Abstract
What are the molecular events that occur when a peptide inserts across a membrane or exits from it? Using the pH-triggered insertion of the pH low insertion peptide to enable kinetic analysis, we show that insertion occurs in several steps, with rapid (0.1 sec) interfacial helix formation, followed by a much slower (100 sec) insertion pathway to give a transmembrane helix. The reverse process of unfolding and peptide exit from the bilayer core, which can be induced by a rapid rise of the pH from acidic to basic, proceeds approximately 400 times faster than folding/insertion and through different intermediate states. In the exit pathway, the helix–coil transition is initiated while the polypeptide is still inside the membrane. The peptide starts to exit when about 30% of the helix is unfolded, and continues a rapid exit as it unfolds inside the membrane. These insights may guide understanding of membrane protein folding/unfolding and the design of medically useful peptides for imaging and drug delivery.
Keywords: folding kinetics, helix formation, lipid–peptide interaction, membranes, transmembrane helix
The stability and folding of membrane proteins are strongly constrained by the formation of secondary structures in the lipid bilayer environment, driven by the hydrophobic effect and hydrogen bonding. Consideration of these factors has led to versions of a thermodynamic framework model for the folding and unfolding of helical membrane proteins (1–5). One concept is that spontaneous insertion and folding includes the formation of helical intermediates at the bilayer surface, followed by insertion, and that unfolding includes the same steps, but in reverse order. Because folding to form a helix is coupled to insertion, a significant experimental challenge in testing the concepts is to separate the process of peptide partitioning into a membrane from the folding events leading to secondary structure.
The pHLIP (pH low insertion peptide) gives an opportunity to observe membrane-associated transitions between surface coil and transmembrane helix and vice versa. At neutral and high pHs pHLIP is monomeric at concentrations < 8 - 10 μM, and equilibrates between unstructured forms in aqueous solution (state I) and bound to the surface of a lipid bilayer if one is available (state II) (6, 7) (Fig. 1A). In an acidic environment the equilibrium is shifted toward a monomeric transmembrane helical form (state III) (6, 7), and the process of insertion is accompanied by an energy release of about 1.8 kcal/mol in addition to the binding energy (6–7 kcal/mol) locating the peptide at the surface (8). The pKa of the transition from state II to state III is 6.0 (6, 8). The insertion is driven by the protonation of two Asp residues in the transmembrane region, leading to an increase of hydrophobicity that results in the folding of the peptide across a membrane (9). Increasing the pH promotes the unfolding and exit of the peptide from the core of the lipid bilayer. The insertion of pHLIP across a membrane is unidirectional: The C-terminus goes inside a cell or vesicle, and the N-terminus stays outside (7, 10). Neither partitioning of an unstructured peptide onto the bilayer surface at neutral pH, nor insertion as a transmembrane helix at low pH promotes membrane fusion nor leakage of vesicles, red blood cells, or cancer cells (7, 9, 10).
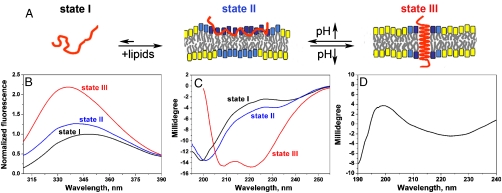
Fig. 1.
(A) A schematic representation of pHLIP in solution and interacting with a lipid bilayer at neutral and low pHs is shown. State I refers to the peptide in solution at normal and basic pHs. Upon addition of vesicles at pH 8, the unstructured peptide is adsorbed on the membrane surface (State II). A drop of pH leads to the protonation of Asp residues, increasing peptide hydrophobicity, and resulting in the insertion and formation of a transmembrane α-helix (State II). Lipids interacting with the peptide directly are marked with blue head groups, lipids influenced by the interaction but not interacting with the peptide directly have cyan head groups, and lipids that are not involved in the interaction with pHLIP have yellow head groups (8). Transitions between states can be monitored by (B) changes of fluorescence and (C) circular dichroism (CD) spectral signals. The fluorescence and CD spectra of pHLIP at pH8 (Black Lines) indicate an unstructured configuration with tryptophan residues fully exposed to solvent. Incubation of pHLIP with liposomes at pH8 (Blue Lines) induces the partial burial of tryptophan residues inside the lipid bilayer without helix formation. Decreasing the pH to 4.0 by the addition of HCl (Red Lines) induces the insertion of pHLIP and helix formation. (D) The transmembrane orientation of the helix has been confirmed by OCD (see text and ref. 11 for discussions of OCD spectroscopy).
Because pH changes can be accomplished by rapid mixing, it is possible to study the kinetics of the insertion and exit processes, and to examine the sequences of events that are involved. Here we present the results of such a kinetic investigation of pHLIP insertion/folding and exit/unfolding, which lead to new insights on the pathways.
Results
Steady-state fluorescence and CD spectroscopy are useful ways to monitor conformational and environmental changes as pHLIP experiences transitions between its three principal states (Fig. 1B and C). Spectroscopic signals in solution provide a good assessment of the peptide helicity as a function of pH and in the absence and presence of the lipid bilayer, but they do not allow a distinction between membrane-surface and transmembrane orientation of the helical form. We previously used FTIR to make this distiction, and now add to that evidence using oriented circular dichroism (OCD) measurements. It has been predicted and confirmed experimentally (11) that, for planar samples with the incident light perpendicular to the planes of the bilayers,, a membrane-surface orientation of the helix gives a CD signal similar to the α-helical CD signal in solution (similar to the red line in Fig. 1C). However, if the helix axis has a transmembrane orientation, the incident light is parallel to the axis and several transitions are degenerate giving a characteristic CD spectrum with a positive amplitude around 200 nm and a shift of the negative amplitude to longer wavelengths (225–230 nm) (Fig. 1D) (for more information about OCD spectroscopy see (11) and references therein). CD spectroscopy on oriented supported bilayers has been successfully applied to distinguish membrane-surface and transmembrane helix orientation (12). We obtained a characteristic OCD spectrum for transmembrane helix orientation using direct insertion of pHLIP peptides into supported bilayers at low pH (in contrast to earlier experiments, where bilayers were assembled with membrane peptides). The results are in excellent agreement with our previous FTIR data demonstrating transmembrane helical orientation of the pHLIP at low pH (6), and with a number of studies indicating that the C-terminus of pHLIP inserts across the lipid bilayer (7, 10). Transmembrane helix formation is accompanied by the appearance of a characteristic helical CD signal, by a shift of the fluorescence spectrum maximum and by an increase of fluorescence intensity (Fig. 1B and C). These signals are the basis of our kinetic studies.
An advantage of our system is that the initial states in the process of folding/entry or unfolding/exit are well defined. For the folding/insertion study, the initial state is the peptide bound to the surface of a membrane as an unstructured monomer; for the unfolding/exit experiments the peptide starts as a transmembrane helix. The choice of experimental conditions (peptide: POPC (1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine) molar ratio at approximately 1∶140) is based on the results of our previous studies of peptide interaction with POPC vesicles at different temperatures (8), and is chosen to avoid crowding of the peptide on the surface of vesicles (the “parking problem”). Given the high pH surface binding energy of 6–7 kcal/mol observed for temperatures in the range from 15 to 37 °C (8), the bound to free ratio is approximately 104–105, so the initial state in the insertion experiments at all temperatures is predominantly an unstructured surface bound state of the peptide. In the unfolding experiments, the initial state, at low pH, is predominantly the transmembrane helical configuration of the peptide (> 95% state III).
Transitions between states can be induced by rapidly changing pH using fast mixing of a aqueous solutions of pHLIP preincubated with POPC at pH 8.0 or pH 4.0 with diluted solutions of HCl or NaOH, respectively. Membrane-associated peptide folding/insertion and unfolding/exit were monitored by changes of CD and fluorescence signals (Fig. 2). For fast fluorescence measurements, a filter was used to capture the emission, but changes of the entire fluorescence spectrum were also recorded in a global mode with use of an emission monochromator (Fig. 2C and F). The fluorescence spectra obtained in stopped-flow mode clearly show that the increase of fluorescence is accompanied by a shift of the maximum, which indicates peptide insertion into the hydrophobic core of the membrane.
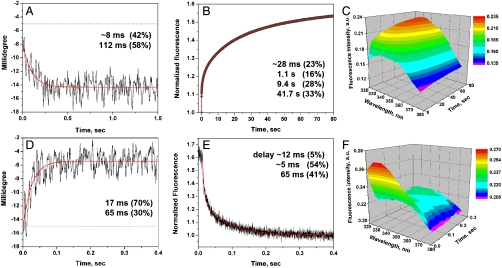
Fig. 2.
Membrane-associated pHLIP peptide folding and insertion across a POPC bilayer (A–C) and unfolding and exit (D–F) from the core of the bilayer were monitored by stopped-flow CD and fluorescence. Polypeptide folding and unfolding were induced by the rapid mixing of pHLIP-POPC solutions with diluted HCl or NaOH to give pH4 or pH8, respectively. The changes of intensity of CD (A, D) and fluorescence (B, E) were recorded at 225 nm (CD) and through a 320 nm cutoff filter using an excitation wavelength of 280 nm (fluorescence) at 25 °C. The fluorescence signal recorded over 80 sec was corrected for photobleaching. The CD and fluorescence data were fitted by kinetic models with one or three intermediate states by using Eqs. S3 and 4 (the fitting curves are red). In the case of the helix–coil transition (D) the experimental noise did not allow to reveal statistically significant differences between solutions with no (Blue Line) or a single intermediate (Red Line), thus both fitting curves and calculated parameters are shown. The changes in the entire fluorescence spectrum during folding (C) and unfolding (F) were recorded in a global mode with the emission monochromator at the excitation wavelength of 275 nm to minimize the contribution of the scattered light component at short wavelengths (spectra were corrected for the instrument sensitivity). The details of the experimental protocol can be found in SI Text.
We find that the process of helix formation, triggered by a pH drop, occurs on the bilayer surface within 1 sec (about 90% of total changes of CD signal), whereas insertion of the helix across the membrane takes approximately an additional 100 sec (Fig. 2A–C). Intriguing results were also obtained for the unfolding/exit pathway, which proceeds much faster than folding/insertion (Fig. 2D). Starting with the inserted helix, a rapid increase of pH leads to a short delay (about 12 msec), followed by a decrease of about 90% of the fluorescence signal (Fig. 2E) and the helix–coil transition, all within the next 0.3 sec, followed by the final equilibration of the peptide at the surface of the lipid bilayer until the signal reaches a stable level (10–20 sec). To estimate activation energy parameters for the process of peptide insertion into the lipid bilayer, we recorded the kinetics curves at five different temperatures (Fig. 3A). Increasing the temperature speeds up the process of insertion, presumably as a result of increased thermal fluctuations. Steady-state fluorescence spectroscopy was used to measure the total changes of the fluorescence signal at various temperatures as a result of the transition from states II to III and vice versa. The total fluorescence changes were relatively unchanged over a range from 11 to 37 °C, allowing us to conclude that the process of peptide insertion is completed to the same extent at all temperatures in our study.
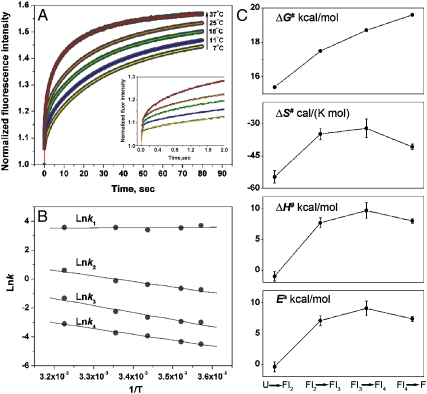
Fig. 3.
(A) The membrane-associated pHLIP insertion across the lipid bilayer of POPC vesicles was monitored by changes of the fluorescence signal through a 320 nm cutoff filter at an excitation wavelength of 280 nm and at various temperatures. The fitting curves obtained at various temperatures are color coded. The data were fitted by the kinetic model with three intermediates by using function (Eq. S3). (B) The Arrhenius plot was constructed according to the Arrhenius equation. (C) The changes of the activation energy, 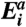 , enthalpy
, enthalpy 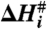 , entropy
, entropy 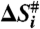 and Gibbs free energy,
and Gibbs free energy, 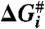 for each transition are plotted. The values of the thermodynamic activation parameters are presented in Table S2.
for each transition are plotted. The values of the thermodynamic activation parameters are presented in Table S2.
Our experimental data clearly show differences between the pHLIP folding and insertion and unfolding/exit pathways. To gain further insights, we fit the data with a sequential kinetic model. In general, parallel pathways may exist in heterogeneous systems, where peptide isomers or oligomers may present, so we selected initial experimental conditions where pHLIP is monomeric and predominantly bound to the bilayer surface in an unstructured configuration or inserted as a helix across the lipid bilayer (7, 8). Another potential source of heterogeneity might be associated with isomerization of the Pro located in the middle of the transmembrane part of the peptide. To examine this possibility, a pHLIP variant was synthesized with Pro replaced by Ala. Unfortunately, the variant shows some helical structure in solution at neutral pH (Fig. S1), and binding of the variant to a vesicle at high pH further promotes the coil–helix transition. Because replacement of Pro led to conformational changes of the peptide in solution and in peptide–bilayer interactions, we asked whether the presence of prolyl isomerase (cyclophilin A), which promotes Pro isomerization (13), might affect the folding and insertion of the nonmutated peptide. No differences were observed in membrane-associated folding in the absence and presence of prolyl isomerase at various temperatures. Thus, we conclude that the proline is simply acting as a helix breaker in solution and on the bilayer surface.
We found that the fluorescence changes over the entire process of pH-triggered insertion from State II to State III (Figs. 2B and 3A) can be well described by a pseudo-first order kinetics model with 4 consecutive steps involving State II (I1) , three intermediates, and State III (I5):
All attempts to fit the fluorescence curves obtained at various temperatures using < 4 exponential functions led to significant discrepancies between experimental and theoretical curves, especially within the first 2 sec. Our experimental data also imply a fast component at the beginning of the process of insertion (Fig. 3A Insert), which must be included in the fit. The mathematical complexity of the fit increases dramatically with each additional step that is included in the model. To solve the systems of differential equations and find analytical solutions, the simplified approach of sequential reactions in one direction was implemented (SI Text). To establish the best fit of fluorescence and CD curves, kinetic models with zero, one, two, or three intermediate states were tested, and the models chosen were those giving an adequate fit to the data using the minimum number of states. Kinetic parameters obtained from the fitting of fluorescence kinetic data recorded at various temperatures are presented in Table S1. Kinetic measurements at various temperatures allow thermodynamic activation parameters to be calculated (Table S2), and their changes during the transition from one state to another are presented in Fig. 3C. The coil–helix transition (Fig. 1A) was well fit by a model with a single intermediate (2 component solution), including a fast component that was established by recording initial and final signals without triggering (see Methods for more details about the experimental set up of CD stopped-flow measurements).
The CD and fluorescence decays reporting the helix-coil exit pathway (Fig. 2D and E) were well described by a kinetic model with two intermediates, where the first intermediate is formed within the first 12 msec and the transition to it is not accompanied by any significant change of fluorescence signal (delay of intensity changes on Fig. 2E). The CD signal was noisy and could be fit with models containing no intermediates (Fig. 2D, Blue Line) or one intermediate (Fig. 2D, Red Line). The model with one intermediate gave a slightly better solution, and it was chosen because the rates are in very good agreement with rates obtained from the fluorescence decay analysis.
Discussion
The pH-triggered bilayer insertion and exit of the pHLIP allow kinetic studies of these processes to be performed. Further, the folding from an unstructured state at the bilayer surface to the transmembrane helical configuration allows secondary structure formation to be followed. Fluorescence and CD spectroscopy have proven useful for monitoring pHLIP folding/entry across and unfolding/exit from the bilayer, and we used them to follow the pathways in real time.
Earlier pH-jump experiments monitored by changes of pHLIP fluorescence were reported by Hunt et al. in 1997 (6), but these did not include measures of secondary structure. More recently, a fluorescence kinetic study was reported by Tang and Gai (14), who followed peptide binding to the surface and insertion into a bilayer. As in the Hunt study, the CD signal was not measured and only the intensity of tryptophan fluorescence was recorded. Moreover, the kinetic process was monitored for only the first 5 sec out of 100 sec, so > 50% of the fluorescence signal changes were missed. We have expanded on these earlier efforts using stopped-flow measurements of both the fluorescence parameters (intensity and wavelength position of maxima of spectra) and the CD signal. The results clearly demonstrate differences in the processes of (i) coil–helix transition and peptide insertion into membrane, and (ii) peptide folding/entry and unfolding/exit from the membrane. Our main findings are that the entry pathway begins with rapid helix formation, followed by slow transbilayer insertion, and that the exit pathway is distinctly different.
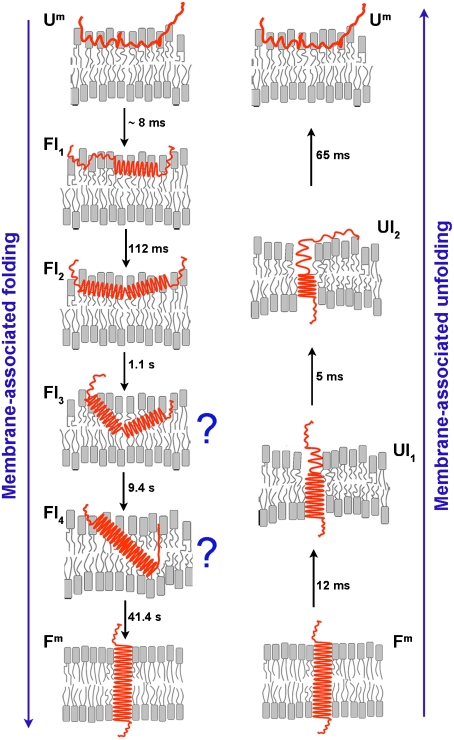
In an attempt to gain more insights on the folding and unfolding pathways we introduced a kinetic model to fit the experimental curves, resulting in the mechanistic model shown in Fig. 4. Some features of the model are viewed with confidence whereas others are much less certain, and we can only formulate educated guesses as to the nature of several intermediate states. An important caveat is that the model assumes single pathways of insertion and exit. Starting with the surface bound peptide of State II, and following the drop in pH, we see a rapid formation of helix, in two steps (folding intermediates 1 and 2, forming at 8 msec and 112 msec), each producing about half of the total helix. The helix might be a single straight helix, or it might consist of several short helices with breaks in between (these possibilities are experimentally indistinguishable). This part of the interpretation is secure: Regardless of the kinetic model, helix formation clearly is faster than most of the fluorescence changes. It is possible that slight sinking of a polypeptide during helix formation or helix rotation at the interface account for the fast first (23%) fluorescence change. Following the rapid helix formation, the rest of the insertion process is reported by fluorescence, and is about 1,500 times slower, describable as several kinetically distinct steps over approximately the next 100 sec.
Fig. 4.
A schematic interpretation of the membrane-associated folding and unfolding events suggested by the kinetic data. The pHLIP starts as a surface-bound coil at pH8. A rapid drop of pH to 4 causes a rapid formation of helical structure on the surface, followed by a much slower insertion of the helix across the bilayer, with several kinetically distinct intermediates for which speculative models are shown. A sudden elevation of the pH from 4 to 8 results in the rapid unfolding and exit of the peptide via a different pathway with different intermediates (see discussion in the text).
We cannot be certain of the exact nature of the kinetic intermediates during insertion, but we suggest a few thoughts as working ideas. From the surface helix formation to the fully inserted transmembrane helix, two intermediate states (FI3 and FI4) followed from our analysis, and are the rate limiting steps in the insertion process. We assume that at least one of these intermediates is related to the translocation of the polar C-terminus of pHLIP across the bilayer. It may be that the fourth intermediate is a transbilayer inserted form with the C-terminus across the bilayer, or that the C-terminus crosses the bilayer during the last transition. We have no direct evidence yet to choose one of these, but we know that the C-terminus must cross the bilayer at some point. Each step from the unfolded to the folded state is associated with a decrease of entropy (decrease of disorder) that reflects the process of folding (ordering). It is possible that both the peptide and the lipids undergo a disorder–order transition.
In contrast with previously proposed theoretical models suggesting that entry and exit pathways mirror each other, our data reveal that the unfolding and exit of the peptide in response to a sudden pH increase occurs within 100 msec, orders of magnitude faster than insertion. Assuming that the increase of pH leads to the deprotonation of the Asp residues [perhaps involving proton transfer from Asp86 to Arg83, which is known to happen in bacteriorhodopsin, from which the pHLIP is derived (15, 16)], followed by the helix–coil transition and a rapid exit of the polypeptide from the bilayer interior. Surprisingly, about 30% of the helix–coil transition is completed within approximately 12 msec, while practically no change of the fluorescence signal is observed, indicating that the polypeptide remains in the membrane interior as an unfolding intermediate (UI1). Thus, partial unfolding of the TM helix, involving about 5–6 residues out of 20–22 residues, might occur inside the membrane and the peptide subsequently moves out of the bilayer interior. Another possibility is unfolding of part of the helix, followed by immediate exit of the polypeptide from the membrane, which leads to a thinning of the lipid bilayer around the shortened helical part of the peptide. This suggestion is based on recent findings that the lipid bilayer can accommodate short helices with a minimal energetic cost (17, 18). Further propagation of the helix-coil transition is accompanied by a rapid exit of part of the polypeptide within the next 5 msec (UI2). The remaining 30% of the membrane-embedded helical structure unfolds and exits within 65 msec (including the C-terminus), and equilibrium is established between pHLIP in its unfolded membrane-bound (Um) and soluble forms within the final 10–20 sec. Unfolding experiments carried out at 7 °C showed that the peptide exit also occurs in two steps with characteristic times of 17.6 and 152 msec. Because the C-terminus, which must cross the bilayer, has several carboxyl groups, an interesting possibility is that protonation and deprotonation steps might account for steps in the entry and exit pathways.
Several authors have reported peptide coil–helix transitions induced by binding to bilayers and the existence of interfacial folded intermediates, mostly using fluorescence approaches (19–24). Although interesting, the results of molecular dynamics simulations of membrane-associated folding and insertion of various peptides are not definitive (25–28). The pHLIP presents a case where the binding of the pHLIP to the membrane itself does not promote folding, so a stable, unstructured state can be established at a bilayer surface. The protonation of charged Asp residues, which leads to increased hydrophobicity of the peptide, induces a coil–helix transition and peptide insertion. We show that the formation of an interfacial helical intermediate is a step in the pathway of pHLIP insertion into a membrane. Helix formation reduces the free-energy penalty associated with the partition of the peptide backbone into the low dielectric environment of the bilayer, despite the fact that the coil–helix transition is associated with a loss of entropy. Helix insertion, most probably, is accompanied by a significant perturbation of lipids beyond that of the surface bound state, although the lipid perturbation is reduced in the overall process. The insertion of pHLIP is slow, because the peptide is initially located on the outer leaflet of the bilayer, and it takes time for the polar C-terminus to cross the membrane and to reorganize lipids around the transmembrane helix. In contrast to the insertion, unfolding and exit occur much faster, perhaps because the peptide can be conceptualized as occupying a small channel across the lipid, so that the peptide can quickly exit without the significant lipid reorganization needed for the intermediate states of insertion. Such a channel would close immediately after exit of the peptide.
We have demonstrated in vivo that pHLIP can target diseased tissues with elevated levels of extracellular acidity, such as tumors (9, 29, 30) and that the energy of the insertion events can be used for the selective translocation of polar cell-impermeable cargo molecules across the membranes of liposomes and cells (10, 30). Our kinetic data provide insights on the mechanisms of membrane-associated polypeptide folding and unfolding, and may be useful in improving the design of peptide based transmembrane delivery agents.
Methods
pHLIP Peptide.
The pHLIP sequences AEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT and its Pro to Ala variant AEQNPIYWARYADWLFTTALLLLDLALLVDADEGT were prepared by solid-phase peptide synthesis using Fmoc (9-fluorenylmethyloxycarbonyl) chemistry and purified by reverse phase chromatography at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. For use of the peptide, the lyophilized powder is dissolved in a solution containing 3 M urea and transferred to 10 mM phosphate buffer, pH8.0 using a G-10 size-exclusion spin column. The concentration of the peptide was determined by absorbance (ε280 = 13,940 M-1 cm-1).
Vesicle Preparations.
Large unilamellar vesicles were prepared by extrusion. 1 ml of 25 mg POPC (1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine, Avanti Polar Lipids, Inc.) in chloroform was desolvated on a rotary evaporator and dried under vacuum for several hours. The phospholipid film was rehydrated in 10 mM phosphate buffer, pH 8.0, vortexed for 2 h, and repeatedly extruded using a 50 nm membrane pore size.
Steady-State Measurements.
Steady-state fluorescence measurements were carried out on a PC1 spectrofluorometer (ISS, Inc.) under temperature control. Peptide fluorescence spectra were recorded from 310 nm to 400 nm with the spectral widths of excitation and emission slits set at 2–4 nm and 2 nm, respectively, using excitation wavelengths of 275, 280, and 295 nm. Changes of the fluorescence signal in the process of transition from one state to another are independent of the excitation wavelength, indicating that the spectral contribution of Tyr fluorescence is minimal. The polarizers in the excitation and emission paths were set at the “magic” angle (54.7° from the vertical orientation) and vertically (0°), respectively, to reduce Wood’s anomalies from the reflecting holographic grating. Steady-state CD and OCD measurements were carried out on a MOS-450 spectrometer (BioLogic, Inc.) under temperature control. All measurements were performed at 25 °C unless otherwise indicated. The detailed description of the OCD measurements including preparation of samples can be found in SI Text.
Stopped-Flow Measurements.
Stopped-flow fluorescence and CD measurements were carried out on a SFM-300 mixing apparatus connected to a MOS-450 spectrometer (BioLogic, Inc.) under temperature control. The FC-20 and TS-100/15 observation cuvettes were used for the fluorescence and CD measurements, respectively. All solutions were degassed several minutes under vacuum before loading into the syringes to minimize air bubbles. pHLIP (7 μM) was preincubated with POPC (1 mM) at pH8.0 to reach binding equilibrium and folding/insertion was induced by fast mixing (5.7 ms dead time) of equal volumes of pHLIP-POPC pH8 and appropriately diluted HCl, to obtain a drop of pH from 8 to 4. In the unfolding experiments, pHLIP was preincubated with POPC at pH8.0, then HCl was added to lower the pH to 4.0, and time was allowed for equilibration (minutes). Unfolding was induced by mixing equal volumes of pHLIP-POPC pH4 and NaOH diluted to increase the pH from 4 to 8. In majority of cases, samples were collected after the shots and steady-state fluorescence were recorded on a PC1 spectrofluorometer. In the case of experiments in the presence of prolyl isomerase (cyclophilin A), which catalyses the isomerization of Pro residues in peptides, pHLIP was preincubated with the cyclophilin A (Sigma) at a ratio of 1∶100 for 2 h before mixing with liposomes, followed by fluorescence (at various temperatures) and CD kinetics measurements. Additional information on the stopped-flow measurements can be found in SI Text.
Data Analysis.
The kinetic equations were solved by integration (Eq. S1) in Mathematica 5 (Wolfram Research). Nonlinear least squares curve fitting procedures were carried out in Origin 7 and MatLab. The goodness-of-fit was assessed by the adjusted R-square statistics (adjR2) and rms error according to the standard formula. The details of the kinetic model used in the study and description of the calculated thermodynamic activation parameters can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank all members of the Andreev, Engelman, and Reshetnyak laboratories for fruitful discussions. This work was supported in part by National Institutes of Health, National Cancer Institute R01 133890.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914330107/DCSupplemental.
References
- 1.Jacobs RE, White SH. The nature of the hydrophobic binding of small peptides at the bilayer interface: Implications for the insertion of transbilayer helices. Biochemistry. 1989;28:3421–3437. doi: 10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- 2.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: The helical hairpin hypothesis. Cell. 1981:41l–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 3.Popot JL, Engelman DM. Membrane protein folding and oligomerization: The two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 4.White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 5.Engelman DM, et al. Membrane protein folding: Beyond the two stage model. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 6.Hunt JF, Rath P, Rothschild KJ, Engelman DM. Spontaneous pH-dependent membrane insertion of a transbilayer alpha-helix. Biochemistry. 1997;36:15177–151792. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 7.Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: In solution attached to and inserted across lipid bilayers. Biophys J. 2007;93:2363–2672. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reshetnyak YK, Andreev OA, Segala M, Markin V, Engelman DM. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc Natl Acad Sci USA. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreev OA, et al. Mechanism and uses of a peptide that targets tumors and other acidic tissue in vivo. Proc Natl Acad Sci USA. 2007;104:893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci USA. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Huang HW, Olah GA. Method of oriented circular dichroism. Biophys J. 1990;57:797–806. doi: 10.1016/S0006-3495(90)82599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merzlyakov M, Li E, Hristova K. Directed assembly of surface-supported bilayers with transmembrane helices. Langmuir. 2006;22:1247–1253. doi: 10.1021/la051933h. [DOI] [PubMed] [Google Scholar]
- 13.Reader JS, et al. A partially folded intermediate species of the β-sheet protein apo-pseudoazurin is trapped during proline-limited folding. Protein Sci. 2001;10:1216–1224. doi: 10.1110/ps.52801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang J, Gai F. Dissecting the membrane binding and insertion kinetics of a pHLIP peptide. Biochemistry. 2008;47:8250–8252. doi: 10.1021/bi801103x. [DOI] [PubMed] [Google Scholar]
- 15.Balashov SP, et al. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation proton release and the photochemical cycle. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 16.Balashov SP, et al. The two pKa’s of aspartate-85 and control of thermal isomerization and proton release in the arginine-82 to lysine mutant of bacteriorhodopsin. Biochemistry. 1995;34:8820–8834. doi: 10.1021/bi00027a034. [DOI] [PubMed] [Google Scholar]
- 17.London E, Shahidullah K. Transmembrane vs non-transmembrane hydrophobic helix topography in model and natural membranes. Curr Opin Struct Biol. 2009 doi: 10.1016/j.sbi.2009.07.007. (in press) [DOI] [PubMed] [Google Scholar]
- 18.Andersen OS, Bruno MJ, Sun H, Koeppe RE., II Single-molecule methods for monitoring changes in bilayer elastic properties. Methods Mol Biol. 2007;400:543–570. doi: 10.1007/978-1-59745-519-0_37. [DOI] [PubMed] [Google Scholar]
- 19.Constantinescu I, Lafleur M. Influence of the lipid composition on the kinetics of concerted insertion and folding of melittin in bilayers. Biochim Biophys Acta. 2004;1667:26–37. doi: 10.1016/j.bbamem.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Ladokhin AS, White SH. Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry. 2004;43:5782–5791. doi: 10.1021/bi0361259. [DOI] [PubMed] [Google Scholar]
- 21.Tucker MJ, Tang J, Gai F. Probing the kinetics of membrane-mediated helix folding. J Phys Chem B. 2006;110:8105–8109. doi: 10.1021/jp060900n. [DOI] [PubMed] [Google Scholar]
- 22.Tang J, Signarvic RS, DeGrado WF, Gai F. Role of helix nucleation in the kinetics of binding of mastoparan X to phospholipid bilayers. Biochemistry. 2007;46:13856–13863. doi: 10.1021/bi7018404. [DOI] [PubMed] [Google Scholar]
- 23.Zakharov SD, et al. Membrane-bound state of the colicin E1 channel domain as an extended two-dimensional helical array. Proc Natl Acad Sci USA. 1998;95:4282–4287. doi: 10.1073/pnas.95.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyrychenko A, Posokhov YO, Rodnin MV, Ladokhin AS. Kinetic intermediate reveals staggered pH-dependent transitions along the membrane insertion pathway of the diphtheria toxin T-domain. Biochemistry. 2009;48:7584–7594. doi: 10.1021/bi9009264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milik M, Skolnick J. Insertion of peptide chains into lipid membranes: An off-lattice Monte Carlo dynamics model. Proteins. 1993;15:10–25. doi: 10.1002/prot.340150104. [DOI] [PubMed] [Google Scholar]
- 26.Im W, Brooks CL. Interfacial folding and membrane insertion of designed peptides studied by molecular dynamics simulations. Proc Natl Acad Sci USA. 2005;102:6771–6776. doi: 10.1073/pnas.0408135102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nymeyer H, Woolf TB, Garcia AE. Folding is not required for bilayer insertion: Replica exchange simulations of an alpha-helical peptide with an explicit lipid bilayer. Proteins: Struct, Funct, Bioinf. 2005;59:783–790. doi: 10.1002/prot.20460. [DOI] [PubMed] [Google Scholar]
- 28.Ulmschneider MB, Ulmschneider JP. Membrane adsorption folding insertion and translocation of synthetic trans-membrane peptides. Mol Membr Biol. 2008;25:245–257. doi: 10.1080/09687680802020313. [DOI] [PubMed] [Google Scholar]
- 29.Vavere AL, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreev OA, Engelman DM, Reshetnyak YK. Targeting acidic diseased tissue: New technology based on use of the pH (Low) Insertion Peptide (pHLIP) Chim Oggi. 2009;27:34–37. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.