightly reduced the HBV RNA levels and the packaging of the pgRNA (Fig. 4). However, it significantly suppressed HBV DNA replication. This observation indicates that autophagy enhances HBV replication mostly at the step of viral DNA replication. How autophagy may enhance HBV DNA replication remains unresolved. On the basis of the observation that HBV core/e antigens and surface antigens partially colocalized with LC3 puncta and PI3P (Figs. 1, 2, and 6), which are associated with autophagic vacuoles, it is tempting to speculate that, similar to poliovirus, autophagic vacuoles may serve as the sites for viral DNA replication and morphogenesis. Note that because BAF, which suppresses the formation of autolysosomes, does not abolish HBV DNA synthesis (Fig. S3), it is likely that late autophagic vacuoles do not play a major role in HBV DNA replication.
Our observation that wild-type HBV could induce autophagy, whereas the HBVX− mutant could not, indicates that HBx is critical for HBV-induced autophagy (Fig. 5). Indeed, the expression of HBx by itself was sufficient to induce autophagy (Fig. 5C). This was apparently due to the ability of HBx to bind to PI3KC3 to enhance its activity. Note that the interaction between HBx and PI3KC3 was demonstrated using HA-tagged HBx. Recently, it was reported that HBx could induce the expression of beclin-1 by activating its promoter and sensitize cells to starvation-induced autophagy (19). Beclin-1 is a component of the activated PI3KC3. However, we were not able to detect the induction of beclin-1 by HBV or HBx in cell cultures and the mouse liver (Fig. S5). The reason for this discrepancy is unclear. Because HBx can directly bind to PI3KC3 and enhance its activity (Fig. 6), the need for HBx to also induce the expression of beclin-1 would appear to be redundant. In any case, our results indicate that HBx could induce autophagy in the absence of nutrient starvation.
HBx can enhance HBV replication (8, 20). However, the molecular mechanism of this enhancement has been a subject of much debate. As shown in Fig. 5E, the suppression of PI3K activity in HepG2 cells with 3-MA reduced HBV DNA replication to a level similar to that of the HBVX− mutant. This result indicates that most of the effects of HBx on HBV DNA replication in HepG2 cells may be mediated by PI3KC3 and autophagy. A surprise finding of ours is that 3-MA as well as Atg7 knockdown also suppressed the replication of wild-type HBV DNA but not that of the HBVX− mutant in Huh7 cells (Fig. 5D and Fig. S4). These results indicate that, in spite of the observation that abolishing the expression of HBx had no apparent effect on HBV DNA replication in Huh7 cells (Fig. 5D, also see refs. 9, 15), HBx in fact still stimulates HBV DNA replication in Huh7 cells via the induction of autophagy. The high replication efficiency of the HBVX− mutant, which does not induce autophagy, may be caused by other factors, which stimulate HBV DNA replication via an autophagy-independent manner. For example, membrane structures may be altered in Huh7 cells to allow HBV replication in an autophagy-independent manner in the absence of HBx. These membranes may be further restructured in the presence of HBx, due to its interaction with PI3KC3. This may explain why the replication of wild-type HBV is sensitive to 3-MA whereas the replication of the HBx-negative mutant is not. Alternatively, HBx has multiple activities and can activate signaling pathways in the cytoplasm and gene expression in the nucleus (21). It is unclear at present whether any of these other activities of HBx also affect autophagy. However, it is conceivable that HBx may also enhance HBV DNA replication via the combination of its multiple activities including the induction of autophagy. When autophagy is suppressed such as by 3-MA or Atg7 knockdown, other activities of HBx may instead exert a negative effect on HBV DNA replication. Further research will be required to resolve this question.
In conclusion, our studies demonstrate that HBV induces autophagy to enhance its DNA replication. This induction of autophagy by HBV requires HBx, which binds to PI3KC3 to enhance its activity. Our studies thus delineated the molecular pathway by which HBV induces autophagy and also provide a unique example of a DNA virus that uses autophagy to enhance its own replication. Our studies also raise the possibility of targeting the autophagic pathway for the treatment of HBV patients.
Materials and Methods
DNA Plasmids and Transgenic Mice.
The plasmids p1.3merHBV and p1.3merHBVX− contain 1.3mer of the wild-type HBV genome and the HBx-negative genome, respectively, in the pUC19 vector (8). The plasmid pECE1–HAX expresses the HA-tagged HBx (22). p40PX–EGFP and pFlag–PI3KC3 are the expression plasmids for p40phox–EFGP and Flag–Vps34, respectively (23). The HBV transgenic mice Tg05, Tg08, and TgX have been described before (8).
siRNA Knockdown and DNA Transfection.
Duplex siRNAs with a two-nucleotide overhang at the 3′-end of the sequence were used. Atg7 and Vsp34 siRNAs were purchased from Qiagen and the negative control siRNA was purchased from Invitrogen. The target sequences were as follows: sense Atg7, 5′- GCA UCA UCU UCG AAG UGA Att-3′, antisense Atg7, 5′-UUC ACU UCG AAG AUG AUG Ctg-3′; sense hVsp34, 5′-CGC GAA AGU GGA AAU CGU Att-3′, antisense Vsp34 5′-UAC GAU UUC CAC UUU CGC Gtt-3′. The lowercase letters indicate deoxyribonucleotides. The siRNAs and DNA transfections were performed using Lipofectamine 2000 (Invitrogen) per the manufacturer’s instructions.
Northern and Southern Blot Analyses.
For total RNA isolation, cells were rinsed with ice-cold PBS twice, followed by RNA isolation using TRIzol (Invitrogen). The RNA was subjected to Northern blot analysis using 32P-labeled HBV DNA as the probe. For DNA isolation, cells in a 10-cm dish were washed and lysed, and HBV DNA packaged in core particles were isolated using the protocols described in ref. 9. The DNA was then subjected to Southern blot analysis using the 32P-labeled HBV DNA probe.
Supplementary Material
Acknowledgments
The authors would like to dedicate this paper to Dr. Ben Yen who passed away while this manuscript was being prepared. We thank Ms. Michelle McVeigh at the University of Southern California (USC) Research Center for Liver Diseases and Dr. Omar Khalid at the USC Department of Biochemistry and Molecular Biology for help with confocal microscopy. We also thank Drs. Chengyu Liang and Jae Jung for the plasmids p40PX–EGFP and pFLAG–PI3KC3. This research was supported by National Institutes of Health Grants CA123328 (to J.-h.J.O. and T.-S.B.Y.) and CA055578 (to T.-S.B.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
1Deceased August 31, 2009.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911373107/DCSupplemental.
References
- 1.Schmid D, Dengjel J, Schoor O, Stevanovic S, Münz C. Autophagy in innate and adaptive immunity against intracellular pathogens. J Mol Med. 2006;84:194–202. doi: 10.1007/s00109-005-0014-4. [DOI] [PubMed] [Google Scholar]
- 2.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sir D, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol. 2009;90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- 7.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Woters Kluwer Lippincott Williams and Wilkins; 2007. pp. 2977–3029. [Google Scholar]
- 8.Xu Z, et al. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J Virol. 2002;76:2579–2584. doi: 10.1128/jvi.76.5.2579-2584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, Li J, Johnson DL, Ou JH. Regulation of hepatitis B virus replication by the ras-mitogen-activated protein kinase signaling pathway. J Virol. 2003;77:7707–7712. doi: 10.1128/JVI.77.14.7707-7712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto A, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 11.Koike K, Takada S. Biochemistry and functions of hepatitis B virus X protein. Intervirology. 1995;38:89–99. doi: 10.1159/000150417. [DOI] [PubMed] [Google Scholar]
- 12.Blum HE, et al. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein NP, Bouchard MJ, Wang LH, Kobarg C, Schneider RJ. Src kinases involved in hepatitis B virus replication. EMBO J. 1999;18:5019–5027. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouchard MJ, Wang L, Schneider RJ. Activation of focal adhesion kinase by hepatitis B virus HBx protein: Multiple functions in viral replication. J Virol. 2006;80:4406–4414. doi: 10.1128/JVI.80.9.4406-4414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang YC, Thoman M, Linton PJ, Deisseroth A. Use of CD40L immunoconjugates to overcome the defective immune response to vaccines for infections and cancer in the aged. Cancer Immunol Immunother. 2009;58:1949–1957. doi: 10.1007/s00262-009-0718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlomai A, Paran N, Shaul Y. PGC-1alpha controls hepatitis B virus through nutritional signals. Proc Natl Acad Sci USA. 2006;103:16003–16008. doi: 10.1073/pnas.0607837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang H, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 20.Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology. 2009;390:122–129. doi: 10.1016/j.virol.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Xu Z, Zheng Y, Johnson DL, Ou JH. Regulation of hepatocyte nuclear factor 1 activity by wild-type and mutant hepatitis B virus X proteins. J Virol. 2002;76:5875–5881. doi: 10.1128/JVI.76.12.5875-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 24.Guo WT, Wang J, Tam G, Yen TS, Ou JS. Leaky transcription termination produces larger and smaller than genome size hepatitis B virus X gene transcripts. Virology. 1991;181:630–636. doi: 10.1016/0042-6822(91)90896-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.