Abstract
Homeotic (Hox) genes encode transcription factors that confer segmental identity along the anteroposterior axis of the embryo. However the molecular mechanisms underlying Hox-mediated transcription and the differential requirements for specificity in the regulation of the vast number of Hox-target genes remain ill-defined. Here we show that synthetic Sex combs reduced (Scr) genes that encode the Scr C terminus containing the homedomain (HD) and YPWM motif (Scr-HD) are functional in vivo. Synthetic Scr-HD peptides can induce ectopic salivary glands in the embryo and homeotic transformations in the adult fly, act as transcriptional activators and repressors during development, and participate in protein-protein interactions. Their transformation capacity was found to be enhanced over their full-length counterpart and mutations known to transform the full-length protein into constitutively active or inactive variants behaved accordingly in the synthetic peptides. Our results show that synthetic Scr-HD genes are sufficient for homeotic function in Drosophila and suggest that the N terminus of Scr has a role in transcriptional potency, rather than specificity. We also demonstrate that synthetic peptides behave largely in a predictable way, by exhibiting Scr-specific phenotypes throughout development, which makes them an important tool for synthetic biology.
Keywords: synthetic genes, transcriptional specificity, Hox genes, Sex combs reduced, homeotic transformations
Homeotic genes code for transcription factors that play an instrumental role in animal development by specifying the identity of body segments along the anteroposterior axis of the embryo (1–4). Hox genes have persisted in the animal kingdom; they are found in animals as diverse as worms and humans (5, 6) and the Homeodomain (HD), a helix-turn-helix DNA-binding domain, has been strikingly conserved in animals since before the bilaterian split (1, 7, 8). Sequence-specific binding of Hox proteins has been studied for the Drosophila Sex combs reduced (Scr) (9, 10), Antennapedia (Antp) (11, 12) and Ultrabithorax (Ubx) (11, 12) HDs. A consensus sequence TAATC/GC/G recognition core was identified in all of them, which alone is obviously not sufficient to confer transcriptional specificity, because it occurs statistically every kilobase in the genome. Similar sequence preferences have been identified for Deformed (Dfd) and Abdominal-B (Abd-B) (11), raising the question of how target specificity is achieved among different Hox paralogs.
A closer look into conserved residues outside the HD identified its amino-terminal YPWM motif that is present in almost all Hox proteins, from flies to vertebrates [with the exception of Abdominal-B (Abd-B), which has conserved only the tryptophan at position 3] (13). Extradenticle (Exd) and its mammalian homolog Pbx1 (14) were found to interact specifically with the YPWM motif of Hox proteins in vitro (15) and crystallographic analysis of a Ubx-Exd complex determined the topology of this interaction (16). Recently the link between the Antp YPWM motif and the transcriptional machinery was made through the identification of Bric-à-Brac interacting protein 2 (Bip2) (17). However, Hox cofactors do not suffice to entirely explain Hox specificity. The finding of cofactor independent Hox function (18) contributed to the realization that further sequences residing in the N terminus of Hox proteins might be the link for increased specificity in vivo.
In the present work we have derived synthetic Scr genes that encode the YPWM, HD and C terminus of the Drosophila Scr. The synthetic Scr-HD retained many functions of the full-length protein in vivo by participating in homeotic transformations, transcriptional regulation, and protein-protein interactions, thus reflecting to a great extent the properties of the native Scr protein. Constitutively active and inactive variants, in which threonine-6 and serine-7 of the Scr-HD have been substituted by alanines (Scr-HDAA) or aspartates (Scr-HDDD), respectively (19), behaved in the synthetic peptides similar as in the full-length protein, but the synthetic peptides exhibited stronger homeotic function as compared to their full-length counterpart. Moreover, we show that the synthetic peptides bind specifically to native Scr and consensus HD-binding sites in vitro and accumulate at sites of loose chromatin conformation in live salivary gland nuclei, where Scr is normally expressed during development (20). Taken together our results indicate that synthetic Scr peptides are functional in vivo and thus challenge the role of the N-terminal part of the endogenous fly peptide in transcriptional specificity.
Results
Synthetic Scr Genes are Capable of Inducing Ectopic Salivary Glands in the Embryo.
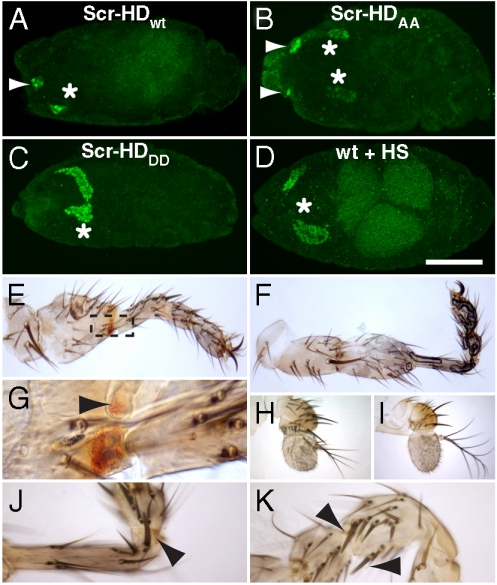
Scr acts as a master control regulator of salivary gland morphogenesis during embryonic development (20–25). When expressed throughout the embryo, Scr is able to induce an additional pair of salivary glands anterior to parasegment 2 (20), where the normal salivary glands form. Posteriorly its function is restricted by teashirt (tsh) and Abdominal-B (Abd-B) (26). In accordance with these findings, the wild type (Scr-HDwt) and the constitutively active (Scr-HDAA) synthetic genes could also induce ectopic salivary glands in the embryonic head, while the constitutively inactive variant (Scr-HDDD) could not (Fig. 1A–C) and displayed a normal pair of salivary glands, similar to the control embryos, also treated with heat shock (Fig. 1D).
Fig. 1.
Synthetic Scr peptides induce homeotic transformations in the fly (A–C). Expression of the synthetic genes throughout the embryo results in the formation of additional salivary glands in the cephalic region for Scr-HDwt (A) and Scr-HDAA (B), but not Scr-HDDD (C). (D) Wild type embryo also treated with heat shock allows the development of one normal pair of salivary glands. Arrowheads show the ectopic—and asterisks, the normal—salivary glands. All constructs were induced using Heat-Shock-Gal4. Stainings are on stage 16 embryos for dCREB-A (19), a salivary gland luminal marker. Scale bar 100 μm. (E–I) Expression of Scr-HDwt (E) and Scr-HDAA (F) in the antennal disc results in complete antenna-to-tarsus transformations. (G) Magnification of the outlined area in (E) shows pigmented cells (Arrowhead) at the distal part of the transformed A3. (H) Scr-HDDD only confers a small reduction in the size of the arista. (I) Wild type antenna. (J–K) Sex comb teeth on antennal tarsi generated by ectopic expression of Scr-HDwt (J) and Scr-HDAA (K) (Arrowheads). All transformations were generated using a Dll-Gal4 driver.
Misexpression of Synthetic Scr Genes Causes Homeotic Transformations in the Adult Head.
Both the fly Scr gene and its functional mouse homolog Hox-a5 (previously described as Hox-1.3) have been shown to induce partial antenna-to-tarsus transformations in the fly head (27, 28). To examine the ability of Scr-HDwt and Scr-HDAA to trigger homeotic transformations in vivo we expressed them ectopically in the antennal portion of the eye-antennal disc using the Distalless (Dll) enhancer (29). Transformation of the third antennal segment (A3) and the arista to a fully grown tarsus (Fig. 1E and F, respectively) was observed. The inactive mutant (Scr-HDDD) could only induce a reduction in the size of the arista (Fig. 1H) as compared to the wt antenna (Fig. 1I). The ectopic tarsi generated by gain-of-function of Scr-HDwt displayed a patch of red pigmentation, an indication that a group of cells might have been transformed to ectopic eye-cells (Fig. 1G). Moreover, the presence of one to two sex comb teeth on the ectopic tarsi mediated by Scr-HDwt (Fig. 1J) and Scr-HDAA (Fig. 1K) indicated that the ectopic tarsi are prothoracic (T1), normally specified by Scr (28).
Synthetic Scr Peptides Participate in Protein-Protein Interactions with Pax Transcription Factors in Vivo.
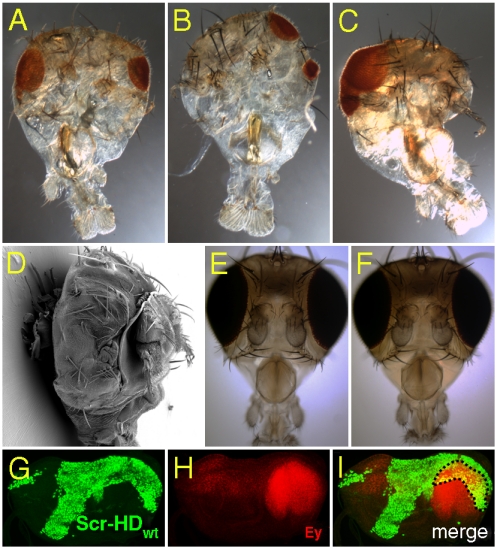
At the molecular level, Hox transcription factors are known to participate in protein-protein interactions. Such interactions have been demonstrated to take place between two HDs or a HD and a PAIRED domain (PD), resulting in mutual inhibition of DNA-binding properties, which leads to defects that resemble mutant phenotypes, caused by absence of either gene product. A phenotypic manifestation of such a negative posttranslational regulatory inhibition is the eye reduction caused by ectopic expression of various Hox proteins in the eye-antennal disc, shown to be triggered by binding of the HD of Hox proteins to the PD of eyeless (ey) (30, 31). In accordance with these findings ectopic expression of Scr-HDwt and Scr-HDAA using dppblink-Gal4 exhibited strong eye phenotypes (Fig. 2A–D) as compared to the full-length peptide (30), ranging from reduction to the complete absence of eyes. No abnormal eye phenotype was observed with Scr-HDDD (Fig. 2E), and the eyes in this case were indistinguishable from the control (Fig. 2F). Colocalization of Scr-HDwt (Fig. 2G) with endogenous Ey (Fig. 2H) suggested their interaction at the posttranslational level in the region of coexpression (Fig. 2I, Dashed Line).
Fig. 2.
Synthetic Scr peptides interact with Pax transcription factors in vivo (A–D). Eye-reduction phenotypes exhibited by ectopic expression of Scr-HDwt (A, B, and D) and Scr-HDAA (C). According to the strength of expression, different lines exhibited phenotypes ranging from eye-reduction (A–C) to eye-absence (D). (E) Ectopic expression of Scr-HDDD resulted in no detectable phenotype. (F) Wild type head. (G–I) Ectopic expression of Scr-HDwt in the eye-disc (G) does not repress ey (H). Dashed lines show the domain of colocalization of Scr-HDwt and Ey (I). The dppblink-Gal4 driver has been used throughout.
Synthetic Scr-HD Peptides Act as Transcriptional Activators and Repressors in the Eye-Antennal Disc.
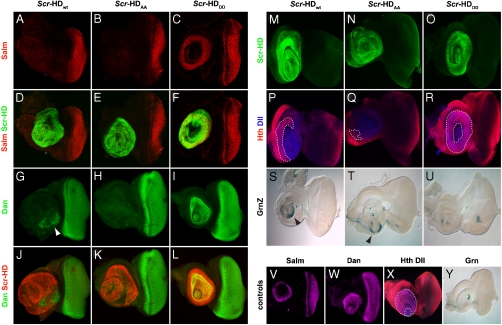
If Scr-HD peptides can cause antenna-to-tarsus transformations, genes responsible for antennal development must be repressed and leg determination genes must be activated in the antennal disc (32). Ectopic expression of Antp or Scr in the antennal disc is able to repress Spalt major (Salm) at the transcriptional level, thus preventing the differentiation to an antenna and allowing the initiation of the leg determination program (21, 33). In agreement with this principle, both Scr-HDwt (Fig. 3A and D) and Scr-HDAA (Fig. 3B and E) repressed Salm. As expected, no repression was observed with Scr-HDDD (Fig. 3C and F).
Fig. 3.
Synthetic Scr-HD peptides act as transcriptional activators and repressors in the antennal primordium. (A–F) Repression of Spalt major (Salm) in the antennal disc by ectopic expression of Scr-HDwt (A and D) and Scr-HDAA (B and E). No repression was observed with Scr-HDDD (C and F). (G–L) Repression of distal antenna (dan) in the antennal disc is complete with Scr-HDAA (H and K) and incomplete with Scr-HDwt (G and J), leaving a patch of cells that retain Dan activity (Arrowhead in G). These cells do not express ectopic Scr-HDwt. (I and L) Scr-HDDD does not repress dan. (M–R) Repression of Hth results in a shift of the Hth-Dll boundary. Partial repression of Hth by Scr-HDwt (M and P) and Scr-HDAA (N and Q), as compared to Scr-HDDD (O and R), where no repression is observed. (S–U) X-gal stainings showing the activation of grain (grn) by Scr-HDwt (S) and Scr-HDAA (T) (Arrowheads). Scr-HDDD (U) fails to activate ectopic grn expression. (V–Y) Normal expression of Salm (V), dan (W), Hth-Dll (X), and grn (Y) in eye-antennal discs. Dll-Gal4 was used to drive expression of all constructs (A-Y).
The same paradigm was found to apply for distal antenna (dan), another antenna determination gene normally expressed in the eye-antennal but not in the leg disc (34). Ectopic Antp in the antennal portion of the disc, induced in a mutant spineless (ss) background, results in the repression of dan, thus transforming the antenna into leg structures, a result suggesting that dan is repressed in discs undergoing tarsal transformations (34). Our findings show that this also occurs with Scr. Scr-HDwt partially repressed dan (Fig. 3G, arrowhead, and J). In the case of Scr-HDAA repression of dan was complete (Fig. 3H and K), while no repression was detected with Scr-HDDD (Fig. 3I and L).
Similar behavior was expected to apply in the repression of Homothorax (Hth), the function of which in the antennal determination program has been described extensively (35). Although expressed both in the antennal and the leg disc during development, its expression domain in the antennal disc largely overlaps with Dll, whereas in the leg disc the two gene products are distributed to distinct, nonoverlapping regions (36, 37). Because ectopic Antp in the antennal disc represses Hth (36) we assumed a general mechanism, according to which tarsal transformations in the antenna repress and thus restrict Hth outside of the Dll expression domain, resulting in a shift of the Hth-Dll boundary. Indeed, ectopic expression of Scr-HDwt (Fig. 3M and P) and Scr-HDAA (Fig. 3N and Q) resulted in partial or complete repression of Hth respectively, while no repression was seen with the inactive construct Scr-HDDD (Fig. 3O and R).
Inversely, to probe the transformed antennal discs for ectopically activated leg-specific genes, we tested the synthetic Scr-HD variants in their ability to activate grain (grn). grn encodes a GATA transcription factor (GATAc) that plays an important role in cell rearrangement during morphogenesis. It is expressed in the Central Nervous System (CNS), midgut, and lateral ectoderm during development (38). grn is normally expressed in the leg disc but not in the antennal disc; however, it is activated in the antennal disc upon ectopic expression of Antp [grn enhancer trap lines have been published as rK781 in (33) and as klecks in (39)]. Fig. 3S–U show the ectopic activation of grn mediated by the wild type and active constructs, whereas Scr-HDDD failed to activate transcription of the reporter (Fig. 3U). Taken together, these results demonstrate that the synthetic Scr peptides participate, directly or indirectly, in both transcriptional repression and activation in vivo.
Cells that Maintain dan Activity in the Antennal Primordium Give Rise to Ectopic Eyes on the Antennal Tarsi.
We were next interested to provide an explanation as to whether the pigmented cells observed in the transformed antennae (Fig. 1B and C) are indeed eye structures. We hypothesized that cells that retained dan activity in the antennal disc (Fig. 3G, Arrowhead) might account for the formation of ectopic eyes on the transformed tarsi upon misexpression of Scr-HDwt. Therefore, we tested the presence of photoreceptor cell markers in Dan protein positive cells in the antennal disc (Fig. 4A–D). Ectopic expression of Embryonic lethal abnormal visual system (Elav) colocalized with cells that still expressed dan but not ectopic Scr-HDwt (Fig. 4D), suggesting that these cells have acquired neuronal fate, similar to the differentiated photoreceptors in the eye portion of the disc. dan participates in both antennal and eye development (34, 40), and because these cells cannot differentiate into an antenna, they are able to form ectopic eyes in the transformed A3 segment. Scanning Electron Microscopy (SEM) of adult antennae confirmed that the pigmented cells observed previously (Fig. 1B and C) are indeed compound eyes, comprising several ommatidia (Fig. 4E).
Fig. 4.
Cells with no ectopic Scr-HDwt activity fail to repress dan but activate Elav and thus differentiate into compound eyes. (A) Elav gain-of-function, (B) Dan (Arrowheads), and (C) merge of (A) and (B) with Scr-HDwt. (D) Magnification of outlined area in (C). Arrow points at cells that express both dan and Elav but not ectopic Scr-HDwt. (E) Adult antenna transformed into a tarsus, which bears a small ectopic eye in the A3. Note the presence of ommatidia and interommatidial bristles (Red Asterisk). Scale bars in (C) 100 μm and in (D, E) 50 μm.
Molecular Interactions of Synthetic Scr Peptides in Live Salivary Gland Cells Using High-Resolution Quantitative APD Imaging.
In order to understand at the molecular level differences between the active and inactive peptide variants in their ability to interact with nuclear DNA, we studied their interactions with chromosomal DNA in live cells using advanced fluorescence imaging with Avalanche Photo Detectors (APD imaging) (41). Here we substantiate the phenotypic findings by molecular imaging. Elaborate quantitative study of Scr-HD–DNA interactions in live salivary gland cells is presented in the following paper (42).
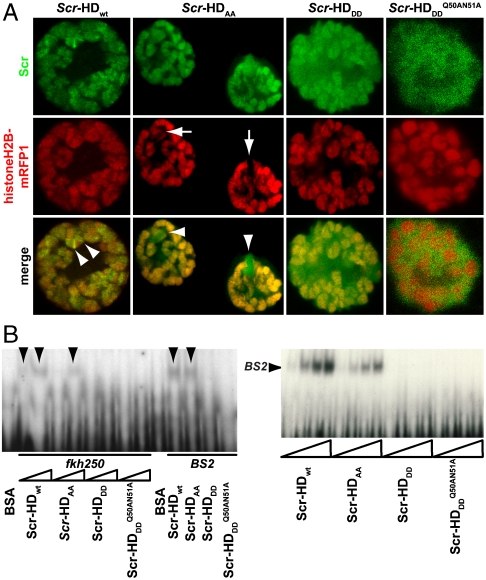
To visualize chromatin, a ubiquitously expressed histone-H2B-mRFP1 line was used. mCitrine-Scr-HD fusions expressed in salivary gland polytene nuclei showed that Scr-HDwt and Scr-HDAA peptides associate significantly with the DNA (Fig. 5). As observed for Scr-HDwt and Scr-HDAA, the transcription factor did not associate uniformly with the DNA but accumulated at sites of loose chromatin conformation where the histone signal was almost absent (Fig. 5A, first and second column). These sites should correspond to transcriptionally active regions. In contrast, Scr-HDDD showed some association with chromatin, but the transcription factor was also observed in the nucleoplasm among polytene chromosomes (Fig. 5A, third column). To understand this weaker yet substantial association of Scr-HDDD with chromatin we additionally mutated residues 50 and 51 of the Scr-HD to alanines (Scr-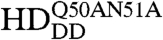 ). Glutamine-50 and asparagine-51 participate in third helix binding of the Antp HD to the DNA (43). We assumed that these mutations would behave accordingly in the synthetic Scr-HDDD peptide, acting in synergy with T6D and S7D and rendering the peptide interactions with chromatin even weaker. As expected, the synthetic Scr-
). Glutamine-50 and asparagine-51 participate in third helix binding of the Antp HD to the DNA (43). We assumed that these mutations would behave accordingly in the synthetic Scr-HDDD peptide, acting in synergy with T6D and S7D and rendering the peptide interactions with chromatin even weaker. As expected, the synthetic Scr-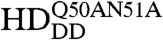 peptides appeared markedly dispersed in the nucleoplasm (Fig. 5A, fourth column) and excluded from chromatin. Flies expressing Scr-
peptides appeared markedly dispersed in the nucleoplasm (Fig. 5A, fourth column) and excluded from chromatin. Flies expressing Scr-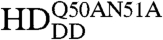 constitutively in the embryo, or ectopically in the antennal disc, did not display any of the embryonic or adult phenotypes.
constitutively in the embryo, or ectopically in the antennal disc, did not display any of the embryonic or adult phenotypes.
Fig. 5.
High-resolution APD imaging of DNA-Scr-HD interactions in live cells. (A) Third instar salivary gland polytene nuclei expressing Scr-HDwt, Scr-HDAA, Scr-HDDD, and Scr- under the control of dppblink-Gal4. Ubiquitously expressed mRFP1-tagged histone H2B was used to visualize chromatin. Scr-HDwt and Scr-HDAA readily associate with the chromosomes (as shown in the Green Channel) but also show sites of accumulation along the chromosome where loose chromatin compaction is shown as a low histone signal (Arrows). Arrowheads point at sites of high accumulation observed for Scr-HDwt and Scr-HDAA. The nucleus expressing the inactive Scr-HDDD shows some association of the transcription factor with the DNA, but it is also dispersed in the nucleoplasm. There is no pronounced banding pattern observed in this case, which suggests absence of specific binding. Scr-
under the control of dppblink-Gal4. Ubiquitously expressed mRFP1-tagged histone H2B was used to visualize chromatin. Scr-HDwt and Scr-HDAA readily associate with the chromosomes (as shown in the Green Channel) but also show sites of accumulation along the chromosome where loose chromatin compaction is shown as a low histone signal (Arrows). Arrowheads point at sites of high accumulation observed for Scr-HDwt and Scr-HDAA. The nucleus expressing the inactive Scr-HDDD shows some association of the transcription factor with the DNA, but it is also dispersed in the nucleoplasm. There is no pronounced banding pattern observed in this case, which suggests absence of specific binding. Scr-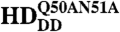 appears almost completely excluded from the chromosomes, mainly residing in the nucleoplasm. Scale bars in all cases are 20 μm. (B) Electrophoretic Mobility Shift Assay (EMSA) shows that only Scr-HDwt and Scr-HDAA bind DNA specifically in vitro. Both variants bound more strongly to BS2 than fkh250 (Left). Titration of peptide concentration (Right) revealed that even at high concentrations of transcription factor, Scr-HDDD and Scr-
appears almost completely excluded from the chromosomes, mainly residing in the nucleoplasm. Scale bars in all cases are 20 μm. (B) Electrophoretic Mobility Shift Assay (EMSA) shows that only Scr-HDwt and Scr-HDAA bind DNA specifically in vitro. Both variants bound more strongly to BS2 than fkh250 (Left). Titration of peptide concentration (Right) revealed that even at high concentrations of transcription factor, Scr-HDDD and Scr-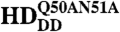 do not bind DNA (BS2) specifically.
do not bind DNA (BS2) specifically.
Synthetic Scr-HD Peptides Bind to DNA Specifically in Vitro.
To further investigate the observed association of Scr-HDDD with chromatin, we studied the specific binding of the synthetic peptides to DNA in vitro by gel-shift assays (Fig. 5B). Using the native Scr binding site fkh250 and the generic HD-binding site BS2, we observed specific binding for both Scr-HDwt and Scr-HDAA but not for the inactive peptides Scr-HDDD and Scr- , not even at high peptide concentrations (Fig. 5B, Left). Binding to BS2 appeared to be stronger for the transcriptionally active variants (Fig. 5B, Right) and was used for the titration. This result suggested that binding of Scr-HDDD to chromosomal DNA in polytene nuclei is largely nonspecific, because Scr-HDDD has additionally no homeotic function in the fly. Thus, the imaging analysis in live cells and the in vitro binding assay support our genetic findings and strengthen the notion that synthetic Scr genes function in a predictable way in vivo.
, not even at high peptide concentrations (Fig. 5B, Left). Binding to BS2 appeared to be stronger for the transcriptionally active variants (Fig. 5B, Right) and was used for the titration. This result suggested that binding of Scr-HDDD to chromosomal DNA in polytene nuclei is largely nonspecific, because Scr-HDDD has additionally no homeotic function in the fly. Thus, the imaging analysis in live cells and the in vitro binding assay support our genetic findings and strengthen the notion that synthetic Scr genes function in a predictable way in vivo.
Discussion
The genetic role of Hox genes has been studied extensively by gain- and loss-of-function experiments (24, 27, 35, 44–46), and the properties of the HD-DNA complex have been elucidated in solution by NMR (43). Nevertheless, the precise mechanisms that orchestrate the transcriptional regulation of the vast number of Hox target genes remain elusive. Here we have derived synthetic Scr genes and analyzed their function throughout development. We demonstrated that they are able to induce homeotic transformations in the adult fly and embryo, repress and activate antenna and leg-specific genes, respectively, and participate in protein-protein interactions. Our results support that they are functional in vivo and thus question the role of the N terminus of Scr in transcriptional specificity.
Hox-mediated antenna-to-tarsus transformations proceed through repression of genes necessary for antennal specification and ectopic activation of leg-specific genes in the antennal disc. This transformation is DNA-binding-dependent for Antp (30), because residues that impair binding of the HD to the minor groove (47) also abolish its transformation capacity in vivo. This suggested a similar requirement for Scr and thus for synthetic Scr peptides. Indeed, the latter bound putative Scr and generic HD-binding sites in vitro (Fig. 4B), repressed antennal-specific genes (Salm, dan, and Hth) and activated leg-specific genes (grn) in the antennal disc (Fig. 3). The inability of Scr-HDDD to trigger any of these phenotypes is in line with substitutions of threonine 6 and serine 7 to aspartates in the full-length protein, which impaired its DNA-binding activity in vitro (19). Our findings suggest that these mutations also abolish the capacity of Scr-HDDD to participate in HD-PD or HD-HD interactions. Although mutation of glutamate 19 to glycine abolished the dimerization capacity of Antp in vivo (30), in the case of Scr other residues of the HD, in addition to glutamate 19, might participate in protein-protein interactions, or, alternatively, the negative charge introduced by the aspartates might be responsible for abolishing these interactions through electrostatic repulsion. The fact that Scr-HDAA exhibited similar phenotypes as Scr-HDwt (compare Fig. 2C to Fig. 2A, B and D) is in favor of this scenario.
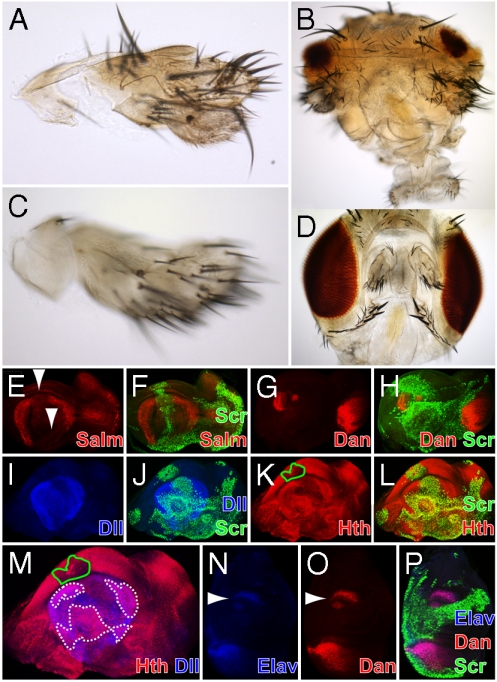
Surprisingly, the homeotic transformations observed in the adult fly (Fig. 1E–I) were considerably stronger than the ones observed with the full-length protein (Fig. 6A and C). Partial antenna-to-tarsus transformations were obtained with both the dppblink (Fig. 6A, as compared to Fig. 4E) and the Dll enhancers (Fig. 6C and D, as compared to Fig. 1E) and the eye-reduction phenotypes were comparable in strength to the full-length protein (compare Fig. 6B to Fig. 2A, B, and D). The same applied in the repression of antennal-specific markers (Salm in Fig. 6E and F, dan in Fig. 6G and H, Dll in Fig. 6I and J, and Hth in Fig. 6K and L) and the greater overlap observed between Hth and Dll (Fig. 6M, areas indicated by white dashed lines, as compared to Fig. 3P) supported the partiality of the tarsal transformation. Finally, ectopic expression of Elav on the antennal disc was found to colocalize with Dan protein positive cells (Fig. 6N–P). Weaker transformations obtained by the full-length Scr (27, 28) support the notion that the synthetic genes exhibit stronger homeotic function in vivo. In the paradigm of salivary gland induction, a morphogenetic process initiated by Scr, the synthetic genes behaved in a predictable way. They triggered the formation of an additional pair of salivary glands in the region of the embryonic head (Fig. 1A), comparable to those induced by the full-length protein (19).
Fig. 6.
The full-length Scr peptide exhibits weaker homeotic function than its synthetic counterparts in vivo. (A) Partial antenna-to-tarsus transformation mediated by Scr full-length using the dppblink enhancer. (B) The same gain-of-function results in eye-reduction in the head. (C) Transformation using the Dll enhancer is also incomplete. (D) Adult head of a fly expressing Scr full-length using Dll-Gal4. (E–F) Repression of Salm by the full-length Scr in the antennal disc (Arrowheads in E) is complete. (G–H) The same applies in the repression of dan. (I–M) Repression of Dll and Hth is incomplete (I, K, and M) and considerable overlap of Hth and Dll is observed in the antennal disc (Dashed White Lines in M). Scr represses Hth also outside the Dll expression domain (Solid Green Line in K and M). (N–P) Cells that maintain Dan activity (Arrowhead in O) in the antennal disc express ectopic Elav (Arrowhead in N). The full-length Scr has been induced by dppblink-Gal4 (E–P).
Four lines of evidence support that the function of the synthetic genes is specific rather than a generic HD effect. First, the sex comb teeth observed on the antennal tarsi indicate prothoracic leg identity (T1), specified by Scr. Second, the ectopic salivary glands in the embryo imply Scr-specific function (26). Third, mutations in the HD of the full-length peptide (19) behaved accordingly in synthetic peptides. Finally, specificity of Antp and Scr is owed to the N-terminal part of the HD (44), also present in the synthetic peptides.
It has been proposed that the large N-terminal part of Ubx and Scr is required for transcriptional activation in vivo (48). Our data show that this need not be the case for several Scr-specific functions. So, if Scr-specificity is achieved by synthetic genes, what is the function of the large N-terminal sequence of Scr in vivo? Sequence comparison of the fly Scr with its insect and vertebrate homologs (Fig. S1) revealed the divergence of the N terminus of the protein in length and sequence, with the exception of the MSSYFVNS, the DYTQL and the SCKYA motifs. Both the fly (48) and the murine MSSYFVNS domain in flies (49) seem to be limited to contributing to transcriptional potency rather than specificity, because its deletion resulted in homeotic transformations (49) and ectopic activation of Scr target genes, albeit weaker than the wild type protein (48, 49). Substitution of serine-10 by leucine in the Scr14 hypomorphic allele only resulted in a mild decrease in the number of sex comb teeth in the adult (50). The same applies to the DYTQL motif, because Scr15, another hypomorphic allele of Scr lacking this motif, displayed partially compromised Scr-activity but no loss of specificity in vivo (50). When parts of the N terminus of the murine Hox-a5 were removed, its transcriptional activity in live cells and its DNA-binding in vitro were considerably weaker, though not completely abolished (49). Although Hox-a5 lacking the complete N terminus could not trigger homeotic transformations in the fly (49), this indicates that the N terminus is responsible for regulating transcriptional levels. Our results show that the HD, YPWM motif and C terminus of Scr are both necessary and sufficient for providing transcriptional specificity in vivo. A change in potency, rather than in specificity, was also observed for Antp lacking fractions of its N terminus (44), suggesting a general requirement of the latter for transcriptional “fine-tuning” rather than specificity among Hox paralogs and homologs. An example of a dose-dependent developmental output of Hox gene products has already been described for Ubx-mediated repression of Dll (48) and might apply to a few Hox peptides. Quantitative studies might help to precisely describe such features of Hox peptides in vivo.
Our work also supports the hypothesis that synthetic genes and/or peptides behave to a large extent in a predictable way. This notion is central to synthetic biology (51–53), not only for engineering artificial processes, but also for developing new techniques for gene and peptide therapy. In the long run, functional synthetic genes/peptides may bear advantages for medical applications. Due to their size, they are expected to be considerably easier to engineer/synthesize and have better penetration efficiency into target cells or tissue—one of the greatest obstacles in peptide therapy. More studies in this direction will help unravel such perspectives. In the following paper (42), we take first steps in studying quantitatively the interactions of synthetic Scr peptides with DNA in live cells.
Materials and Methods
Cloning Procedure and Fly Transgenesis.
Generation of plasmids was performed using standard procedures. Transgenic lines were generated as described (54). Fly stocks used are outlined in SI Materials and Methods.
Immunohistochemistry.
Antibody stainings were performed as outlined in SI Materials and Methods.
High-Resolution APD Imaging.
High-resolution APD imaging was performed on a uniquely modified ConfoCor3 instrument (Carl Zeiss, Jena, Germany) (refer to SI Materials and Methods for more detail).
Electrophoretic Mobility Shift Assay (EMSA).
All mCitrine-Scr-HD fragments were cloned to pCR2.1-TOPO Vector (Invitrogen) and the peptides produced in vitro using TNT® T7 Quick Coupled Transcription/Translation System (Promega). EMSA was then performed as described (31).
Supplementary Material
Acknowledgments.
This work was supported by the Kantons of Basel-Stadt and Basel-Landschaft, a grant from the Swiss National Science Foundation, the European Network of Excellence “Cells into Organs,” the Swedish Brain Foundation, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, and the Ministry of Sciences and Technological Development of Serbia (Grants no. 142025 and 142019).
Footnotes
The authors declare no conflict of interest.
See companion article on page 4093.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914595107/DCSupplemental.
References
- 1.Gehring WJ. Homeo boxes in the study of development. Science. 1987;236(4806):1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- 2.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 3.Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- 4.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 5.Kappen C, Ruddle FH. Evolution of a regulatory gene family: HOM/HOX genes. Curr Opin Genet Dev. 1993;3(6):931–938. doi: 10.1016/0959-437x(93)90016-i. [DOI] [PubMed] [Google Scholar]
- 6.Kmita-Cunisse M, Loosli F, Bierne J, Gehring WJ. Homeobox genes in the ribbonworm Lineus sanguineus: Evolutionary implications. Proc Natl Acad Sci USA. 1998;95(6):3030–3035. doi: 10.1073/pnas.95.6.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Affolter M, Schier A, Gehring WJ. Homeodomain proteins and the regulation of gene expression. Curr Opin Cell Biol. 1990;2(3):485–495. doi: 10.1016/0955-0674(90)90132-x. [DOI] [PubMed] [Google Scholar]
- 8.Scott MP, Tamkun JW, Hartzell GW., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 9.Ryoo HD, Mann RS. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev. 1999;13(13):1704–1716. doi: 10.1101/gad.13.13.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odenwald WF, Garbern J, Arnheiter H, Tournier-Lasserve E, Lazzarini RA. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989;3(2):158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- 11.Ekker SC, et al. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994;13(15):3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller M, et al. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 1988;7(13):4299–4304. doi: 10.1002/j.1460-2075.1988.tb03328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian YQ, et al. NMR structure determination reveals that the homeodomain is connected through a flexible linker to the main body in the Drosophila Antennapedia protein. Proc Natl Acad Sci USA. 1992;89(22):10738–10742. doi: 10.1073/pnas.89.22.10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15(8):3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson FB, Parker E, Krasnow MA. Extradenticle protein is a selective cofactor for the Drosophila homeotics: Role of the homeodomain and YPWM amino acid motif in the interaction. Proc Natl Acad Sci USA. 1995;92(3):739–743. doi: 10.1073/pnas.92.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397(6721):714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 17.Prince F, et al. The YPWM motif links Antennapedia to the basal transcriptional machinery. Development. 2008;135(9):1669–1679. doi: 10.1242/dev.018028. [DOI] [PubMed] [Google Scholar]
- 18.Galant R, Walsh CM, Carroll SB. Hox repression of a target gene: Extradenticle-independent, additive action through multiple monomer binding sites. Development. 2002;129(13):3115–3126. doi: 10.1242/dev.129.13.3115. [DOI] [PubMed] [Google Scholar]
- 19.Berry M, Gehring W. Phosphorylation status of the SCR homeodomain determines its functional activity: Essential role for protein phosphatase 2A,B'. EMBO J. 2000;19(12):2946–2957. doi: 10.1093/emboj/19.12.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panzer S, Weigel D, Beckendorf SK. Organogenesis in Drosophila melanogaster: Embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development. 1992;114(1):49–57. doi: 10.1242/dev.114.1.49. [DOI] [PubMed] [Google Scholar]
- 21.Abzhanov A, Holtzman S, Kaufman TC. The Drosophila proboscis is specified by two Hox genes, proboscipedia and Sex combs reduced, via repression of leg and antennal appendage genes. Development. 2001;128(14):2803–2814. doi: 10.1242/dev.128.14.2803. [DOI] [PubMed] [Google Scholar]
- 22.Andrew DJ, Henderson KD, Seshaiah P. Salivary gland development in Drosophila melanogaster. Mech Dev. 2000;92(1):5–17. doi: 10.1016/s0925-4773(99)00321-4. [DOI] [PubMed] [Google Scholar]
- 23.Haberman AS, Isaac DD, Andrew DJ. Specification of cell fates within the salivary gland primordium. Dev Biol. 2003;258(2):443–453. doi: 10.1016/s0012-1606(03)00140-4. [DOI] [PubMed] [Google Scholar]
- 24.Percival-Smith A, Weber J, Gilfoyle E, Wilson P. Genetic characterization of the role of the two HOX proteins, Proboscipedia and Sex Combs Reduced, in determination of adult antennal, tarsal, maxillary palp and proboscis identities in Drosophila melanogaster. Development. 1997;124(24):5049–5062. doi: 10.1242/dev.124.24.5049. [DOI] [PubMed] [Google Scholar]
- 25.Kerman BE, Cheshire AM, Andrew DJ. From fate to function: The Drosophila trachea and salivary gland as models for tubulogenesis. Differentiation. 2006;74(7):326–348. doi: 10.1111/j.1432-0436.2006.00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrew DJ, Horner MA, Petitt MG, Smolik SM, Scott MP. Setting limits on homeotic gene function: Restraint of Sex combs reduced activity by teashirt and other homeotic genes. EMBO J. 1994;13(5):1132–1144. doi: 10.1002/j.1460-2075.1994.tb06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao LC, Liaw GJ, Pai CY, Sun YH. A common mechanism for antenna-to-Leg transformation in Drosophila: Suppression of homothorax transcription by four HOM-C genes. Dev Biol. 1999;211(2):268–276. doi: 10.1006/dbio.1999.9309. [DOI] [PubMed] [Google Scholar]
- 28.Zhao JJ, Lazzarini RA, Pick L. The mouse Hox-1.3 gene is functionally equivalent to the Drosophila Sex combs reduced gene. Genes Dev. 1993;7(3):343–354. doi: 10.1101/gad.7.3.343. [DOI] [PubMed] [Google Scholar]
- 29.Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274(5285):252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- 30.Plaza S, et al. Cross-regulatory protein-protein interactions between Hox and Pax transcription factors. Proc Natl Acad Sci USA. 2008;105(36):13439–13444. doi: 10.1073/pnas.0806106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plaza S, et al. Molecular basis for the inhibition of Drosophila eye development by Antennapedia. EMBO J. 2001;20(4):802–811. doi: 10.1093/emboj/20.4.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gehring WJ, Kloter U, Suga H. Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol. 2009;88:35–61. doi: 10.1016/S0070-2153(09)88002-2. [DOI] [PubMed] [Google Scholar]
- 33.Wagner-Bernholz JT, Wilson C, Gibson G, Schuh R, Gehring WJ. Identification of target genes of the homeotic gene Antennapedia by enhancer detection. Genes Dev. 1991;5(12b):2467–2480. doi: 10.1101/gad.5.12b.2467. [DOI] [PubMed] [Google Scholar]
- 34.Emerald BS, Curtiss J, Mlodzik M, Cohen SM. Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development. 2003;130(6):1171–1180. doi: 10.1242/dev.00323. [DOI] [PubMed] [Google Scholar]
- 35.Casares F, Mann RS. The ground state of the ventral appendage in Drosophila. Science. 2001;293(5534):1477–1480. doi: 10.1126/science.1062542. [DOI] [PubMed] [Google Scholar]
- 36.Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392(6677):723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- 37.Dong PD, Chu J, Panganiban G. Coexpression of the homeobox genes Distal-less and homothorax determines Drosophila antennal identity. Development. 2000;127(2):209–216. doi: 10.1242/dev.127.2.209. [DOI] [PubMed] [Google Scholar]
- 38.Brown S, Castelli-Gair Hombria J. Drosophila grain encodes a GATA transcription factor required for cell rearrangement during morphogenesis. Development. 2000;127(22):4867–4876. doi: 10.1242/dev.127.22.4867. [DOI] [PubMed] [Google Scholar]
- 39.Grieder NC, Morata G, Affolter M, Gehring WJ. Spalt major controls the development of the notum and of wing hinge primordia of the Drosophila melanogaster wing imaginal disc. Dev Biol. 2009;329(2):315–326. doi: 10.1016/j.ydbio.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Curtiss J, Burnett M, Mlodzik M. Distal antenna and distal antenna-related function in the retinal determination network during eye development in Drosophila. Dev Biol. 2007;306(2):685–702. doi: 10.1016/j.ydbio.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vukojevic V, et al. Quantitative single-molecule imaging by confocal laser scanning microscopy. Proc Natl Acad Sci USA. 2008;105(47):18176–18181. doi: 10.1073/pnas.0809250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vukojević V, Papadopoulos DK, Terenius L, Gehring WJ, Rigler R. Quantitative study of synthetic Hox transcription factor-DNA interactions in live cells. Proc Natl Acad Sci USA. doi: 10.1073/pnas.0914612107. doi:10.XXXX/XXXXXXXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehring WJ, et al. Homeodomain-DNA recognition. Cell. 1994;78(2):211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 44.Gibson G, Schier A, LeMotte P, Gehring WJ. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell. 1990;62(6):1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- 45.Morata G, Sanchez-Herrero E. Patterning mechanisms in the body trunk and the appendages of Drosophila. Development. 1999;126(13):2823–2828. doi: 10.1242/dev.126.13.2823. [DOI] [PubMed] [Google Scholar]
- 46.Wakimoto BT, Kaufman TC. Analysis of larval segmentation in lethal genotypes associated with the antennapedia gene complex in Drosophila melanogaster. Dev Biol. 1981;81(1):51–64. doi: 10.1016/0012-1606(81)90347-x. [DOI] [PubMed] [Google Scholar]
- 47.Otting G, et al. Protein–DNA contacts in the structure of a homeodomain–DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tour E, Hittinger CT, McGinnis W. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development. 2005;132(23):5271–5281. doi: 10.1242/dev.02138. [DOI] [PubMed] [Google Scholar]
- 49.Zhao JJ, Lazzarini RA, Pick L. Functional dissection of the mouse Hox-a5 gene. EMBO J. 1996;15(6):1313–1322. [PMC free article] [PubMed] [Google Scholar]
- 50.Sivanantharajah L, Percival-Smith A. Analysis of the sequence and phenotype of Drosophila Sex combs reduced alleles reveals potential functions of conserved protein motifs of the Sex combs reduced protein. Genetics. 2009;182(1):191–203. doi: 10.1534/genetics.109.100438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6(7):533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaznessis YN. Models for synthetic biology. BMC Syst Biol. 2007;1(47) doi: 10.1186/1752-0509-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 2006;4(4):e45. doi: 10.1371/journal.pbio.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.