Abstract
Transcription in eukaryotic nuclei is carried out by DNA-dependent RNA polymerases I, II, and III. Human RNA polymerase III (Pol III) transcribes small untranslated RNAs that include tRNAs, 5S RNA, U6 RNA, and some microRNAs. Increased Pol III transcription has been reported to accompany or cause cell transformation. Here we describe a Pol III subunit (RPC32β) that led to the demonstration of two human Pol III isoforms (Pol IIIα and Pol IIIβ). RPC32β-containing Pol IIIβ is ubiquitously expressed and essential for growth of human cells. RPC32α-containing Pol IIIα is dispensable for cell survival, with expression being restricted to undifferentiated ES cells and to tumor cells. In this regard, and most importantly, suppression of RPC32α expression impedes anchorage-independent growth of HeLa cells, whereas ectopic expression of RPC32α in IMR90 fibroblasts enhances cell transformation and dramatically changes the expression of several tumor-related mRNAs and that of a subset of Pol III RNAs. These results identify a human Pol III isoform and isoform-specific functions in the regulation of cell growth and transformation.
Keywords: RPC32α, RPC32β, ES cells, differentiation, transcription
Transcription in eukaryotes is mediated by three nuclear DNA-dependent RNA polymerases (Pol I, Pol II, and Pol III) (1, 2). Pol III directs transcription of small noncoding RNAs that are involved in translation, splicing, and other cellular processes. Transcription by Pol III is directed by at least three distinct promoter types. Type 1 (5S RNA) and type 2 [tRNA, Alu RNA, and adenoviral viral-associated (VA) RNA] promoters are internal to the gene. Type 3 (U6 and 7SK RNA) promoters are located 5′ to the transcription initiation site (3). The transcription factors that directly recognize these promoters [type 1 by gene-specific TFIIIA and general initiation factor TFIIIC; type 2 by TFIIIC; and type 3 by gene-specific PSE-binding transcription factor/small nuclear RNA–activating protein complex (PTF/SNAPc)] have been well characterized and shown to recruit general initiation factor TFIIIB to their cognate promoters (reviewed in ref. 4). Overall, the multisubunit compositions of TFIIIC and TFIIIB have been conserved from yeast to human (5, 6), but two distinct isoforms of TFIIIB have been identified in human cells—one (TFIIIB-β) active in transcription of type 1 and type 2 promoters and one (TFIIIB-α) active in transcription of type 3 promoters (7). This functional difference reflects the presence of BRF1 in TFIIIB-β and of its paralogue BRF2 in TFIIIB-α (8, 9).
Pol III is highly conserved from yeast to humans and composed of 17 subunits. Of these subunits, five are common to all three polymerases, two are shared by Pol I and Pol III, and five are paralogous to subunits found in Pol I and Pol II. However, five subunits are specific to Pol III without a counterpart in Pol I or Pol II (reviewed in refs. 5 and 10). Three of these five Pol III-specific subunits (RPC32, RPC39, and RPC62) form a dissociable ternary subcomplex that is specifically required for transcription initiation (11). This ternary complex is evolutionary conserved from yeast to human and has been shown to interact with RPC160 and BRF1 (12, 13) and to be positioned near the transcription initiation site of the SUP4t RNATyr gene (14). Thus far, isozymes for Pol I, II, or III have been documented only for Arabidopsis Pol II (15). However, the possibility of Pol III isozymes was raised by the earlier purification, from mouse myeloma cells, of two forms of Pol III that were identical in subunit composition except for the presence of a 32-kDa (Pol IIIA) vs. a 33-kDa (Pol IIIB) subunit (16, 17). However, no further characterization of these enzymes was reported.
Transcription by Pol III is tightly regulated in normal cells, but this regulation is lost during tumorigenesis. Thus, Pol III transcription is negatively regulated in normal cells by tumor suppressor gene products (e.g., Rb, p53, or PTEN) or other factors (MAF1) and activated via signal transduction cascades such as the MAP kinase or the PI3 kinase pathways (reviewed in ref. 18). The loss of tumor suppressor protein activity or deregulated activation of signal transduction cascades leads to enhanced Pol III transcription. Interestingly, recent studies have indicated that enhanced Pol III transcription is required for cell transformation by the MYC oncogene (19) and that ectopic expression of the TFIIIB-β subunit BRF1 or the initiator tRNAiMet gene leads to transformation of mouse 3T3 fibroblasts (20). However, despite the knowledge that increased Pol III transcription is correlated with tumor development, only limited information is available regarding the underlying molecular mechanisms. Here, after identification of a unique human Pol III subunit, we demonstrate that human cells contain two Pol III isoforms with at least partially distinct functions and, importantly, that one isoform selectively contributes to transformation of human cells.
Results
Identification of RPC32 Paralogues RPC32α and RPC32β and Functional Characterization of the Corresponding Polymerase Isoforms Pol IIIα and Pol IIIβ.
A database search initially identified a paralogue, RPC32β, of the previously described human Pol III subunit RPC32 (10, 11) that is hereafter referred to as RPC32α (Fig. S1). We then established cell lines that stably express epitope-tagged RPC32α or RPC32β and, from derived nuclear extracts, affinity-purified Pol IIIα, containing RPC32α, and Pol IIIβ, containing RPC32β (Fig. 1A). The two Pol III preparations show very similar polypeptide patterns after SDS/PAGE and silver staining and probably differ only in the presence of RPC32α vs. RPC32β. Both forms of Pol III localize primarily to the nucleus (Fig. S2), and both are active in transcription of all known Pol III promoter types in vitro, as evidenced by transcription of 7SK, 5S, VA1, and tRNA genes in reconstituted Pol III-depleted nuclear extracts (Fig. S3) and by transcription of the VA1 gene in a system reconstituted with recombinant and highly purified components (Figs. 1 A–C).
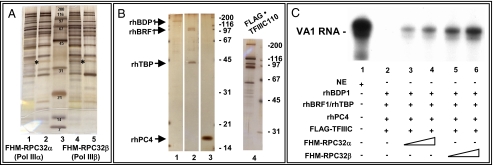
Fig. 1.
RPC32α and RPC32β assemble into distinct isoforms of human Pol III active in transcription of class III genes. (A) SDS/PAGE (4–20%) analysis of affinity-purified human Pol IIIα and IIIβ preparations. Purification of FLAG-HA-Myc (FHM)-RPC32α-associated Pol IIIα in the presence of 0.5% Nonidet P-40 and either 100 (lane1) or 300 mM KCl (lane 2). Purification of FHM-RPC32β-associated Pol IIIβ in the presence of 0.5% Nonidet P-40 and either 100 (lane 4) or 300 mM KCl (lane 5). The migration of FHM-RPC32α (lanes 1 and 2) or FHM-RPC32β (lanes 4 and 5) is indicated by an asterisk. Lane 3: Molecular weight standards with indicated masses. (B) SDS/PAGE (4–20%) analysis of purified Pol III transcription factors. Lane 1: 10 ng recombinant human (rh) BDP1 (TFIIIB150; ref 9). Lane 2: 12 ng rhBRF1 and 8 ng rhTBP. Lane 3: 50 ng rhPC4. Lane 4: 3 μL affinity-purified TFIIIC (Flag-TFIIIC110 cell line). (C) Transcription of the VA1 gene in a system reconstituted from highly purified transcription factors (shown in B). Lanes 2–6: 1 μL TFIIIC, 150 ng rhPC4, 6 ng rhBDP1 (TFIIIB150), 4 ng rhTBP, and 6 ng rhBRF1. Lanes 3 and 4: 10 and 20 ng Pol IIIα (A, lane 2). Lanes 5 and 6: 10 and 20 ng of Pol IIIβ (A, lane 5). Lane 1: 10 μg HeLa S3 nuclear extract.
RPC32α and RPC32β Display Distinct Expression Patterns During Embryonic Stem Cell Differentiation and Cellular Transformation.
In a further search for possible differences between Pol IIIα and Pol IIIβ, we found that RPC32α and RPC32β mRNAs are both expressed in human H1 ES cells. Notably, however, there is a strong decline in the level of RPC32α mRNA, which parallels the decline in ES cell marker mRNAs, during ES cell differentiation (Fig. 2A). In contrast, the level of RPC32β mRNA does not change substantially during ES cell differentiation (Fig. 2A).
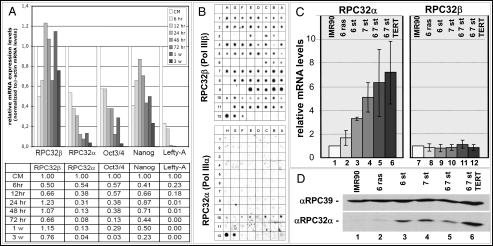
Fig. 2.
RPC32α mRNA expression is down-regulated during human stem cell differentiation and up-regulated during cell transformation. (A) Relative mRNA levels determined by RT-qPCR (Materials and Methods) before induction of differentiation and at specific time points during 3 weeks of differentiation of human H1 ES cells. mRNAs analyzed are indicated below the respective diagrams. (B) Dot blot analysis (Materials and Methods) of RPC32α and RPC32β mRNA levels in 73 different human tissues or cell lines. Representation of RNAs from different tissues is shown in Table S1. (C) Schematic representation of relative mRNA levels of RPC32α (Left) or RPC32β (Right) in human IMR90 fibroblasts (lanes 1 and 7) and IMR90 fibroblasts that stably express the proteins indicated at the top: E6 and ras (lanes 2 and 8), E6 and st (lanes 3 and 9), E7 and st (lanes 4 and 10), E6/E7/st (lanes 5 and 11), or E6/E7/st and TERT (lanes 6 and 12). 6, HPV E6; 7, HPV E7; st, SV40 small t; TERT, catalytic subunit of telomerase. Relative mRNA levels determined by RT-qPCR (Materials and Methods) are depicted on the left. (D) Western blot analysis of nuclear extracts derived from IMR90 cells (lane 1) or derivatives of IMR90 cells expressing E6 and ras (lane 2), E6 and st (lane 3), E7 and st (lane 4), E6/E7/st (lane 5), or E6/E7/st/TERT (lane 6). The same blot was probed with anti-RPC39 antibodies (Top) or affinity-purified anti-RPC32α antibodies (Bottom; the specificity of the anti-RPC32α antibodies is shown in Fig. S8 A and B).
We next analyzed expression levels of RPC32α and RPC32β mRNAs in differentiated human tissues. RPC32α mRNA was barely detectable in differentiated tissues and could only be detected in some tumor cell lines (Fig. 2B, Bottom). In contrast, RPC32β mRNA was found in all tissues analyzed, with slightly lower expression levels in certain tumor cell lines (Fig. 2B, Top). To determine in more detail whether expression of RPC32α or RPC32β mRNA changes during cell transformation, we analyzed IMR90 cells displaying increasingly transformed phenotypes in response to retrovirus vector-mediated expression of combinations of G12V-HRAS, HPV E6, E7, SV40 small T-antigen, and TERT (21, 22). We observed a dramatic increase of RPC32α mRNA (Fig. 2C) and protein levels (Fig. 2D and Fig. S4A) upon cellular transformation, whereas the levels of RPC32β mRNA remained unchanged (Fig. 2C). As a consequence, relative RPC32α:RPC32β mRNA levels changed from 1:4 in IMR90 cells to 2:1 in IMR90/E6/E7/st/TERT cells (Fig. S4B). To analyze whether altered RPC32α expression levels during cellular transformation simply reflect changes in proliferation rates, we arrested IMR90 cells in G0 phase of the cell cycle by serum starvation and released them from arrest thereafter by splitting cells into serum-containing growth media. Surprisingly, RPC32α expression levels increased 2.6-fold upon arrest in G0 and a further 5.6-fold upon return to midlog phase proliferation (Fig. S5, Bottom), demonstrating that RPC32α expression, although being influenced by proliferation, does not strictly correlate with proliferation rate. The mRNA expression of RPC32β did not significantly change during growth arrest and return to midlog phase growth (Fig. S5, Top).
Suppression of RPC32β by siRNAs Inhibits General Cell Growth, Whereas Suppression of RPC32α by siRNAs Inhibits Anchorage-Independent Growth.
We next asked whether the expression of RPC32α or RPC32β is essential for the survival of human cells and whether either influences tumor growth properties. For this purpose, we identified siRNAs that specifically suppress either RPC32α or RPC32β expression in transiently transfected Huh7 human hepatocarcinoma cells (Fig. S6A). Further analyses showed that reduction of RPC32β mRNA by a cognate siRNA, but not a control siRNA, substantially reduced growth of these cells and increased the number of floating dead cells (Fig. S6B). Seven independent attempts to establish HeLa cell lines that stably express an RPC32β siRNA yielded one extremely slow-growing line that still expressed RPC32β mRNA at approximately half the normal level but none that lacked RPC32β—thus indicating that RPC32β may be essential for cell viability and cannot be compensated by RPC32α. Importantly, analysis of mRNA expression levels showed an approximately 2.6-fold higher level of RPC32α mRNA than RPC32β mRNA in HeLa cells that stably express FLAG-RPC53 (Fig. S7, lane 1), suggesting that the suppression of RPC32β does not lead to a general lack of of Pol III activity in these cells. Western blot analyses of purified Pol III preparations confirmed that Pol IIIα is present in at least equivalent quantities as Pol IIIβ in HeLa cells (Fig. S8C). In contrast to siRNA-mediated suppression of RPC32β, siRNA-mediated suppression of RPC32α in Huh7 cells was readily tolerated, and the cells did not show slowed growth (Fig. S6B). Furthermore, suppression of RPC32α in HeLa cells by a stably expressed RPC32α siRNA (Fig. 3A and Fig. S7) did not affect growth on Petri dishes. Surprisingly, however, stable suppression of RPC32α by siRNAs dramatically reduced the formation of colonies in soft-agar assays (Fig. 3B).
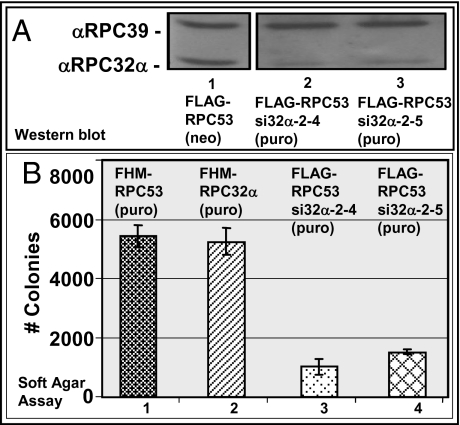
Fig. 3.
siRNA-mediated suppression of RPC32α impedes colony formation of HeLa cells in soft-agar assays. (A) Western blot. HeLa cells (BN51) that stably express FLAG-RPC53/neo (11) were transfected with pSuper-si32α-2/puro, and individual clones were selected after addition of 2 μg/mL puromycin. Nuclear extracts were used for affinity purification of Pol III (via FLAG-RPC53), and the eluates were analyzed with anti-RPC39 or anti-RPC32α antibodies. Purifications from 500 μg nuclear extract of FLAG-RPC53/neo HeLa cells (lane 1), FLAG-RPC53-si32α-2 clone 4 (lane 2), or clone 5 (lane 3) HeLa cells. (B) Soft agar assay, in the presence of 2 μg/mL puromycin, of HeLa cells stably expressing FHM-RPC53/puro (lane 1), FHM-RPC32α/puro (lane 2), FLAG-RPC53/neo (lanes 3 and 4), and RPC32α siRNA 32α-2 (lane 3, clone 4 and lane 4, clone 5). The numbers of colonies formed are indicated to the left.
Ectopic Expression of RPC32α Induces Anchorage-Independent Growth in Partially Transformed Human IMR90 Fibroblasts.
Because suppression of RPC32α inhibited colony formation in soft-agar assays, we asked whether ectopic expression of RPC32α would induce colony formation in such assays. For this purpose, we used IMR90 fibroblasts and established partially transformed IMR90 derivatives (expressing HPV E6 and E7 proteins) (21, 22) that stably express epitope-tagged forms of either RPC32α or RPC32β (at least three independent clones for each cell line). Expression levels of ectopic RPC32α or RPC32β were monitored by Western blot (Fig. S9). The partially transformed IMR90 cell lines expressing ectopic RPC32α showed enhanced growth properties at ≈10–15 doubling times after selection by puromycin. Importantly, ectopic expression of RPC32α also enhanced the colony formation capability of the partially transformed IMR90 fibroblasts, whereas ectopic expression of RPC32β, by contrast, slightly inhibited the colony formation capability of these cells (Fig. 4A and Fig. S10).
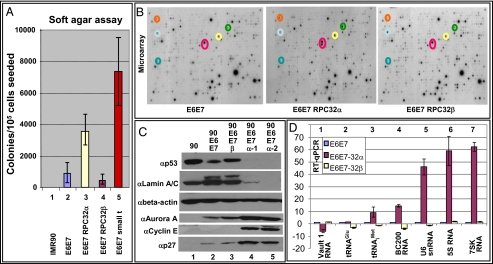
Fig. 4.
RPC32α contributes to transformation of IMR90 fibroblasts and changes the expression of several transformation-associated Pol II genes. (A) Soft-agar assay of human IMR90 fibroblasts (lane 1) and IMR90 fibroblasts that stably express the proteins indicated at the bottom (lanes 2–5). The numbers of colonies formed are indicated on the left. (B) Atlas Cancer 1.2 microarray (Clontech). Membranes were incubated with radioactively labeled cDNAs from IMR90 cells expressing E6 and E7 (Left), E6, E7, and RPC32α (Middle), or E6, E7, and RPC32β (Right). Table S2 summarizes quantitative results obtained by using the Image Quant program. Some of the regulated RNAs are encircled (S100A4: lower dot in the red oval; circles: RFC40/green; ezrin/orange; rac1/yellow; prefoldin/light blue; KLF6/turquoise. (C) Western blot. Twenty micrograms of nuclear extract, derived from IMR90 fibroblasts (lane 1) or IMR90 fibroblasts expressing E6 and E7 (lane 2), E6, E7, and RPC32β (lane 3), or E6, E7, and RPC32α (lanes 4 and 5; two distinct clones) were separated by SDS-10% PAGE, transferred to nitrocellulose membranes, and probed with the antibodies indicated on the left. (D) RT–qPCR. Expression of individual RNAs was determined in IMR90 fibroblasts expressing E6 and E7 (blue bars), E6, E7, and RPC32α (purple bars), or E6, E7, and RPC32β (light yellow bars). The following Pol III-transcribed genes were analyzed: Vault 1 RNA (lane1); tRNAGlu (lane 2); Initiator tRNAiMet (lane 3); BC200 RNA (lane 4); U6 RNA (lane 5); 5S RNA (lane 6); and 7SK RNA (lane 7). The height of the bars indicates the relative expression level compared with the expression in IMR90, E6, and E7, which was set as 1.
Ectopic Expression of RPC32α Strongly Influences the Expression of Tumor- and Invasion-Related mRNAs.
To investigate the molecular basis for the observed differences between ectopically expressed RPC32α and RPC32β effects on the growth of transformed IMR90 fibroblasts, RNAs from the three cell lines were subjected to Atlas Cancer 1.2 microarray analyses (Fig. 4B). Strikingly, ectopic expression of RPC32α in the partially transformed IMR90 cell lines increased the expression of genes, including S100A4 (23), replication factor C subunit RFC40 (24), EZRIN (25) and RAC1 (26), that previously have been associated with cell survival, tumor growth and metastasis. Furthermore, ectopic expression of RPC32α reduced expression of genes with reported tumor suppressor activity, such as PREFOLDIN 5 (MM-1) (27) or KLF6 (28), when compared either with partially transformed IMR90 cells or with partially transformed IMR90 cells with ectopically expressed RPC32β (Fig. 4B and Table S2). These data led us to analyze (by Western blot) whether the expression of other tumor-related proteins was altered upon ectopic expression of RPC32α or RPC32β in partially transformed cells. In this regard, cyclin E, aurora A, and P27KIP1 levels were increased by ectopic RPC32α expression but remained unchanged by ectopic RPC32β expression (Fig. 4C). In contrast, the levels of p53 and lamin A/C were dramatically down-regulated by ectopic RPC32α expression, but either unchanged (lamin A/C) or moderately increased (p53) by ectopic RPC32β expression (Fig. 4C). It should be noted that lamin A/C is not expressed in mouse cells before day 9 of development (29) and is thus absent in ES cells that contain high levels of Pol IIIα, consistent with the possibility that RPC32α may be involved in the regulation of lamin A/C expression. Along with the above-described effects of ectopically expressed RPC32α and RPC32β on the growth properties of cells, these gene expression results suggest that RPC32α exhibits oncogenic potential, whereas RPC32β instead may show tumor-suppressive activity under our experimental procedures.
Even though we did not detect clear differences in the transcriptional activities of Pol IIIα and Pol IIIβ in our DNA-templated in vitro assays (Fig. 1C and Fig. S3 B–E), where normal constraints may be missing, we used RT–quantitative PCR (qPCR) to analyze the expression of several Pol III-transcribed genes in partially transformed IMR90 cells and in derivative cell lines that ectopically express either RPC32α or RPC32β. As shown in Fig. 4D, expression of 5S RNA, U6 RNA, and 7SK RNA was strongly enhanced (lanes 5-7), and that of initiator tRNAMet and of BC200 RNA moderately enhanced (lanes 3 and 4), upon stable ectopic expression of RPC32α. In contrast, tRNAGlu expression remained unaffected, and vault1 RNA expression decreased upon ectopic expression of RPC32α (lanes 1 and 2). Remarkably, ectopic expression of RPC32β did not change considerably the expression of any of these Pol III genes (Fig. 4D). These data indicate that the observed dramatic changes in 5S RNA, U6 RNA, and 7SK RNA expression are specifically caused by ectopic expression of RPC32α and not due to a general deregulation of transcription.
Discussion
In this report we describe a human Pol III isoform that contains the newly identified RPC32β subunit instead of the paralogue RPC32α subunit and further compare the expression and function of RPC32α and RPC32β and the corresponding isozymes. RPC32β is widely expressed in all tissues analyzed and seems to be indispensable for cell survival, leading to the conclusion that RPC32β-containing Pol IIIβ represents the general form of human Pol III. In contrast, RPC32α shows a restricted expression pattern and is predominantly found in undifferentiated human ES cells and in transformed cells. Furthermore, and most importantly, ectopic expression of RPC32α in partially transformed cells enhances transformation in association with gene-specific transcription events, whereas its suppression in tumor cells impedes tumor cell growth, underscoring an important function of RPC32α in the establishment and maintenance of tumor cell growth.
Isoform-Specific Functions of Pol IIIα and Pol IIIβ.
On the basis of our findings that the distinguishing subunit, RPC32β, is expressed in all tissues tested and in both undifferentiated and differentiated human ES cells, Pol IIIβ seems to be ubiquitous. These results suggest that Pol IIIβ may be critical for transcription of class III genes that are essential for the growth and homeostasis of both undifferentiated and differentiated cells. This hypothesis is further supported by the apparently lethal phenotype of siRNA-mediated suppression of RPC32β, which also indicates that Pol IIIα cannot replace all Pol IIIβ functions in vivo. Hence, and given the dispensability of Pol IIIα for cell viability, Pol IIIβ seems to be the more general form of Pol III.
In contrast to Pol IIIβ, the originally identified form of human Pol III (Pol IIIα) does not seem to be essential for the survival of all cells, but it may exert important functions in only a subset of cells. The elevated expression of RPC32α mRNA—and thus Pol IIIα—in undifferentiated human H1 ES cells and in tumor cells, as well as a clear increase in RPC32α mRNA levels during transformation of IMR90 fibroblasts with defined genetic elements, indicate that RPC32α mRNA expression may be related to the differentiation status of a cell. Although siRNA-mediated knockdown of RPC32α is not lethal for HeLa cells, suggesting that Pol IIIβ can provide all essential Pol III functions in these tumor cells, it does result in loss of colony formation in soft-agar assays, underscoring an important function for RPC32α in sustaining anchorage-independent growth of transformed cells. Because RPC32α, and thus Pol IIIα, is predominantly found in undifferentiated or transformed cells, the functions of Pol IIIα may include the transcription of noncoding genes that help keep these cells in an undifferentiated or transformed state.
Target Gene Specificity of Pol IIIα and Pol IIIβ In Vitro and In Vivo.
In vitro transcription assays with naked DNA templates and either Pol III-depleted nuclear extract or purified initiation factors have shown nearly comparable abilities of purified Pol IIIα and Pol IIIβ to accurately transcribe 5S RNA, tRNA, VA1 RNA, and 7SK RNA genes (Fig. 1C and Fig. S3). Thus, in conjunction with general transcription initiation factors, Pol IIIα and Pol IIIβ both have intrinsic abilities for accurate transcription directed by each of the three class III promoter types. Although target gene specificity for Pol IIIα vs. Pol IIIβ was not evident from the limited number of class III genes tested in vitro, ectopic expression of RPC32α in partially transformed human fibroblasts resulted in enhanced levels of some, but not all, of the analyzed Pol III transcripts in vivo (Fig. 4D).
Isoform-specific effects on different class III genes in vivo clearly suggest isoform-specific interactions with accessory transcription factors or other gene regulatory proteins. In this regard, the differential effects of ectopically expressed Pol IIIα on transcription of two distinct type 2 genes (tRNAiMet and tRNAGlu) that commonly involve promoter recognition by TFIIIC and TFIIIB are most intriguing, because they also imply the existence of additional, hitherto unsuspected regulatory transcription factors for these genes. Regulatory factors could counteract intracellular constraints to transcription that may include (i) repressive chromatin structures that impose requirements for chromatin remodeling factors that are directly recruited or stabilized by the Pol III transcription machinery (30), (ii) limiting concentrations of general initiation or gene-specific Pol III factors, or (iii) various general repressors of Pol III transcription, such as p53, Rb, ARF, PTEN, and MAF-1 (18). Alternatively, such regulatory factors could be involved in the assembly of subnuclear architectures, as may be evidenced by perinuclear compartments that depend on Pol III transcription and are predominant in certain cancer cells (31).
Some of the Pol III genes that are differentially regulated in vivo, most notably initiator tRNAiMet, have been reported to be associated with cell proliferation and transformation. Specifically, it was shown that initiator tRNAiMet expression increases, whereas elongator tRNAMet expression decreases, during cell cycle progression in proliferating liver cells (32). This is consistent with our observation of a selective effect of ectopic RPC32α expression on tRNAiMet synthesis relative to tRNAGlu synthesis. Furthermore, enhanced initiator tRNAiMet levels were reported to accompany or cause cell proliferation and transformation in experimental systems (19, 20). Although we observed the highest increases in 7SK, U6, and 5S RNA expression, rather than in initiator tRNAiMet expression, it remains to be determined whether these ubiquitously expressed RNAs actively intervene in cell transformation or whether their ectopic expression simply accompanies this process. It also remains to be determined whether, as seems likely, there are other, hitherto unknown Pol III-transcribed genes that contribute to the regulation of ES cell differentiation or to cell transformation and that are differentially transcribed by Pol IIIα and Pol IIIβ.
Interestingly, ectopic RPC32α expression in partially transformed fibroblasts also increased the expression of several Pol II-transcribed genes associated with cell survival, tumor growth, and metastasis, while reducing expression of several genes with tumor suppressor activity. These effects were also specific for ectopically expressed RPC32α relative to RPC32β and must result from indirect effects of Pol IIIα transcription events on the corresponding Pol II-transcribed genes. Because it was recently reported that Pol III is involved in transcription of several human micro RNAs (33, 34), it is conceivable that Pol IIIα mediates transcription of certain miRNAs or other types of regulatory RNAs that target genes contributing to the induction of differentiation or to the inhibition of cell transformation-related events. Given our identification of profound effects of Pol IIIα on cell differentiation and transformation, future studies must be directed toward identification of the direct Pol IIIα target genes involved in these effects and the mechanisms involved in their activation and function.
Materials and Methods
Plasmids and Protein Purification.
Plasmids ph7SK, ptRNA, pVA1, and pH5S8544, in vitro transcription conditions, purification of recombinant proteins, establishment of cell lines that stably express epitope-tagged proteins and their purification, SDS/PAGE, Western blot, and silver staining procedures were as previously described (refs. 7, 9, 35; SI Materials and Methods).
Suppression of RPC32α and of RPC32β by siRNAs.
Transfection of Huh7 and of HeLa cells was performed with jetSI-ENDO (Eurogentec). The establishment of cell lines stably expressing siRNAs from pSuper vectors was performed as described for the stable expression of epitope-tagged proteins (9). The siRNA sequences are available on request.
Cell Culture.
The growth of HeLa cells was as previously described (7). Conditions for growth and differentiation of human H1 ES cells were as previously described (36). IMR90 cells were grown in the presence of 15% FBS. Nuclear extracts were obtained from cells in midlog phase growth at maximally 70% confluence. pBabe-ras (37), pBabe-TERT (21), and pZIPSV40 (38) were kindly provided, respectively, by Scott Lowe, Robert Weinberg, and Parmjit Jat. The gene encoding SV40 small t was amplified by PCR from pZIPSV40 and cloned into the pBabe-puro vector. Growth of cells in soft agar was as previously described (21).
Dot Blot Analyses.
The BD MTE (multiple tissue expression) Array was sequentially hybridized with radioactive RPC32β and RPC32α probes. The probes were amplified by PCR with appropriate cDNAs as template and with primers specific for RPC32α or RPC32β, respectively (sequences are available on request). PCR products (25 ng) were radio-labeled with α32P-dCTP by random priming labeling (Invitrogen Random Primer DNA Labeling System). The probes were incubated with the MTE Array for 16 h at 42 °C in 50% formamide, 5× standard saline phosphate/EDTA (SSPE) buffer, 5× Denhardt’s solution, and 0,5% SDS. Subsequent washes were performed as recommended by the supplier (Clontech).
RT Quantitative Real-Time PCR.
For each sample, 2 μg of total RNA were reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) and random hexamer primers (Roche). Reverse transcribed RNA (5 ng) was analyzed by SYBR Green PCR analysis using the Mx4000 Multiplex Quantitative PCR System and Mx4000 software, version 4.2 (Stratagene). Results were normalized as previously described (39).
Supplementary Material
Acknowledgments
Supported by a start-up grant from the Conseil Régional d’Aquitaine and the European Regional Development Fund for (to M.T.); and by grants from the Agence Nationale de la Recherche (ANR) “REGPOLSTRESS” (to M.T.), the Ligue Contre le Cancer–Comités Gironde and Dordogne (to M.T and M.P.), and the National Institutes of Health (to R.G.R.). V.H. was supported by a postdoctoral fellowship from the Conseil Régional d’Aquitaine. D.B. was supported by Grant BE3257/1-1 from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RPC32β cDNA and protein sequences reported in this article have been submitted to GenBank (accession no. DQ418461).
This article contains supporting information online at www.pnas.org/cgi/content/full/0914980107/DCSupplemental.
References
- 1.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 2.Roeder RG. Lasker Basic Medical Research Award. The eukaryotic transcriptional machinery: Complexities and mechanisms unforeseen. Nat Med. 2003;9:1239–1244. doi: 10.1038/nm938. [DOI] [PubMed] [Google Scholar]
- 3.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumay-Odelot H, et al. Identification, molecular cloning, and characterization of the sixth subunit of human transcription factor TFIIIC. J Biol Chem. 2007;282:17179–17189. doi: 10.1074/jbc.M611542200. [DOI] [PubMed] [Google Scholar]
- 7.Teichmann M, Seifart KH. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14:5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schramm L, Pendergrast PS, Sun Y, Hernandez N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000;14:2650–2663. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teichmann M, Wang Z, Roeder RG. A stable complex of a novel transcription factor IIB- related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci USA. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu P, et al. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 12.Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner M, Chaussivert N, Willis IM, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 14.Bartholomew B, Durkovich D, Kassavetis GA, Geiduschek EP. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ream TS, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sklar VE, Schwartz LB, Roeder RG. Distinct molecular structures of nuclear class I, II, and III DNA-dependent RNA polymerases. Proc Natl Acad Sci USA. 1975;72:348–352. doi: 10.1073/pnas.72.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sklar VE, Roeder RG. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase III from the mouse plasmacytoma, MOPC 315. J Biol Chem. 1976;251:1064–1073. [PubMed] [Google Scholar]
- 18.Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8:911–914. doi: 10.1038/nrc2539. [DOI] [PubMed] [Google Scholar]
- 19.Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 21.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 22.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 24.Gupte RS, Weng Y, Liu L, Lee MY. The second subunit of the replication factor C complex (RFC40) and the regulatory subunit (RIalpha) of protein kinase A form a protein complex promoting cell survival. Cell Cycle. 2005;4:323–329. [PubMed] [Google Scholar]
- 25.Fais S. Cannibalism: A way to feed on metastatic tumors. Cancer Lett. 2007;258:155–164. doi: 10.1016/j.canlet.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Bauer NN, Chen YW, Samant RS, Shevde LA, Fodstad O. Rac1 activity regulates proliferation of aggressive metastatic melanoma. Exp Cell Res. 2007;313:3832–3839. doi: 10.1016/j.yexcr.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Fujioka Y, et al. MM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J Biol Chem. 2001;276:45137–45144. doi: 10.1074/jbc.M106127200. [DOI] [PubMed] [Google Scholar]
- 28.Camacho-Vanegas O, et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–1395. doi: 10.1002/ijc.22809. [DOI] [PubMed] [Google Scholar]
- 29.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 30.Mertens C, Roeder RG. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol. 2008;28:5764–5776. doi: 10.1128/MCB.01262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollock C, Huang S. The perinucleolar compartment. J Cell Biochem. 2009;107:189–193. doi: 10.1002/jcb.22107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanduc D. Changes of tRNA population during compensatory cell proliferation: Differential expression of methionine-tRNA species. Arch Biochem Biophys. 1997;342:1–5. doi: 10.1006/abbi.1996.9869. [DOI] [PubMed] [Google Scholar]
- 33.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 34.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Luo T, Roeder RG. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J Biol Chem. 2004;279:45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- 37.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 38.Jat PS, Cepko CL, Mulligan RC, Sharp PA. Recombinant retroviruses encoding simian virus 40 large T antigen and polyomavirus large and middle T antigens. Mol Cell Biol. 1986;6:1204–1217. doi: 10.1128/mcb.6.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.