Abstract
In an effort to add to the versatility of three-dimensional scaffolds for tissue engineering applications, recent experimental designs are incorporating biological molecules such as plasmids and proteins within the scaffold structure. Such scaffolds act as reservoirs for the biological molecules of interest while regulating their release over various durations of time. Here, we describe the use of coaxial electrospinning as a means for the fabrication of fiber mesh scaffolds and the encapsulation and subsequent release of a non-viral gene delivery vector over a period of up to 60 days. Various fiber mesh scaffolds containing plasmid DNA (pDNA) within the core and the non-viral gene delivery vector poly(ethylenimine)-hyaluronic acid (PEI-HA) within the sheath of coaxial fibers were fabricated based on a fractional factorial design that investigated the effects of four processing parameters at two levels. Poly(ε-caprolactone) sheath polymer concentration, poly(ethylene glycol) core polymer molecular weight and concentration, and the concentration of pDNA were investigated for their effects on average fiber diameter, release kinetics of PEI-HA, and transfection efficiency. It was determined that increasing the values of each of the investigated parameters caused an increase in the average diameter of the fibers. The release kinetics of PEI-HA from the fibers were affected by the loading concentration of pDNA (with PEI-HA concentration adjusted accordingly to maintain a constant nitrogen to phosphorous (N:P) ratio within the complexes). Two-dimensional cell culture experiments with model fibroblast-like cells demonstrated that complexes of pDNA with PEI-HA released from fiber mesh scaffolds could successfully transfect cells and induce expression of enhanced green fluorescent protein (EGFP). Peak EGFP expression varied with the investigated processing parameters, and the average transfection observed was a function of poly(ethylene glycol) (core) molecular weight and concentration. Furthermore, fibroblast-like cells seeded directly onto coaxial fiber mesh scaffolds containing PEI-HA and pDNA showed EGFP expression over 60 days, which was significantly greater than the EGFP expression observed with scaffolds containing pDNA alone. Hence, variable transfection activity can be achieved over extended periods of time upon release of pDNA and non-viral gene delivery vectors from electrospun coaxial fiber mesh scaffolds, with release and subsequent transfection controlled by tunable coaxial fiber mesh fabrication parameters.
1. Introduction
Traditionally, the role of tissue engineering scaffolds has been to provide mechanical support to damaged or excised tissue while facilitating the infiltration and attachment of cells. However, the development of novel processing techniques has significantly broadened their scope by allowing the incorporation and subsequent release of bioactive molecules, thus transforming the scaffolds into multifunctional bioactive factor delivery units. The scaffold can operate as a reservoir for biological molecules, the release of which can be modulated by controlling the scaffold processing parameters. Such scaffolds have shown sustained release of a variety of proteins [1] as well as plasmids [2, 3]. It has become increasingly feasible to deliver plasmid DNA (pDNA) to cells so as to facilitate in situ production of the encoded growth factors, signaling molecules and insoluble bioactive molecules of interest. This approach carries a significant advantage over the direct delivery of these biological agents, as intracellular expression of the delivered plasmids can be sustained over a period of days, thus mitigating the drawbacks of limited bioactivity associated with short half-lives of most biological factors. Furthermore, concerns related to gene delivery, such as low transfection efficiencies and the general requirement of high plasmid doses are gradually being mitigated with the development of new non-viral vectors and improved delivery strategies.
Tissue engineering scaffolds that entrap and release plasmid DNA have been adapted by various groups [2, 4–8], and such scaffolds are popularly referred to as gene activated matrices (GAMs). The release of pDNA encoding a protein from three-dimensional biodegradable scaffolds has resulted in greater expression of the encoded protein than a similar amount of pDNA delivered to two-dimensional cell culture systems [9–11]. The enhancement in expression has been attributed to the close proximity of the cells to the gene delivery reservoir, as well as the sustained release of the plasmid over time [10, 12]. Scaffolds similar in concept to GAMs created by gas foaming [2], emulsion [13, 14], or electrospinning [11] have all been shown to successfully incorporate pDNA, release it over an extended period of days to weeks, and preserve plasmid bioactivity over the duration of release. Successful expression of pDNA released from biodegradable scaffolds has been demonstrated using plasmids encoding reporter proteins such as luciferase [9, 15, 16], beta-galactosidase [2, 16, 17] and enhanced green fluorescent protein [10, 13] as well as functional genes such as parathyroid horomone-1 [18], vascular endothelial growth factor [19], bone morphogenetic protein-2 [20, 21] and bone morphogenetic protein-4 [22].
Coaxial electrospinning has previously not been employed to produce scaffolds for gene delivery in the context of tissue engineering. Coaxial fiber mesh scaffolds have a sheath/core fiber morphology where individual fibers can be fabricated from two separate immiscible polymer solutions, which allows for physical separation of aqueous-based biological molecules from the organic solvents essential for scaffold fabrication and minimizes the interaction between the two to the order of microseconds [23, 24]. Furthermore, electrospinning allows for the fabrication of multi-layered scaffolds, as demonstrated by previous experiments in our laboratory [25], where each layer can potentially incorporate and release a plasmid encoding a unique protein. Hence it is essential to determine the processing parameters that control the incorporation of pDNA into and release kinetics from such coaxial electrospun fiber meshes.
In this study we have incorporated a non-viral gene delivery vector previously developed in our laboratory, a hyaluronic acid (HA) derivative of poly(ethylenimine) (PEI) (PEI-HA) into non-woven coaxial electrospun fiber meshes. We incorporated pDNA into an aqueous poly(ethylene glycol) (PEG) solution to fabricate the core section of the fiber and the gene delivery vector PEI-HA into an organic sheath polymer solution of poly(ε-caprolactone) (PCL) in chloroform and methanol. The coaxial electrospinning method not only minimized the interaction of the plasmid with the organic solvents, but also allowed the integration of pDNA without the need to process it through methods such as lyophilization, which in some cases has been shown to reduce the plasmid bioactivity [26, 27]. Furthermore, the volatile sheath polymer solution facilitated the processing and solidification of the fibers into non-woven fiber meshes. The plasmid was incorporated within the core of the fibers and the gene delivery vector was contained within the sheath. The hypothesis in generating these scaffolds was that, as the electronegative plasmids diffused out of the fiber cores, they would complex with the positively charged PEI-HA released from the fiber sheath and transfect cells present on the fiber surface.
To this end, we formulated a fractional factorial design to investigate the effects of various processing parameters, including (a) core polymer concentration and (b) molecular weight, (c) sheath polymer concentration, and (d) pDNA concentration, on fiber diameter distribution, PEI-HA release kinetics, and transfection efficiency. The gene delivery vector was tagged with a fluorescent molecule, rhodamine-B-isothiocyanate, to monitor its release, whereas the plasmid release was indirectly monitored through its reporter protein (EGFP) activity.
2. Materials and Methods
2.1 Materials
Chemicals for PEI-HA synthesis, namely, sodium borate, PEI (Mw = 25kDa) and sodium cyanoborohydrate, were purchased from Sigma-Aldrich (St. Louis, MO). Rhodamine-B-isothiocyanate for fluorescence tagging of PEI-HA was also purchased from Sigma. Sodium hyaluronate (Mw= 2.3 kDa) was generously provided by Genzyme Corp. (Cambridge, MA). Solvents used for electrospinning, namely, chloroform and methanol, were purchased at ACS grade from Fisher Scientific (Pittsburgh, PA). Chemicals used for tissue culture purposes such as Phosphate Buffered Saline (PBS), Dulbecco’s Modified Eagle Medium (DMEM) with high glucose, Minimum Essential Medium (MEM) Amino Acid Solution (50X), MEM Non Essential Amino Acids (NEAA) (100X), L-Glutamine (200 mM), MEM Vitamin solution (100X) and sodium pyruvate (100 mM) were purchased from Gibco (Carlsbad, CA). Plasmid DNA encoding enhanced green fluorescent protein (EGFP) with a cytomegalovirus (CMV) promoter (pCMV-EGFP) was generously donated by Dr. Michael Barry from Mayo Clinic (Rochester, MN). Qiagen Plasmid Giga Kits for pCMV-EGFP amplification and purification were purchased from Qiagen (Valencia, CA).
2.2 Synthesis of Rhodamine Tagged PEI-HA
The synthesis of PEI-HA has previously been described by our laboratory [28]. For the current study, the published protocol was adapted for increased quantities of reactants. 500 mg of PEI and 1 g of HA were mixed in a 3-neck flask in the presence of 0.2 M sodium borate buffer (pH 8.5) maintained at 40°C. The solution was stirred continuously after addition of 0.27 g of sodium cyanoborohydrate on initiation of the reaction and an additional 0.20 g 30 hrs after the initiation of the reaction. The reaction was carried out over 120 hrs, after which the product was dialyzed against 0.02 M of sodium borate buffer, and the dialysis solution was gradually transitioned to water. The product was lyophilized, and the chemical structure was confirmed with 1H NMR as previously described. Based on NMR analysis, 11.67 ± 0.45% of the amine groups within PEI were chemically modified due to reductive amination with HA.
Lyophilized PEI-HA was dissolved in 0.2 M sodium bicarbonate buffer at 10 mg/mL concentration at pH 9.0. 10 mg of rhodamine-B-isothiocyanate was dissolved in 1 mL dimethyl sulfoxide (DMSO), and the mixture was added to 50 mL of the PEI-HA solution. The reaction solution was placed on a rotating table for 2 hrs, after which the solutions were dialyzed initially with 0.1 M sodium bicarbonate solution. The dialysis process was repeated until the dialysate showed a steady fluorescence reading during three consecutive dialysis cycles. The dialysis solution was then switched to water for three dialysis cycles. The product obtained at the end of dialysis, rhodamine-PEI-HA (r-PEI-HA), was lyophilized and stored at 4°C.
2.3 Plasmid Amplification
Plasmid DNA encoding EGFP with a CMV promoter was amplified as described previously [28]. Briefly, pCMV-EGFP (4.7 kb) was amplified in E. Coli bacterial cultures. Plasmids were extracted and purified using standard protocols with the Qiagen Plasmid Giga Kit. The total plasmid yield was determined from the UV absorbance at a wavelength of 260 nm (A260). The plasmid was dissolved in Millipore water at 5 mg/mL and stored at −20°C. The ratio of A260/A280 was determined to be between 1.8 and 2.0 to assess purity of the plasmid produced.
2.4 Fabrication of Coaxial Electrospun Scaffolds
2.4.1 Experimental Design
A two-level fractional factorial design with 4 parameters was formulated to evaluate the release and the related transfection efficiency of r-PEI-HA and pDNA from electrospun coaxial fiber mesh scaffolds. The parameters tested were (1) PCL concentration (Conc.), (2) PEG molecular weight (MW), (3) PEG Conc., and (4) pDNA Conc. within the core fiber. The concentration of r-PEI-HA was also modified with changes in pDNA concentration so as to maintain the same N:P ratio across all formulations. Table 1 summarizes the combination of parameters examined in this study. The range of values used with each of these parameters was predetermined by an elimination process, where a variety of combinations of PEG and PCL polymer concentrations and molecular weights were electrospun together, to determine the values that produced a stable Taylor cone over a period of several minutes. r-PEI-HA release from the scaffolds as well as transfection ability of the released supernatant containing r-PEI-HA and pDNA were studied over a period of 60 days.
Table 1.
| Parameters | PCL Concentration | PEG Molecular Weight | PEG Concentration | pDNA Concentration |
|---|---|---|---|---|
| Group 1 | −1 | −1 | −1 | −1 |
| Group 2 | −1 | −1 | 1 | 1 |
| Group 3 | −1 | 1 | −1 | 1 |
| Group 4 | −1 | 1 | 1 | −1 |
| Group 5 | 1 | −1 | −1 | 1 |
| Group 6 | 1 | −1 | 1 | −1 |
| Group 7 | 1 | 1 | −1 | −1 |
| Group 8 | 1 | 1 | 1 | 1 |
| High (+1) | 16 wt% | 4.6 kDa | 300 mg/mL | 4 mg/mL |
| Low (−1) | 14 wt% | 3.3 kDa | 150 mg/mL | 2 mg/mL |
2.4.2 Coaxial Electrospinning Setup
The setup for coaxial electrospinning was developed and described in detail previously [29]. The syringes containing PCL/r-PEI-HA in the setup for the present study were covered to prevent fluorescence bleaching.
2.4.3 Fabrication of Coaxial Electrospun Scaffolds
PCL sheath and PEG core solutions were made according to parameters described in Table 1. PCL was dissolved in 2:1 chloroform: methanol solution (v/v), whereas PEG solution was made in 150 mM of NaCl solution prepared with Millipore water. r-PEI-HA was ground into a fine powder in the dark using a mortar and pestle and added to the PCL solutions. The amount of r-PEI-HA to be incorporated was calculated at a 7.5:1 N:P ratio of vector polymer to pDNA, assuming that the electrospinning process would be carried out for 50 min with each scaffold type. Hence,
The r-PEI-HA/PCL solution was protected from light with an aluminum foil wrap, vortexed thoroughly and left on an orbital shaker overnight. Similarly, aqueous PEG solution was added drop-wise to pDNA solution to a final pDNA concentration of either 4 mg/mL or 2 mg/mL, based on the spinning parameters. An outer (sheath) flow rate of 8 mL/hr and an inner (core) flow rate of 0.1 mL/hr were used for the fabrication of the scaffolds for a period of 50 min.
2.5 Scanning Electron Microscopy Analysis of Fiber Diameters
Three scaffolds of 1 mm diameter each were punched out from the coaxial fiber mesh mats and mounted on a steel stage above insulating tape. Samples were sputter coated with gold for 1 min at 100 mA, after which the scaffolds were observed with SEM (FEI Quanta 400, Hillsboro, OR). A total of 90 fibers were measured from 3 scaffolds from each group, as described previously by our laboratory [25, 29].
2.6 Release and Quantification
2.6.1Release and Quantification of r-PEI-HA
Circular scaffolds of 10 mm diameter were punched out of each fiber mat and weighed. Amount of r-PEI-HA and pDNA within each scaffold of 10 mm diameter was estimated from the theoretical amount of pDNA incorporated within the fiber mat and the fractional weight of the scaffold compared to that of the fiber mat.
Amount of r-PEI-HA per scaffold was further determined with the assumption that the N:P ratio between r-PEI-HA and pDNA was maintained at 7.5:1. Scaffolds of weights between 10.5 and 12.8 mg were selected from each fiber mat group and placed in 5 mL polypropylene tubes covered with aluminum foil. The scaffolds were sterilized with ethylene oxide over a period of 14 hrs. The samples were then individually submerged in 1 mL of PBS and placed on a shaker table rotating at 115 rpm at 37 °C. The PBS supernatant was collected at predetermined time points and replaced with fresh PBS. The collected supernatant was lyophilized over 48 hrs, and the residue was resuspended in either 1.0 mL or 0.5 mL of PBS, determined by sampling the amount of fluorescence present in the sample. 100 μL of the resuspended solution was added into opaque 96-well plates and fluorescence was measured using a plate reader, Spectra Max M2 (Molecular Devices, Downingtown, PA) at Ab/Em 555/592 nm, with the emission cut-off wavelength of 590 nm. These wavelengths were determined as optimal based on a spectral frequency sweep of wavelengths ranging from 350 nm to 700 nm. Fluorescence values were compared to a standard curve generated using known concentrations of r-PEI-HA/DNA complexes of 7.5:1 N:P ratio to determine the concentration of r-PEI-HA/DNA complexes in solution, with the assumption that there was no significant difference between the fluoresce emitted by r-PEI-HA and r-PEI-HA/pDNA complexes of different N:P ratios. Data obtained was analyzed using Softmax Pro (v 4.6, Molecular Devices, Downingtown, PA).
2.6.2 Release and Preparation of Solutions for Transfection
To assess the transfection ability of the pDNA in the release solution of the scaffolds, additional 10 mm diameter scaffolds of weights between 10.5 and 12.8 mg were placed individually in 5 mL polypropylene tubes. The samples were similarly submerged in 1 mL of PBS and placed on a shaker table at 115 rpm at 37°C. The PBS supernatant was obtained at predetermined time points and replaced with fresh PBS. The supernatant was lyophilized over 48 hrs and resuspended in transfection medium composed of MEM Amino Acid solution, MEM NEAA and MEM Vitamins diluted to 1X concentration, 1000 mg/L of glucose, sodium pyruvate (final concentration 1 mM) and L-glutamine (final concentration 2 mM), approximately 1 hr before transfection. The resuspended solutions were centrifuged at 2000 rpm for 10 min, and the samples were allowed to stand for 50 min.
2.7 Cell Line and Cell Culture
Fischer rat fibroblast cell line (CRL 1764) was obtained from American Type Culture Collection (Manassas, VA). Cells were expanded in T75 flasks with complete medium, (DMEM supplemented with 10 vol. % FBS, 100 μg/mL penicillin, 100 U/mL streptomycin, and 0.5 μg/mL amphotericin B) and cultured at 37°C in 5% CO2 and 95% humidity.
2.7.1 Cell Transfection and Reporter Gene Expression
After expansion of the cells in T75 flasks, cells were trypsinized with 0.025% trypsin ethylenediaminetetraacetic acid (EDTA), centrifuged, quantified, and replated onto 6-well cell culture plates at 250,000 cells per well. The cells were suspended in 2 mL of tissue culture media overnight to facilitate attachment, after which the media was removed and replaced with the transfection solutions described above. The plates were covered in foil and allowed to sit in the incubator overnight. The next morning 1.5 mL of cell culture medium was added to the cells, and the cells were incubated at 37°C for 48 hrs. To assess transfection by flow cytometry, the cells on the 6-well plates were treated with 0.5% trypsin EDTA for 5 min, after which the cells were collected in their respective 5 mL tubes and centrifuged at 2000 rpm for 10 min. The supernatant was decanted, and the cells were washed with PBS. The PBS was further replaced with 2% chilled formaldehyde solution in PBS, and the cells were allowed to fix on ice for 1 hr. The cell solution was further centrifuged (2000 rpm, 10 min), and the formaldehyde was replaced with PBS solution. Cell fluorescence was quantified using a Becton Dickenson FACS Scan instrument (BD Biosciences, San Jose, CA) at high flow and CellQuest Pro software (BD Biosciences, San Jose, CA, v. 5.1). Base-line fluorescence was quantified using cells treated with complete medium alone throughout the cell culture and transfection duration. The number of transfected cells was normalized automatically with respect to the total number of cells through the introduction of gates (isolating the viable cell population) and limiting the total number of cells (both transfected and untransfected) included in the analysis of transfection efficiencies in the population. A total of 2000 events were counted for each sample, and fluorescent cells were determined using a marker at 5% of the untreated cell population.
2.7.2 Reporter Gene Expression of Cells Seeded on Coaxial Fiber Meshes
Cells expanded in T75 flasks were trypsinized as described above. A cell suspension of 100,000 cells/mL was prepared in cell culture medium and 1 mL added to each 5 mL polypropylene tube. Ethylene oxide-sterilized 10 mm scaffolds were added to the tubes, and negative pressure was applied until no bubbles were observed to facilitate infiltration of the cells into the scaffolds. The cell suspension and the scaffold were then placed into 24-well plates and incubated up to predetermined time points, with media changes every 2–3 days. Expression of EGFP within CRL 1764 cells was observed using confocal microscopy at 10X and 20X magnification using a Zeiss LSM 510 confocal microscope (Thornwood, NY). Scaffolds were excited with an argon laser (488 nm, 6% power). Emission wavelengths were monitored between 510–550 nm. Images thus obtained were further visualized using LSM 5 Image Browser (v 3,2,0,115).
2.8 Statistics
The influence of changing values of the main parameters PCL Conc., PEG MW, PEG Conc., and DNA Conc. on fiber diameter, release kinetics and transfection efficiency were analyzed using analysis of variance with SAS JMP software (v. 7.0.1, Cary, NC). The analysis evaluated means as well as least squares mean (LSM) values with the standard error associated with the computations. Further differences between specific results were evaluated using Tukey’s honestly significant differences (HSD) test. To determine the influence of the main parameters at the 2 levels evaluated, LSM values at the low parameter value (−1) were subtracted from the high value (+1). Significance was determined at p < 0.05 unless otherwise specified.
3. Results
3.1 Fiber Distribution of Electrospun Coaxial Scaffolds
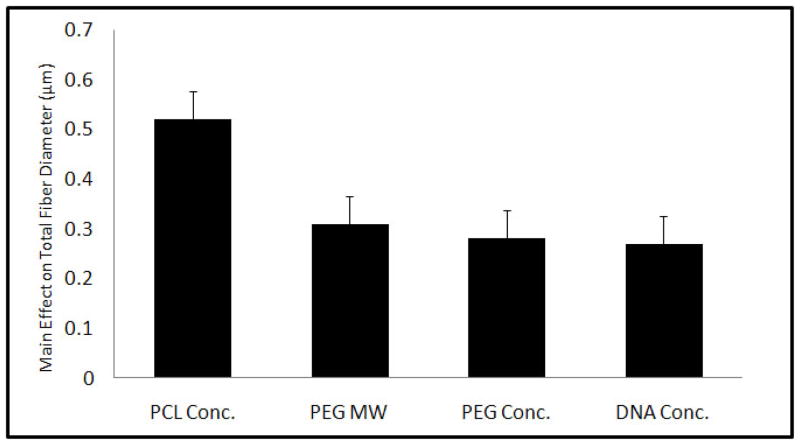
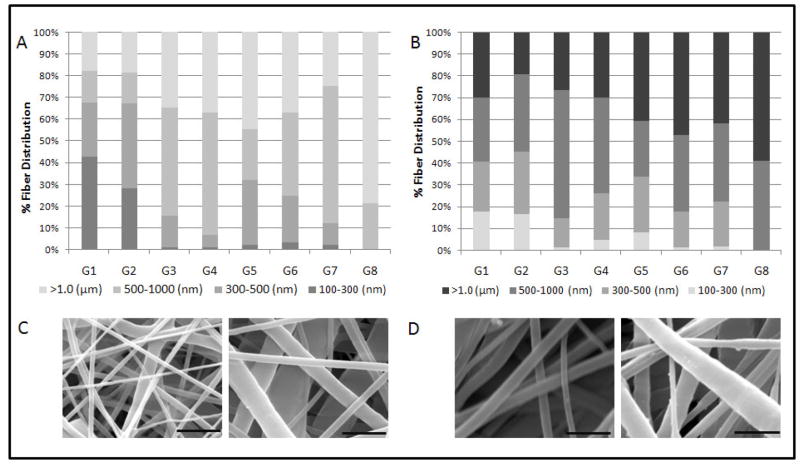
Eight different scaffold types were formulated based on a fractional factorial design with parameters summarized in Table 1. Electrospun coaxial fiber mesh mats had approximate dimensions of 10 × 10.8 cm. Theoretical calculations estimated that the amount of pDNA present per scaffold in the form of a disk of 1 mm diameter was approximately 2 μg for Groups 2, 3, 5 and 8, which contained the high pDNA loading concentrations and approximately 1 μg for the remaining groups that had low pDNA loading concentrations. Fiber diameters were assessed with SEM, as described in Section 2.5. The formulations showed a distribution of fiber diameters ranging from about 200 nm to 4 μm across 8 groups of scaffolds. Assessing the main effects of the parameters on fiber diameter showed that all four parameters, namely, PCL Conc., PEG MW, PEG Conc., and DNA Conc. significantly increased total fiber diameter when the value was increased from low (−1) to high (+1) as shown in Figure 1. The maximum effect on fiber diameter was observed with PCL Conc. (0.52 ± 0.06 μm), followed by PEG MW (0.31 ± 0.06 μm), PEG Conc. (0.28 ± 0.06 μm), and pDNA Conc. (0.27 ± 0.06 μm). A previous study involving electrospun coaxial fibers that did not contain any bioactive molecules suggested that none of the fibers were composed only of the sheath polymer and that some of the fibers within the meshes were composed predominately of the core polymer, which is hydrophilic and easily soluble in aqueous medium [29]. To test if the meshes fabricated with the non-viral vector and pDNA showed fibers with similar properties, the meshes were immersed in PBS with agitation at 115 rpm at 37 °C for 7 days. Figure 2A shows the fiber diameter distribution of fibers directly after fabrication and 2B after immersion in PBS for 7 days. Groups that had a significant percentage of fibers within the smallest fiber diameter distribution range (100–300 nm), i.e., Groups 1 and 2, showed a decrease of 25.1 and 11.5%, respectively, in the percentage distribution of these fibers after 7 days in PBS. Other groups showed a decrease in fiber diameter across different fiber distributions. Taken together, these results suggest that, based on the values of parameters used, some fibers composed predominantly of the core polymer PEG were distributed at various diameters within the coaxial fiber meshes.
Figure 1.
Figure 2.
3.2 Release of r-PEI-HA
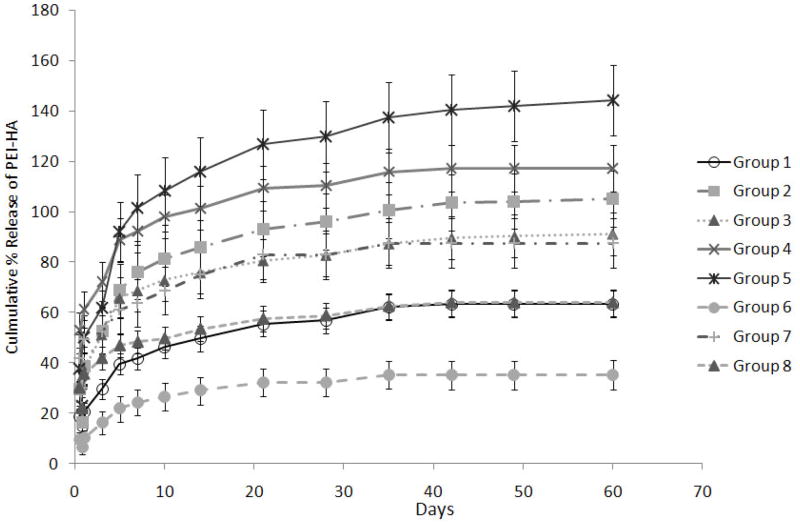
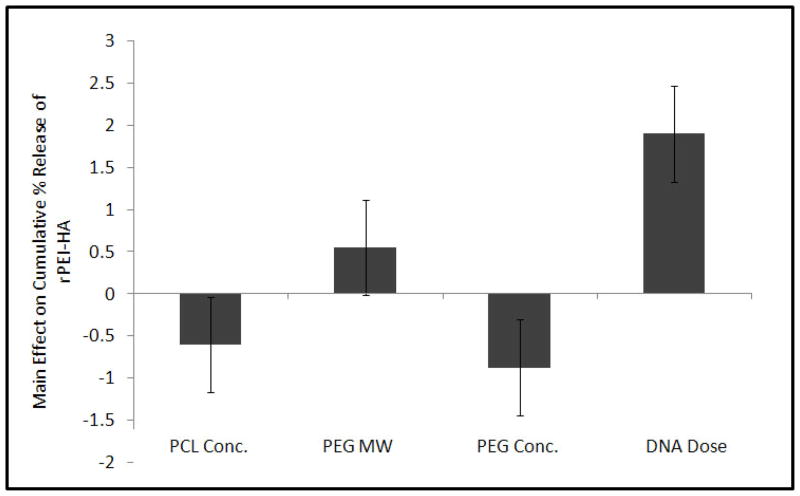
The release of fluorescence tagged r-PEI-HA was monitored with a fluorescence plate reader over a period of 60 days. The 60 day duration for monitoring r-PEI-HA release was further divided into four groups; burst release (0–24hrs), Phase 1 (2–10 days), Phase 2 (11–28 days), and Phase 3 (35–60 days). Burst release during the first 24 hrs ranged from 9.3 ± 1.8% for Group 6 to 47.3 ± 13.3% for Group 7, as described in Table 2 and Figure 3. The main effects of the parameters on the average release of r-PEI-HA are illustrated in Figure 4. The concentration of pDNA (with the concentration of r-PEI-HA scaled accordingly, to maintain the same N:P ratio) was the only parameter that significantly affected the kinetics of r-PEI-HA released. Sheath PCL Conc., core PEG Conc., and PEG MW did not significantly affect the release kinetics of r-PEI-HA. Cumulative release at the end of 60 days ranged from 35.2 ± 5.7% of theoretical loading for Group 6 to 144.1 ± 14.0% for Group 5, as stated in Table 2.
Table 2.
| Burst Release (0–24 hrs) | Phase 1 (2–10 days) %/day | Phase 2 (11 – 28 days) %/day | Phase 3 (35 – 60 days) %/day | Cumulative % Theoretical loading | |
|---|---|---|---|---|---|
| Group 1 | 22.6 ± 1.9#‡ | 3.1 ± 0.4 | 0.7 ± 0.2 | 0.2 ± 0.0 | 63.4 ± 5.2‡† |
| Group 2 | 34.5 ± 9.4#‡ | 4.6 ± 1.4 | 0.9 ± 0.1 | 0.3 ± 0.1 | 105.2 ± 11.1# |
| Group 3 | 37.8 ± 7.6* | 4.4 ± 0.7 | 0.6 ± 0.1 | 0.3 ± 0.1 | 91.1 ± 8.6#¶‡ |
| Group 4 | 43.1 ± 14.7* | 4.9 ± 0.3 | 0.7 ± 0.2 | 0.2 ± 0.0 | 117.2 ± 9.2# |
| Group 5 | 46.7 ± 10.4* | 6.3 ± 1.6 | 1.2 ± 0.4 | 0.4 ± 0.2 | 144.1 ± 14.0* |
| Group 6 | 9.3 ± 1.8* | 2.1 ± 0.7 | 0.3 ± 0.0 | 0.1 ± 0.1 | 35.2 ± 5.7† |
| Group 7 | 47.3 ± 13.3* | 2.1 ± 0.5 | 0.8 ± 1.0 | 0.2 ± 0.1 | 87.5 ± 9.8#¶‡ |
| Group 8 | 35.0 ± 0.5*: | 1.8 ± 0.4 | 0.5 ± 0.3 | 0.2 ± 0.1 | 63.9 ± 5.4¶‡† |
Figure 3.
Figure 4.
There were significant differences between groups related to the burst release of r-PEI-HA, as stated in Table 2. However, there were no statistical differences between groups with respect to the r-PEI-HA released per day between Phases 1 to 3.
3.3 Transfection Efficiency of Released pDNA in 2D Cultures
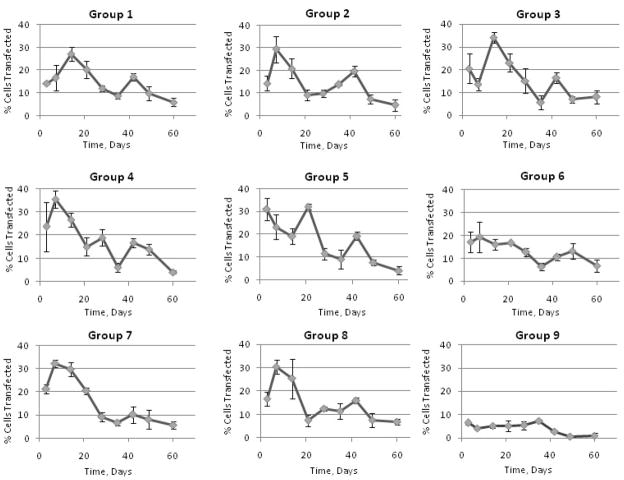
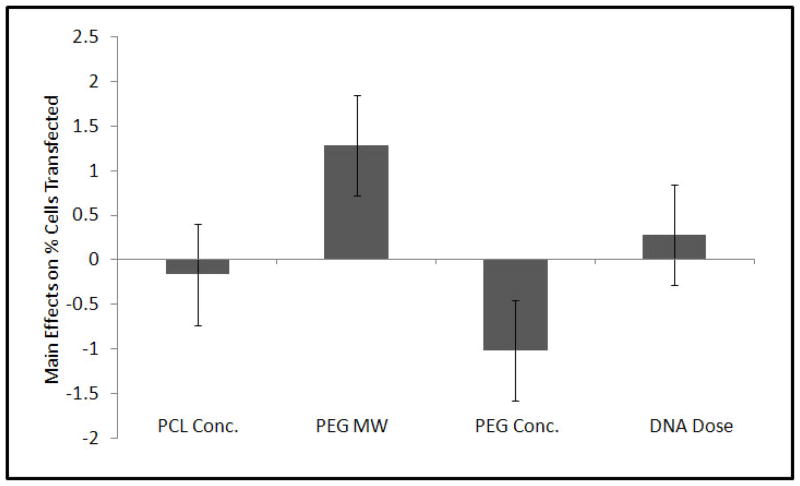
Compared to the control (Group 9) that carried only pDNA within the fiber core and had no r-PEI-HA, all the experimental groups showed a significant increase in transfection efficiency as compared to the cells treated with media alone. A graphical representation of transfection efficiencies across 60 days for all groups is shown in Figure 5. Although there were specific differences in average transfection efficiencies between groups as shown in Table 3, transfection efficiencies were not statistically different from each other after day 21. Across the groups over a period of 60 days, only PEG MW and PEG Conc. had a significant effect on transfection at p = 0.09 and p = 0.10 respectively as represented in Figure 6.
Figure 5.
Table 3.
| Group # | M ± SE |
|---|---|
| Group 1 | 14.45 ± 0.62‡¶ |
| Group 2 | 13.46 ± 0.60‡¶ |
| Group 3 | 16.23 ± 0.60*#‡ |
| Group 4 | 17.54 ± 0.60* |
| Group 5 | 17.37 ± 0.60*# |
| Group 6 | 13.25 ± 0.61¶ |
| Group 7 | 15.45 ± 0.62*#‡ |
| Group 8 | 14.54 ± 0.60#‡¶ |
Figure 6.
3.4 Transfection of Cells Seeded onto Coaxial Electrospun Fiber Mesh Scaffolds
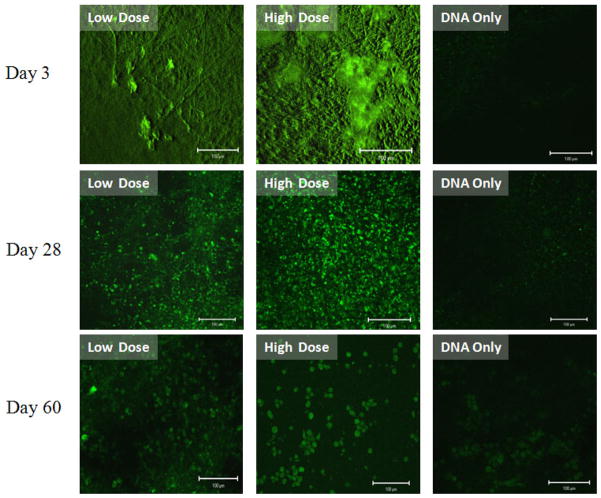
Compared to the pDNA only group, CRL 1764 cells seeded onto scaffolds with both r-PEI-HA within the fiber sheath and pDNA within the fiber core showed a significant number of cells expressing EGFP as shown in Figure 7. EGFP expression by the cells was observed over the duration of the experiment, with qualitative differences in transfection efficiencies observed between groups and time points, as shown in Figure 7.
Figure 7.
4. Discussion
The study described here was designed to determine the effect of certain processing parameters on electrospun fiber diameter distribution, PEI-HA release kinetics, and transfection efficiencies of pDNA released from electrospun coaxial fiber mesh scaffolds incorporating pDNA and PEI-HA, a non-viral gene delivery vector. Coaxial electrospinning has thus far not been employed for delivery of pDNA, and factors influencing the formation of coaxial fiber meshes and their release properties are largely unknown. The experimental plan was formulated with the goal of establishing parameters that allowed for the formation of coaxial electrospun fiber meshes and determining if the examined values of these parameters could dictate the release kinetics of pDNA and r-PEI-HA, as well as the associated transfection efficiencies. Just as the process of electrospinning is dependent on the interaction of multiple factors, including the dielectric properties of the solvents used [30, 31], flow rates of polymer solutions during extrusion [32], the electric potential and quantity of charge circulating through the electrospinning circuit [33], and the distance between the needle and collecting plate [34]; the coaxial electrospinning process has a similar set of complex governing interactions.
In the experimental design implemented here, pDNA was incorporated within the core polymer (PEG) solution, whereas r-PEI-HA was pulverized and dissolved within the sheath polymer (PCL) solution. We had previously described experiments where a set of coaxial electrospun fiber mesh scaffolds were fabricated based on a full factorial design using parameters similar to those described in this study [29]. These common parameters included sheath (PCL) Conc., core (PEG) Conc., and (PEG) MW. It was observed that the range of the parameters tested had to be limited significantly to allow the coaxial electrospinning of polymer solutions incorporating the cationic gene delivery vector and anionic pDNA, thus limiting the versatility of the coaxially electrospun groups. Furthermore, despite the formation of a stable Taylor cone during fabrication of the fiber meshes for all the formulations, the coaxial fibers showed a greater distribution of fiber diameters, as shown in Figure 2 and discussed in Section 3.1. Previously, DNA has been incorporated into uniaxial electrospun fibers by Luu et al. [11], and the fibers obtained had a significant variation in fiber diameters. Both these observations suggest that the inclusion of highly charged moieties, such as a cationic gene delivery vector and anionic pDNA, significantly affect the electrospinning properties of polymer jets.
The assessed parameters showed a similar effect of increasing sheath polymer concentration and core polymer molecular weight on fiber diameter, as was previously observed in the absence of the vector and plasmid. The average fiber diameter increased with the increase in sheath (PCL) Conc. and PEG MW. In addition, an increase in the concentration of PEG, pDNA and r-PEI-HA also caused a similar increase in the average fiber diameter. To determine if all fibers within the mesh were truly coaxial, we immersed them in PBS for 7 days. Analysis showed a significant number of fibers across various groups that completely dissolved in a period of 7 days, which suggested that these fibers were composed predominantly of PEG and were prevalent in various size ranges within different groups. However, there was also a significant and larger population of fibers that did not change in prevalence across subgroups of fiber diameters, suggesting that these fibers were indeed coaxial.
As one of the goals of this experiment, we attempted to characterize the release of incorporated r-PEI-HA from the sheath of coaxial fibers. The direct release of pDNA could not be monitored in this case, as r-PEI-HA significantly decreased binding of pDNA to dyes such as PicoGreen or ethidium bromide during complex formation. Hence, the release of r-PEI-HA was monitored via fluorescence. In the case of uniaxial electrospinning, Luu et al. [11] observed that most of the burst release occurred at 2 hrs, after which pDNA release decreased precipitously. Similar to observations made by Luu et al., r-PEI-HA contained within the sheath fibers of the present study displayed a burst release within 24 hrs after immersion in PBS. However, in the fibers fabricated here, there was also a significant amount of r-PEI-HA released between days 2 to 10, ranging from 1.75 ± 0.39 to 6.30 ± 1.60% of theoretical loading. Only the loading concentration of pDNA (and subsequently that of r-PEI-HA, which was increased to maintain a constant N:P ratio among groups) appeared to significantly influence the release kinetics of r-PEI-HA. Some of the burst release observed here could be attributed to the dissolution of fibers made predominantly from PEG as described above.
The release kinetics were also significantly influenced by the location of PEI-HA and DNA within the coaxial fibers. The initial design of the experiments described here attempted to electrospin r-PEI-HA/pDNA complexes entirely within the core of the coaxial fibers by preassembling the complexes before mixing them with the core polymer solution. To accommodate for the solubility of PEI-HA/pDNA complexes, dextran (instead of PEG) was used as a core polymer and the optimal viscosity for spinning was attained at concentrations noted in the table in the supplemental section (Supplemental Table 1). However, the core polymer containing the complexes showed negligible release of PEI-HA/pDNA complexes (Supplemental Figure 1). Although we could not determine the cause behind the absence of release of PEI-HA/pDNA complexes, a possible reason could be that the interactions between the core and sheath polymer limited the incorporation of the PEI-HA/pDNA complexes within the coaxial fibers.
Some of the coaxial fiber mesh scaffold groups in the study where PEI-HA was incorporated within the polymer sheath and pDNA was incorporated within the fiber core showed greater than 100% cumulative release over the duration of the study. Although, during the fabrication of the scaffolds, the r-PEI-HA and pDNA were loaded such that the N:P ratio between them was constant at 7.5:1, it is feasible that the ratio at which r-PEI-HA and pDNA were released was not constant over the duration of the experiment. The r-PEI-HA/pDNA release values were calculated using a calibration curve generated from known concentrations of r-PEI-HA/pDNA in solution (constant N:P ratio of 7.5:1), with the assumption that there was no significant difference between the fluorescence of r-PEI-HA and r-PEI-HA/pDNA complexes of differing N:P ratios. However, it was found that calibration curves generated by measuring the fluorescence corresponding with known concentrations of r-PEI-HA alone (N:P ratio of 1:0) and r-PEI-HA/pDNA complexes (N:P ratio of 7.5:1) in solution were significantly different from each other, as illustrated in Supplemental Figure 2. The inability of the detection method employed in the release study to differentiate between free r-PEI-HA and r-PEI-HA/pDNA complexes of different N:P ratios in solution was a limitation of the study and taken together with the differences in fluorescence for a given concentration of free r-PEI-HA versus r-PEI-HA/pDNA complexes, may account for the greater than 100% cumulative release observed for some groups.
The temporal differences between peaks of r-PEI-HA/pDNA complex release (Table 2) and maximum EGFP expression in CRL 1764 cell lines (Figure 5) further suggests a potential variation in the ratios of r-PEI-HA and pDNA in the duration of the release. In general, EGFP expression could be more directly correlated to the release of pDNA rather than r-PEI-HA. However, pDNA release could not be directly detected in this experimental design and is a limitation associated with this study. However, transfection efficiencies with scaffolds containing r-PEI-HA were significantly higher than with those containing pDNA alone, suggesting that the presence of r-PEI-HA in the fibers did enhanced transfection, relative to pDNA alone in the fibers. Transfection efficiency seemed to be most influenced by core polymer parameters; PEG MW and concentration. Changing PEG concentration from low to high values decreased the observed transfection of cells. The decrease in transfection could be related to a potential decrease in the amount of pDNA released due to the increase in PEG concentration, as has been observed in other controlled release systems with proteins and peptides [35]. The lower release observed in other studies has been attributed to an increase in the matrix density of the polymer holding the bioactive molecule of interest. However, pDNA release was not directly measured in the present study, so the effects of PEG Conc. on pDNA release are not known in the context of this study.
A similar phenomenon can be expected when the core polymer MW is increased. The increase in PEG MW, however, caused an increase in transfection, which is counter to expectations. However, this effect can be attributed to a number of factors. Increase in the MW of PEG has been reported to increase condensation of pDNA in the presence of NaCl, which is present within the core fiber [36]. pDNA with a more compact structure would potentially have less retention within the coaxial fiber, which could lead to an increase in release and subsequently in transfection. Furthermore, any interaction between the r-PEI-HA and pDNA within the coaxial fiber may also influence release kinetics, due to a differential degree of condensation of pDNA after its interaction and complexation with r-PEI-HA.
We observed a significant effect of PEG Conc. and MW on transfection efficiencies at days 14 and 21. There were significant differences in transfection between groups up to day 21. However, after day 21 there were no significant differences in transfection between groups. It can be surmised that the observed transfection efficiency is dependent on the core polymer properties, i.e., molecular weight and concentration. Further changing the amount of r-PEI-HA loaded within the sheath fibers, which in turn would affect the N:P ratio at which complexes are formed, may give additional insight into the change in release kinetics of r-PEI-HA and the effect on pDNA transfection efficiency.
Lastly, cells directly seeded onto the fabricated coaxial fiber mesh scaffolds showed successful expression of EGFP, and this expression was significantly higher than that observed on meshes containing pDNA alone. The increase in EGFP expression in meshes containing both PEI-HA and pDNA suggests that, despite separating the pDNA and the gene delivery vector (r-PEI-HA) in different components of the coaxial fibers, the pDNA and r-PEI-HA are able to form complexes, be it inside or outside of the coaxial fibers, which are able to transfect cells with a greater efficiency than released pDNA alone.
5. Conclusions
We have successfully designed coaxial electrospun fiber mesh scaffolds containing a non-viral gene delivery vector (r-PEI-HA) and pDNA within the sheath and core of the fiber, respectively. These studies elucidate the role of the processing parameters used to fabricate fiber meshes, i.e., (A) PCL sheath polymer Conc., (B) PEG core polymer MW, (C) PEG core polymer Conc., and (D) Conc. of pDNA within the fiber core, using a fractional factorial design. The results suggest that increasing the parameters above increases the average diameter of the fiber across all groups. Furthermore, the release of r-PEI-HA from the fiber sheath is dependent upon the concentration of pDNA, and the associated concentration of r-PEI-HA, embedded within the fibers. Most of the fabricated scaffolds show extended release of the gene delivery vector over a period of 60 days. Additionally, the transfection efficiency of the pDNA released from scaffolds also incorporating r-PEI-HA was sustained up to 60 days, and the transfection efficiency was dependent upon the concentration and MW of the core polymer, (PEG). Considering the results from the statistical analysis of the release kinetics as well as transfection efficiency, we have fabricated novel scaffolds with variable release kinetics and transfection properties. Furthermore, we have demonstrated that the release kinetics and transfection properties can be modulated by changing the processing parameters of the scaffolds. Such scaffolds with variable and sustained transfection properties can be applied for tissue engineering and other gene delivery applications involving gene therapy.
Supplementary Material
Acknowledgments
We would like to acknowledge funding for the project by the National Institutes of Health (R21 AR56076). We would also like to thank Dr. Jane Grande-Allen for the use of her fluorescence plate reader, Dr. Junghae Suh and Dr. Michael Diehl for the use of their equipment for plasmid amplification and purification, Dr. Joel Moake for the use of his flow cytometer, Dr. Michael Barry for the donation of the pCMV-EGFP plasmid and Genzyme Corp. for the donation of hyaluronic acid. Anita Saraf would also like to thank the Baylor College of Medicine, Medical Scientist Training Program for their support and funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tessmar JK, Gopferich AM. Matrices and scaffolds for protein delivery in tissue engineering. Adv Drug Deliv Rev. 2007;59:274–291. doi: 10.1016/j.addr.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 2.Shea LD, Smiley E, Bonadio J, Mooney DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17:551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 3.Storrie H, Mooney DJ. Sustained delivery of plasmid DNA from polymeric scaffolds for tissue engineering. Adv Drug Deliv Rev. 2006;58:500–514. doi: 10.1016/j.addr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Bonadio J. Tissue engineering via local gene delivery. J Mol Med. 2000;78:303–311. doi: 10.1007/s001090000118. [DOI] [PubMed] [Google Scholar]
- 5.Peng L, Cheng X, Zhuo R, Lan J, Wang Y, Shi B, Li S. Novel gene-activated matrix with embedded chitosan/plasmid DNA nanoparticles encoding PDGF for periodontal tissue engineering. J Biomed Mater Res A. 2009;90:564–576. doi: 10.1002/jbm.a.32117. [DOI] [PubMed] [Google Scholar]
- 6.Holladay C, Keeney M, Greiser U, Murphy M, O’Brien T, Pandit A. A matrix reservoir for improved control of non-viral gene delivery. J Control Release. 2009;136:220–225. doi: 10.1016/j.jconrel.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Kasper FK, Kushibiki T, Kimura Y, Mikos AG, Tabata Y. In vivo release of plasmid DNA from composites of oligo(poly(ethylene glycol)fumarate) and cationized gelatin microspheres. J Control Release. 2005;107:547–561. doi: 10.1016/j.jconrel.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Kasper FK, Seidlits SK, Tang A, Crowther RS, Carney DH, Barry MA, Mikos AG. In vitro release of plasmid DNA from oligo(poly(ethylene glycol) fumarate) hydrogels. J Control Release. 2005;104:521–539. doi: 10.1016/j.jconrel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Heyde M, Partridge KA, Howdle SM, Oreffo RO, Garnett MC, Shakesheff KM. Development of a slow non-viral DNA release system from PDLLA scaffolds fabricated using a supercritical CO2 technique. Biotechnol Bioeng. 2007;98:679–693. doi: 10.1002/bit.21446. [DOI] [PubMed] [Google Scholar]
- 10.Liang D, Luu YK, Kim K, Hsiao BS, Hadjiargyrou M, Chu B. In vitro non-viral gene delivery with nanofibrous scaffolds. Nucleic Acids Res. 2005;33:e170. doi: 10.1093/nar/gni171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luu YK, Kim K, Hsiao BS, Chu B, Hadjiargyrou M. Development of a nanostructured DNA delivery scaffold via electrospinning of PLGA and PLA-PEG block copolymers. J Control Release. 2003;89:341–353. doi: 10.1016/s0168-3659(03)00097-x. [DOI] [PubMed] [Google Scholar]
- 12.Lim SH, Liao IC, Leong KW. Nonviral gene delivery from nonwoven fibrous scaffolds fabricated by interfacial complexation of polyelectrolytes. Mol Ther. 2006;13:1163–1172. doi: 10.1016/j.ymthe.2005.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo D, Woodrow-Mumford K, Belcheva N, Saltzman WM. Controlled DNA delivery systems. Pharm Res. 1999;16:1300–1308. doi: 10.1023/a:1014870102295. [DOI] [PubMed] [Google Scholar]
- 14.Tinsley-Bown AM, Fretwell R, Dowsett AB, Davis SL, Farrar GH. Formulation of poly(D,L-lactic-co-glycolic acid) microparticles for rapid plasmid DNA delivery. J Control Release. 2000;66:229–241. doi: 10.1016/s0168-3659(99)00275-8. [DOI] [PubMed] [Google Scholar]
- 15.Cohen-Sacks H, Elazar V, Gao J, Golomb A, Adwan H, Korchov N, Levy RJ, Berger MR, Golomb G. Delivery and expression of pDNA embedded in collagen matrices. J Control Release. 2004;95:309–320. doi: 10.1016/j.jconrel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Jang JH, Bengali Z, Houchin TL, Shea LD. Surface adsorption of DNA to tissue engineering scaffolds for efficient gene delivery. J Biomed Mater Res A. 2006;77:50–58. doi: 10.1002/jbm.a.30643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YC, Riddle K, Rice KG, Mooney DJ. Long-term in vivo gene expression via delivery of PEI-DNA condensates from porous polymer scaffolds. Hum Gene Ther. 2005;16:609–617. doi: 10.1089/hum.2005.16.609. [DOI] [PubMed] [Google Scholar]
- 18.Fang J, Zhu YY, Smiley E, Bonadio J, Rouleau JP, Goldstein SA, McCauley LK, Davidson BL, Roessler BJ. Stimulation of new bone formation by direct transfer of osteogenic plasmid genes. Proc Natl Acad Sci U S A. 1996;93:5753–5758. doi: 10.1073/pnas.93.12.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao Z, Shi H, Guo R, Ma L, Gao C, Han C, Shen J. Enhanced angiogenesis of porous collagen scaffolds by incorporation of TMC/DNA complexes encoding vascular endothelial growth factor. Acta Biomater. 2009;5:2983–2994. doi: 10.1016/j.actbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Nie H, Khew ST, Lee LY, Poh KL, Tong YW, Wang CH. Lysine-based peptide-functionalized PLGA foams for controlled DNA delivery. J Control Release. 2009;138:64–70. doi: 10.1016/j.jconrel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 21.Krebs MD, Salter E, Chen E, Sutter KA, Alsberg E. Calcium phosphate-DNA nanoparticle gene delivery from alginate hydrogels induces in vivo osteogenesis. J Biomed Mater Res A. doi: 10.1002/jbm.a.32441. in press. [DOI] [PubMed] [Google Scholar]
- 22.Huang YC, Simmons C, Kaigler D, Rice KG, Mooney DJ. Bone regeneration in a rat cranial defect with delivery of PEI-condensed plasmid DNA encoding for bone morphogenetic protein-4 (BMP-4) Gene Ther. 2005;12:418–426. doi: 10.1038/sj.gt.3302439. [DOI] [PubMed] [Google Scholar]
- 23.Sun ZC, Zussman E, Yarin AL, Wendorff JH, Greiner A. Compound core-shell polymer nanofibers by co-electrospinning. Advanced Materials. 2003;15:1929–1932. [Google Scholar]
- 24.Yu JH, Fridrikh SV, Rutledge GC. Production of submicrometer diameter fibers by two-fluid electrospinning. Advanced Materials. 2004;16:1562–1566. [Google Scholar]
- 25.Pham QP, Sharma U, Mikos AG. Electrospun poly(epsilon-caprolactone) microfiber and multilayer nanofiber/microfiber scaffolds: characterization of scaffolds and measurement of cellular infiltration. Biomacromolecules. 2006;7:2796–2805. doi: 10.1021/bm060680j. [DOI] [PubMed] [Google Scholar]
- 26.Allison SD, Anchordoquy TJ. Mechanisms of protection of cationic lipid-DNA complexes during lyophilization. J Pharm Sci. 2000;89:682–691. doi: 10.1002/(SICI)1520-6017(200005)89:5<682::AID-JPS14>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Allison SD, Molina MC, Anchordoquy TJ. Stabilization of lipid/DNA complexes during the freezing step of the lyophilization process: the particle isolation hypothesis. Biochim Biophys Acta. 2000;1468:127–138. doi: 10.1016/s0005-2736(00)00251-0. [DOI] [PubMed] [Google Scholar]
- 28.Saraf A, Hacker MC, Sitharaman B, Grande-Allen KJ, Barry MA, Mikos AG. Synthesis and conformational evaluation of a novel gene delivery vector for human mesenchymal stem cells. Biomacromolecules. 2008;9:818–827. doi: 10.1021/bm701146f. [DOI] [PubMed] [Google Scholar]
- 29.Saraf A, Lozier G, Haesslein A, Kasper FK, Raphael RM, Baggett LS, Mikos AG. Fabrication of nonwoven coaxial fiber meshes by electrospinning. Tissue Eng Part C Methods. 2009;15:333–344. doi: 10.1089/ten.tec.2008.0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hohman MM, Shin M, Rutledge G, Brenner MP. Electrospinning and electrically forced jets. I. Stability theory. Physics of Fluids. 2001;13:2201–2220. [Google Scholar]
- 31.Shin YM, Hohman MM, Brenner MP, Rutledge GC. Experimental characterization of electrospinning: the electrically forced jet and instabilities. Polymer. 2001;42:9955–9967. [Google Scholar]
- 32.Katti DS, Robinson KW, Ko FK, Laurencin CT. Bioresorbable nanofiber-based systems for wound healing and drug delivery: optimization of fabrication parameters. J Biomed Mater Res B Appl Biomater. 2004;70:286–296. doi: 10.1002/jbm.b.30041. [DOI] [PubMed] [Google Scholar]
- 33.Deitzel JM, Kleinmeyer J, Harris D, Tan NCB. The effect of processing variables on the morphology of electrospun nanofibers and textiles. Polymer. 2001;42:261–272. [Google Scholar]
- 34.Wang C, Zhang W, Huang ZH, Yan EY, Su YH. Effect of concentration, voltage, take-over distance and diameter of pinhead on precursory poly (phenylene vinylene) electrospinning. Pigment & Resin Technology. 2006;35:278–283. [Google Scholar]
- 35.Caliceti P, Salmaso S, Elvassore N, Bertucco A. Effective protein release from PEG/PLA nano-particles produced by compressed gas anti-solvent precipitation techniques. J Control Release. 2004;94:195–205. doi: 10.1016/j.jconrel.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Ramos JE, Jr, de Vries R, Ruggiero Neto J. DNA psi-condensation and reentrant decondensation: effect of the PEG degree of polymerization. J Phys Chem B. 2005;109:23661–23665. doi: 10.1021/jp0527103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.