Abstract
Epigenetic control of ribosomal RNA (rRNA) gene transcription by cell type-specific regulators, such as the osteogenic transcription factor Runx2, conveys cellular memory of growth and differentiation to progeny cells during mitosis. Here, we examined whether coregulatory proteins contribute to epigenetic functions that are mitotically transmitted by Runx2 in osteoblastic cells. We show that the transcriptional corepressor Transducin Like Enhancer-1 (TLE1) associates with rRNA genes during mitosis and interphase through interaction with Runx2. Mechanistically, depletion of TLE1 relieves Runx2-mediated repression of rRNA genes transcription and selectively increases histone modifications linked to active transcription. Biologically, loss of TLE-dependent rRNA gene repression coincides with increased global protein synthesis and enhanced cell proliferation. Our findings reinforce the epigenetic marking target genes by phenotypic transcription factors in mitosis and demonstrate a requirement for retention of coregulatory factors to sustain physiological control of gene expression during proliferation of lineage committed cells.
Keywords: cell fate, mitotic NORs, cell cycle, interphase nucleoli, coregulatory factors
Fidelity of transcriptional control determines progenitor commitment toward specific lineages and concomitantly excludes options for divergence toward other cell fates (1). In addition to genetically encoded information, epigenetic mechanisms that include DNA methylation (2) and histone modifications (3) play a critical role in physiological regulation of gene expression. During mitosis, many transcription factors are displaced from the chromosomes (4–6). However, recent studies indicate that association of gene regulatory factors with target genes is an epigenetic mechanism to maintain cellular memory of growth and differentiation (7–11).
Epigenetic control applies to RNA polymerase (Pol) II-transcribed protein coding genes as well as to RNA Pol I-mediated transcription of ribosomal RNA (rRNA) genes (12). A complex network of interdependent biochemical events regulates rRNA transcription during interphase to modulate the overall capacity of the protein-synthesis machinery, which is linked to cell growth and cellular phenotype (13–15). For examples, mechanisms that coordinate rRNA transcription with cell growth and differentiation in lineage-committed cells include the association of the osteogenic transcription factor Runx2 with mitotic chromosomes in osteoblasts (9). Runx2 epigenetically marks rRNA and other target genes during mitosis by directly occupying lineage-specific promoter regions (9, 10). Similarly, MyoD in myoblasts and C/EBP in adipocytes epigenetically mark rRNA genes (16). This mitotic retention provides a component of molecular memory that maintains mesenchymal lineage commitment and competency for protein synthesis in progeny cells (17, 18).
Runx factors are scaffolding proteins that interact with a large cohort of coregulatory proteins in interphase (19, 20). However, which cofactors associate with Runx2 during mitosis and are conveyed to the next interphase have not been established. Here, we show that the Runx2 coregulator TLE1 (21) forms a functional complex with Runx2 and the Pol I transcription factor UBF. TLE1, Runx2, and UBF together occupy nucleolar organizing regions (NORs) during mitosis and control histone modifications. This mitotic regulation by a corepressor protein establishes a unique dimension to a Runx2-mediated epigenetic mechanism that reinforces cell fate decisions as well as sustains protein synthesis capacity and cell growth in lineage committed cells.
Results and Discussion
C-Terminal Regulatory Domain of Runx2 Mediates Transcriptional Repression of rRNA Genes.
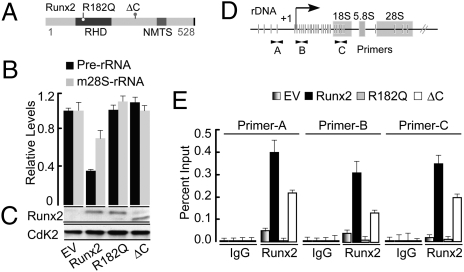
To characterize the mechanisms by which Runx2 controls rRNA gene transcription, we examined rRNA expression in the presence of Runx2 mutant proteins in which DNA binding (R182Q) (22) or transcriptional Delta C terminus (ΔC) functions are abrogated. The R182Q mutation affects the binding of Runx2 to the DNA without affecting nuclear or subnuclear localization of the proteins (23). Removal of the Runx2 C terminus (ΔC) abrogates interactions with several coregulatory proteins and subnuclear targeting without affecting nuclear localization or DNA binding activity (Fig. 1A) (19, 20, 23–25). Human osteoblastic cells were infected with lentiviral vectors expressing epitope-tagged wild type (WT) Runx2 or mutants R182Q and ∆C. In addition, Runx2 down-regulates pre-rRNA transcription levels (Fig. 1A), as has been shown (9). However, both R182Q and ∆C mutants failed to suppress rRNA gene expression (Fig. 1B). Expression of several Runx2 target genes (Cdc6, CyclinH, Cdc 46, CyclinB) (10) that are transcribed by RNA Pol II is up-regulated by Runx2 but not the ΔC mutant or R182Q (Fig. S1A). Western blot analysis revealed that WT Runx2, R182Q and ∆C proteins are expressed at significant and comparable levels after 48 h of infection (Fig. 1C). Chromatin immunoprecipitation (ChIP) assays showed that Runx2 interacts with three representative regions of the rDNA repeat (indicated as primer sets A, B and C; Fig. 1D), but not with the unrelated Phox (GP91) promoter (Fig. S1B). Both WT Runx2 and ∆C exhibited increased rDNA occupancy compared with control cells (Fig. 1E). As expected, the DNA binding mutant R182Q does not interact with the rDNA promoter (Fig. 1E). Taken together, these findings indicate the involvement of the Runx2 C terminus and associated activities in suppression of rRNA expression.
Fig. 1.
Runx2-mediated repression of rRNA gene expression requires the C terminus. (A) A schematic representation of full-length Runx2 protein indicating the DNA binding runt homology domain (RHD) and the nuclear matrix targeting signal (NMTS). The location of the DNA binding point mutation (R182Q) and the C terminus of ΔC truncation protein is also marked. (B) Real-time qPCR demonstrates relative level of precursor-rRNA (pre-rRNA) and mature-rRNA (m28S-rRNA) in response to WT Runx2, R182Q, and ∆C mutants, normalized to mitochondrial Cytochrome oxidase (mCox). (C) Western blot analysis of SaOS-2 cell lysates demonstrating equivalent levels of the Runx2, R182Q, and ∆C proteins in comparison with empty vector (EV) is shown. CdK2 levels were used as a control for equal protein loading. (D) Represents Ribosomal DNA (rDNA) repeat indicating the Runx2 binding sites (vertical lines) are shown. The position of three primer pairs, A, B, and C, used in this study for ChIP analysis are also shown; arrows indicate positions of primers at rDNA repeat. (E) Chromatin immunoprecipitation (ChIP) assay showing increased rDNA occupancy by the WT Runx2 or ∆C mutant, but not the DNA (R182Q) binding mutant.
TLE1/Groucho Corepressor Localizes with Runx2 at NORs During Mitosis.
Runx2 associates with rRNA genes at mitotic NORs and regulates their expression in progeny cells (9), thereby, epigenetically maintaining a molecular memory of the cell growth-related capacity to synthesize rRNA. However, the coregulators that support its epigenetic activity have not been identified. Because the Runx2 C terminus is required for rRNA gene repression (Fig. 1B), we focused on corepressors (i.e., TLE1 and HDAC1) that are known to interact with the Runx2 C terminus and support Pol II-mediated transcription of Runx2 target genes (10, 21, 26, 27). Western blot analysis with lysates from mitotically synchronized cells establishes that both TLE1 and HDAC1 are present in mitotic cells (Fig. 2A). Both proteins are stable upon inhibition of protein synthesis by cycloheximide (Fig. S1C), indicating that preexisting levels of these proteins are retained during mitotic progression.
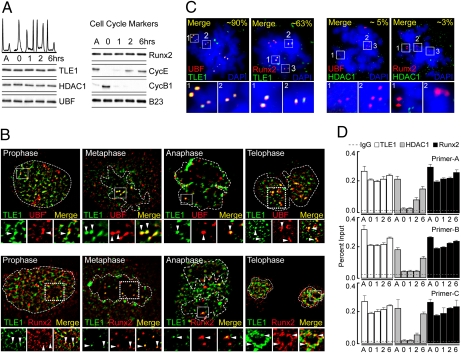
Fig. 2.
Both HDAC1 and TLE1 exhibit stable expression as cell exit mitosis, but only TLE1 associates with mitotic chromosomes. (A) SaOS-2 cells were blocked in mitosis with Nocodazole (100 ng/mL for 18 h) and released into the cell cycle. Progression of SaOS-2 cells through the cell cycle was analyzed by FACS analysis. Western blot analysis shows the temporal expression of cyclins and the stable expression of TLE1, UBF, HDAC1, Runx2, and the control B23 during different time points of the cell cycle. (B) Immunofluorescence of actively proliferating SaOS-2 cells demonstrates that UBF and Runx2 foci colocalize with TLE1 during different stages of mitosis. White dotted lines mark the nuclear or chromosomal boundaries, whereas the dotted white box indicates region where TLE1 colocalizes with UBF. Arrowheads represent the foci where TLE1 colocalizes with Runx2 and UBF. (C) TLE1 also localizes to NORs at mitotic chromosomes (Left), as assessed by immunofluorescence microscopy of metaphase spreads from mitotically blocked SaOS-2 cells. In contrast, HDAC1 foci do not occupy mitotic NORs with UBF or Runx2 (Right). (D) Chromatin immunoprecipitation analysis shows stable rDNA occupancy of TLE1 and Runx2 during stages of the cell cycle; however, HDAC1 rDNA occupancy is decreased during mitosis and gradually increases as cells progress through the cell cycle.
To investigate whether TLE1 and HDAC1 are associated with mitotic chromosomes, we analyzed actively proliferating osteoblastic cells at various mitotic stages by using immunofluorescence microscopy. In addition to Runx2, we included the Pol I transcription factor UBF, a marker for mitotic NORs (28–30), in our analysis. Our key findings are that TLE1 is localized with UBF foci during all stages of mitosis and that TLE1 is present at NORs on metaphase chromosomes (Fig. 2B Upper and Figs. S1D and S2A). Furthermore, TLE1 colocalizes with Runx2 (Fig. 2 B Lower and C Left, and Figs. S2B and S3A). In contrast, the histone modifying enzyme HDAC1, which is a known cofactor of Runx2 (20, 27), is excluded from NORs and does not colocalize with Runx2 or UBF (Fig. 2C Right and Fig. S3B). Our results indicate that TLE1 and Runx2 remain associated with rRNA genes during mitosis, and there is selective loss of HDAC1 from NORs.
We directly addressed whether the detection of TLE1 at NORs reflects association with regulatory regions of rRNA genes by using ChIP assays. Strikingly, TLE1 is constitutively bound to chromatin at the rDNA repeat during mitosis and the ensuing G1 phase of the cell cycle (Fig. 2D). In contrast, HDAC1 occupancy is clearly observed in asynchronous cells and is down-regulated at mitosis, but HDAC1 reassociates with rDNA repeats when cells progress into G1 (Fig. 2D). Consistent with their known epigenetic functions (9, 10), interactions of both Runx2 and UBF with rDNA remain unchanged at all time points (Fig. 2D and Fig. S4A). TLE1, HDAC1, Runx2, or UBF are not bound to the Phox (GP91) gene, establishing the specificity of our ChIP assays (Fig. S4B). These results demonstrate that Runx2 and its corepressor TLE1, but not HDAC1, localize to mitotic NORs and remain bound to rDNA during mitosis. Taken together, our findings suggest that Runx2 recruits TLE1 to the rDNA repeats during interphase and indicate a potential mitotic role for TLE1 in Runx2-mediated epigenetic regulation of rRNA genes.
TLE1 Interaction with UBF and Recruitment to Ribosomal RNA Genes Depend on Runx2.
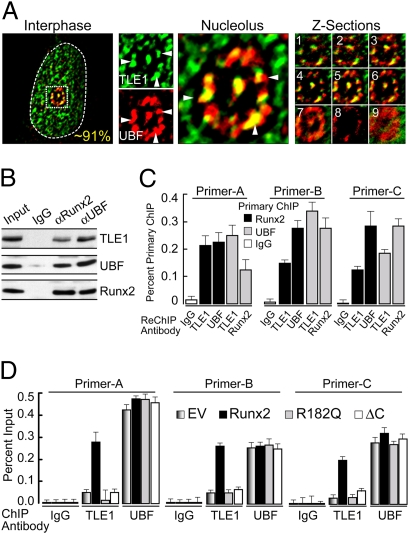
Runx2 colocalizes with UBF at the nucleolar periphery to regulate Pol I transcribed rRNA genes (9) and with TLE1 to suppress Pol II transcribed genes during interphase (21, 24). In addition, immunofluorescence microscopy shows that UBF and TLE1 colocalize at the nucleolar periphery during interphase (Fig. 3A). Therefore, we examined whether endogenous Runx2, UBF, and TLE1 reside in the same complex. Both Runx2 and UBF immunoprecipitate TLE1 (Fig. 3B) and reside on the same rDNA segments based on ChIP-ReChIP analysis (Fig. 3C and Fig. S4C). Because TLE1 lacks DNA binding activity, we addressed whether TLE1 is recruited to rDNA repeats through interactions with Runx2. Forced expression experiments show that TLE1 recruitment to rDNA, but not the unrelated Phox (GP91) gene, is significantly increased in the presence of WT Runx2 (Fig. 3D). Furthermore, no recruitment is observed for Runx2 mutants that are DNA binding defective (R182Q) or lack the VWPRY motif (ΔC) required for TLE1 interaction (Fig. 3D and Fig. S4D). UBF interactions with rDNA repeats are not affected by either WT or mutant Runx2 proteins (Fig. 3D), because UBF can independently recognize rDNA through its cognate motifs. Hence, Runx2 interacts together with TLE1 and UBF at the rDNA repeats and supports recruitment of TLE1 through the Runx2 C terminus.
Fig. 3.
Colocalization and interactions of TLE1 and UBF at rDNA repeats. (A) Immunofluorescence microscopy of SaOS-2 cells during interphase demonstrates that TLE1 localizes to nucleolar periphery and colocalizes with UBF. Average percentage colocalization was calculated by counting the number of nucleoli in 25 nuclei. White dotted line demarks the nuclear boundary, whereas the small white dotted square box marks the nucleolus. Also shown are optical sections of a representative cell, taken by focusing the nucleolus at the top (box 1) and capturing sequential images toward the bottom of the cell (box 9). (B) Coimmunoprecipitation assay shows TLE1 interaction with both UBF and Runx2 in SaOS-2 cells during interphase. (C) ChIP-ReChIP assays were carried out by performing primary ChIP with either Runx2 or UBF antibodies, followed by a secondary ChIP (ReChIP) with IgG, TLE1, UBF, or Runx2 antibodies. The results from a representative experiment are shown here in bar graphs of immunoprecipitated chromatin as a percentage of total chromatin. (D) ChIP assays show increased rDNA promoter occupancy of TLE1 in cells expressing the WT Runx2 but not the R182Q or ∆C mutant when compared to empty vector. The rDNA occupancy of UBF did not show significant change with the expression of Runx2 mutants.
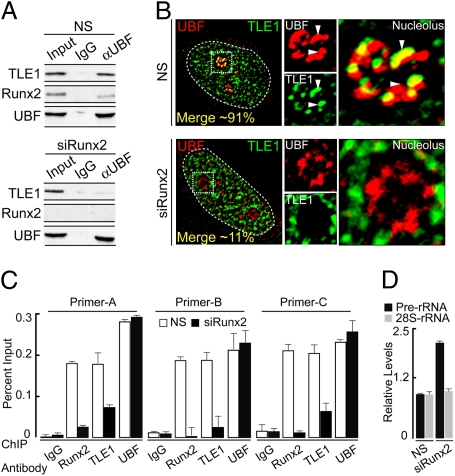
We assessed whether the TLE1 and UBF interaction depends on Runx2. Small interfering RNAs were used to deplete endogenous Runx2 levels (Fig. 4A). Immunoprecipitation analysis shows that depletion of Runx2 abolishes interaction between TLE1 and UBF (Fig. 4A and Fig. S4E). Consistent with these results, we observed decreased colocalization of TLE1 with UBF in siRunx2-treated cells (Fig. 4B). TLE1 and UBF colocalize in 91% of interphase nucleoli (Fig. 4B). In contrast, when Runx2 levels were reduced, TLE1 and UBF exhibited colocalization in only 11% of nucleoli, concomitant with loss of colocalization at mitotic NORs (Fig. S3 C and D). ChIP results show decreased occupancy of rDNA by TLE1 and diminished Runx2–rDNA interactions (Fig. 4C). Furthermore, loss of Runx2 increases pre-rRNA synthesis (Fig. 4D) and does not appreciably change rDNA occupancy by UBF (Fig. 4C and Fig. S4F). Thus, Runx2 mediates TLE1 localization at interphase nucleoli and mitotic NORs, interaction with UBF, and recruitment of TLE1 to rDNA.
Fig. 4.
Runx2 mediates interaction of TLE1 with UBF and its rDNA occupancy. (A) Lysate from SaOS-2 cells, transfected with a nonspecific (NS) or Runx2-specific (siRunx2) small interfering RNA oligonucleotides, were subjected to immunoprecipitation (IP). Results show that UBF interaction with TLE1 decreases significantly when Runx2 is depleted from the cells. (B) Immunofluorescence (IF) microscopy reveals that the TLE1 colocalization with UBF at nucleolar periphery decreases in Runx2 knockdown condition when compared to nonspecific (NS) siRNA oligo nucleotide. (C) Depletion of Runx2 also decreases the occupancy of TLE1 on the rDNA repeats when compared to nonspecific (NS) siRNA oligo nucleotide. However, UBF rDNA occupancy remains unaltered by depleting Runx2 as analyzed by ChIP using primer sets A, B, and C. (D) Relative levels of prerRNA are increased in the absence of Runx2 (siRunx2) when compared with nonspecific (NS) siRNA oligo nucleotide as determined by real-time qPCR. Values are obtained by normalizing the rRNA levels with mitochondrial cytochrome oxidase (mCox) levels.
TLE1 Is Required for Runx2-Mediated Repression of rRNA Gene Transcription.
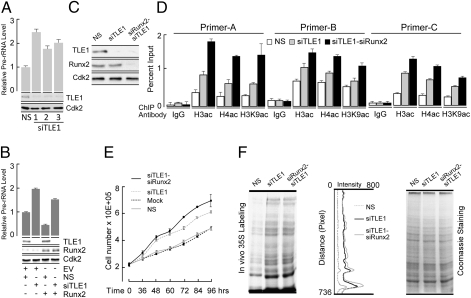
To assess the involvement of TLE1 in regulation of rRNA genes by Runx2, we used RNA interference to down-regulate TLE1 levels in the presence or absence of ectopic Runx2 expression in osteoblastic cells. Depletion of TLE1 by three independent, specific siRNAs resulted in the up-regulation of pre-rRNA levels, a measure of Pol I-mediated rRNA gene transcription (Fig. 5A). Furthermore, TLE1 depletion also relieves the known Runx2-dependent repression of rRNA transcription (Fig. 5B) (9). Loss of TLE1-mediated repression increases acetylation of histone H3 and H4, which represent markers of active transcription, based on ChIP analysis of rDNA (Fig. 5C). Combined depletion of TLE1 and Runx2 further increases histone acetylation (Fig. 5D). To address whether the regulation of RNA gene transcription by a Runx2–TLE1 complex is linked to the regulation of cell growth, we reduced the levels of either TLE1 alone or in combination with Runx2 by using TLE1- and Runx2-specific siRNAs (Fig. 5 D and E). Reduction of TLE1 and/or Runx2 levels results in increased global protein synthesis and significantly increased cell growth (Fig. 5 D and E). Therefore, TLE1 is a unique Runx2-dependent repressor of rRNA gene transcription that modulates cell growth and protein synthesis.
Fig. 5.
TLE1 is required for Runx2-mediated rRNA genes suppression. (A) Real-time qPCR shows that pre-rRNA levels are increased when TLE1 is depleted from cells. Western blots demonstrate specific down-regulation of TLE1 by three different siRNA oligonucleotides (1, 2, and 3). (B) Expression of Runx2 in the presence of a TLE1-specific siRNA does not alter pre-rRNA levels as assessed by qPCR. Western blot shows the knockdown of TLE1 protein in the presence and absence of Runx2 expression. (C) Western blot shows specific down-regulation of TLE1 alone or TLE1 with Runx2 by using siRNA oligonucleotides. (D) Chromatin immunoprecipitation assay demonstrates an increase in active histone modifications on rDNA repeats by knocking down either TLE1 alone or in combination with Runx2. (E) Line graphs represent cell count when either TLE1 or both Runx2 and TLE1 are down-regulated by specific siRNA oligonucleotides. Results indicate that the absence of TLE1 or TLE1 and Runx2 increases cell number. (F) Radioactive metabolic labeling with [35S] methionine shows TLE1 and Runx2 negatively regulate the rate of protein synthesis. Line scan was performed on individual lanes to show the difference in protein synthesis rate. Coomassie stain shows that equal protein samples were loaded in each lane.
Conclusion
In conclusion, our study shows that Runx2, UBF, and the corepressor TLE1 functionally cooperate to modulate rRNA transcription rates and cellular protein synthesis to control cell growth. In addition, we find that TLE1, like Runx2 and UBF, remains associated with the mitotic NORs and mediates selective modifications of histones bound at rDNA repeats in interphase. Thus, the coregulatory factor TLE1 contributes to the epigenetic marking of target genes (e.g., rRNA) during mitosis to maintain physiological regulation in response to the phenotype-related transcription factor Runx2.
Our findings reinforce the emerging concept of architectural epigenetics, which we define as a cellular memory that is reflected by persistent interactions of cell fate-determining transcription factors with Pol I and Pol II target genes in mitotic chromosomes to retain competency for lineage specific gene expression requirements in progeny cells. The demonstration that the corepressor TLE1 is necessary for Runx2-dependent epigenetic control resolves the fundamental question whether mitotically bound gene regulatory factors remain associated with cognate coregulatory proteins during successive cell divisions to ensure fidelity of lineage commitment.
Materials and Methods
In Situ Immunofluorescence Microscopy and Immunoprecipitation Analyses.
Proliferating and mitotically synchronized SaOS-2 osteosarcoma cells were processed for in situ immunofluorescence as described (9, 10) (Table S1). Coimmunoprecipitations, Western blot analysis, and ChIP assays were performed as described (9) (SI Materials and Methods). ChIP Re-ChIP experiments (31) were carried out by eluting immunoprecipitates from the first ChIP by using 10 mM DTT buffer (31) for 30 min at 37 °C. DNA obtained by the second immunoprecipitation was amplified by the indicated primers (Table S1).
RNA Interference and Expression by Using Lentiviral Constructs.
SaOS-2 cells at 60% confluence were transfected for 48 h with siRNAs for Runx2, TLE1, and HDAC1 (40 nmol each; obtained from Dharmacon) by using Oligofactamine reagent (Invitrogen). WT and mutant Runx2 lentiviral vectors were generated by using the Lentiviral Gateway System (Invitrogen) using the pENTR4-FLAG vector and recombined with the pLenti-CMV-Blast-DEST vector using the Gateway LR clonase enzyme mix as described by the manufacturer (Invitrogen). Runx2 constructs and packaging plasmids were transfected into 293T cells to generate lentivirus containing supernatants that were collected after 48 h.
Expression Analysis, Proliferation Assays, and Metabolic Labeling.
Total cellular RNA was analyzed by real-time quantitative PCR according to standard protocols (SI Materials and Methods). Proliferation assays were performed with cells transfected with nonspecific or specific siRNAs for Runx2 and/or TLE1. Trypsinized cell suspensions (20 μl) were loaded onto glass slides and counted by using a Cellometer Auto T4 Cellcounter (Nexcelom Bioscience LLC). Metabolic labeling (32) was carried out by using SaOS-2 cells transfected with siRNAs for Runx2 and/or TLE1 alone or both for 48 h. Cells were incubated in methionine-free RMPI media with 10% dialyzed serum for 1 h followed by incubation with EasyTag Express [35S] protein labeling mix (200 μCu/mL) for 45 min (Perkin-Elmer). Cells were harvested in direct lysis buffer [2 M Urea, 2% SDS, 10 mM DTT, 10% Glycerol, 10 mM Tris·HCl (pH 6.8), 0.2 mg/mL Bromophenol Blue, 1× complete pellet Protease inhibitor (Roche), and 25 μM Mg132]. Proteins were separated by SDS/PAGE (8%). Dried gels were exposed to scientific imaging film MR (Eastman Kodak) overnight at −70 °C.
Supplementary Material
Acknowledgments
We thank the members of our research group, especially Jitesh Pratap, Daniel Young, Mohammad Hassan, Margaretha van der Deen, and Jonathan Gordon, for stimulating discussion and sharing reagents. We also thank Karthiga Gokul, Matthew Mandeville, and Marissa Johnson for technical support and Judy Rask for expert assistance with manuscript preparation. This work was supported by National Institutes of Health Grants P01CA082834 (to G.S.S.), P01AR048818 (to G.S.S.), AR049069 (to A.J.W.), R37DE012528 (to J.B.L.), AR039588 (to G.S.S.), and DK32520 (Core Resources).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000620107/DCSupplemental.
References
- 1.Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Mellor J, Dudek P, Clynes D. A glimpse into the epigenetic landscape of gene regulation. Curr Opin Genet Dev. 2008;18:116–122. doi: 10.1016/j.gde.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 5.Muchardt C, Reyes J-C, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 6.Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: Keeping epigenetic memories from fading. Cell Cycle. 2009;8:818–823. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 8.Xing H, et al. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 9.Young DW, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 10.Young DW, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 12.McStay B, Grummt I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence RJ, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 14.Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 16.Ali SA, et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci USA. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaidi SK, et al. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- 18.Stein GS, et al. Organization, integration, and assembly of genetic and epigenetic regulatory machinery in nuclear microenvironments: Implications for biological control in cancer. Ann N Y Acad Sci. 2009;1155:4–14. doi: 10.1111/j.1749-6632.2009.03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian JB, et al. Runx2/Cbfa1 functions: Diverse regulation of gene transcription by chromatin remodeling and co-regulatory protein interactions. Connect Tissue Res. 2003;44(Suppl 1):141–148. [PubMed] [Google Scholar]
- 20.Jensen ED, Nair AK, Westendorf JJ. Histone deacetylase co-repressor complex control of Runx2 and bone formation. Crit Rev Eukaryot Gene Expr. 2007;17:187–196. doi: 10.1615/critreveukargeneexpr.v17.i3.20. [DOI] [PubMed] [Google Scholar]
- 21.McLarren KW, Theriault FM, Stifani S. Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression activity of the basic helix loop helix factor Hes1. J Biol Chem. 2001;276:1578–1584. doi: 10.1074/jbc.M007629200. [DOI] [PubMed] [Google Scholar]
- 22.Zhou G, et al. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum Mol Genet. 1999;8:2311–2316. doi: 10.1093/hmg/8.12.2311. [DOI] [PubMed] [Google Scholar]
- 23.Zaidi SK, et al. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- 24.Javed A, et al. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(α)/AML/PEBP2(α)) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 25.Choi J-Y, et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Javed A, et al. runt homology domain transcription factors (Runx, Cbfa, and AML) mediate repression of the bone sialoprotein promoter: Evidence for promoter context-dependent activity of Cbfa proteins. Mol Cell Biol. 2001;21:2891–2905. doi: 10.1128/MCB.21.8.2891-2905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HW, et al. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol Endocrinol. 2006;20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 28.Ochs RL, Lischwe MA, Shen E, Carroll RE, Busch H. Nucleologenesis: Composition and fate of prenucleolar bodies. Chromosoma. 1985;92:330–336. doi: 10.1007/BF00327463. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Sanchez JL, Gelpi C, Juarez C, Hardin JA. Anti-NOR 90. A new autoantibody in scleroderma that recognizes a 90-kDa component of the nucleolus-organizing region of chromatin. J Immunol. 1987;139:2579–2584. [PubMed] [Google Scholar]
- 30.Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovhannisyan H, et al. Maintenance of open chromatin and selective genomic occupancy at the cell cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandori C, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.