Abstract
Both Src kinase and membrane type 1 matrix metalloproteinase (MT1-MMP) play critical roles in cancer invasion and metastasis. It is not clear, however, how the spatiotemporal activation of these two critical enzymes is coordinated in response to an oncogenic epithelial growth factor (EGF) stimulation. Here, we have visualized the activities of Src and MT1-MMP concurrently in a single live cell by combining two FRET pairs with distinct spectra: (I) cyan fluorescent protein (CFP) and yellow FP (YFP), and (II) orange FP (mOrange2) and red FP (mCherry). The new FRET pair, mOrange2 and mCherry, was first characterized in vitro and in cultured mammalian cells. When integrated with the CFP/YFP pair, this new pair allowed the revelation of an immediate, rapid, and relatively dispersed Src activity. In contrast, the MT1-MMP activity displayed a slow increase at the cell periphery, although Src was shown to play a role upstream to MT1-MMP globally. This difference in the activation patterns of MT1-MMP and Src in response to EGF is further confirmed using an optimized MT1-MMP biosensor capable of being rapidly cleaved by MT1-MMP. The results indicate that although Src and MT1-MMP act globally in the same signaling pathway, their activations differ in space and time upon EGF stimulation, possibly mediated by different sets of intermediates at different subcellular locations. Our results also demonstrated the potential of mOrange2/mCherry as a new FRET pair, together with the popular variants of CFP and YFP, for the simultaneous visualization of multiple molecular activities in a single live cell.
Keywords: FRET, Fluorescent Protein, Src, MT1-MMP, Live Cell Imaging
Introduction
MT1-MMP is a membrane-anchored enzyme belonging to a matrix metalloproteinase (MMP) family, known to be critical in cancer development by remodeling the extracellular matrix via proteolytic means (1, 2). Since membrane-tethered MMPs can be controlled and concentrated at subcellular locations (3), these membrane-associated MMPs appear to play more significant roles than soluble MMPs during cancer invasion (4). Indeed, MT1-MMP can be detected in a wide range of human cancers in clinical samples (5).
Epithelial growth factor (EGF) signaling and the associated Src activity have been well established to correlate with the invasive potential of a variety of human cancers (6, 7). Recent evidence indicates that Src can regulate MT1-MMP through FAK/endophilin A2 (8) and caveolin (9). Src was also shown to directly phosphorylate MT1-MMP at its cytoplasmic tail and regulate its function (10). However, it remains unclear on how the activities of Src and MT1-MMP are coordinated in space and time to impact on cancer development.
Genetically-encoded biosensors based on fluorescence resonance energy transfer (FRET) have provided powerful tools for the study of molecular signals in live cells (11). To date, the most popular fluorescent protein (FP) pair for FRET is cyan and yellow FPs (CFP and YFP) (12). A variety of genetically-encoded biosensors based on CFP and YFP variants have been successfully developed to visualize molecular signals (13). However, with these biosensors, only one type of FRET signal can be visualized in a single live cell. Several studies have used one FP as the common donor or acceptor for two FRET biosensors to simultaneously visualize different molecular signals in the same cell (8, 14, 15). However, these approaches require more sophisticated means to quantify the signals than a simple donor/acceptor emission ratio. Recently, a newly developed FP Ametrine was elegantly paired with tdTomato to form a second FRET pair (16). Together with the pair of CFP and YFP variants, mTFP1 and mCitrine, this new Ametrine/tdTomato pair has allowed the successful visualization of Caspase-3 activities in different subcellular compartments. While each individual FRET pair in this case can be ratiometrically imaged in the presence of the other, it is difficult to simultaneously visualize these two FRET pairs because of the almost identical emission of mAmetrine and mCitrine (16). Recently, a variety of new FPs with different colors have been developed in which mOrange (or mKO as orange FP) and mCherry (as red FP) appear suitable serving as the donor and acceptor, respectively, for a second FRET pair that is spectrally distinguishable from CFP and YFP variants (12, 17, 18). Either mOrange or mKO has been successfully coupled with mCherry to form a FRET pair with efficiency detectable by fluorescence lifetime imaging microscopy (FLIM) (19). However, the photo-stability of mOrange is relatively poor (18). The CFP excitation also led to a substantial photo-conversion of mKO, which may cause artifacts in cells expressing biosensors containing both CFP/YFP and mKO/mCherry pairs (19). Another two FRET pairs: (I) TagFP and mPlum (20); (II) T-Sapphire and DsRed (21), have been used together with the CFP/YFP pair for dual FRET imaging. However, mPlum has a low quantum yield and poor brightness while DsRed can form artificial tetramers (18). Recently, mOrange2 was developed to have several fold greater photo-stability than both mOrange and mKO (18). The fusion proteins of α-tubulin and connexin-43 also showed correct subcellular localization when conjugated to mOrange2, but not to mKO (18). Hence, mOrange2 appears better suited serving as a FRET donor for mCherry.
In this paper, we have developed genetically-encoded FRET biosensors with mOrange2 and mCherry, and demonstrated that these biosensors are capable of visualizing Src and MT1-MMP activities in live cells. The dynamic activation of Src and MT1-MMP at subcellular levels was then concurrently monitored in a single live cell by combining CFP/YFP- and mOrange2/mCherry-based biosensors. The results suggest that the spatiotemporal activation patterns of Src and MT1-MMP are very different, with Src activity being relatively fast and dispersed while that of MT1-MMP slow and highly localized at cell periphery.
Materials and Methods
Gene construction
The Src and MT1-MMP biosensors based on ECFP and YFP variants, Citrine or YPet, were described previously (22, 23). For the construction of mOrange2/mCherry-based MT1-MMP and Src biosensors, the ECFP and Citrine (or YPet) within the biosensors were replaced by mOrange2 and mCherry, respectively. For the MT1-MMP biosensor, mCherry gene was amplified by polymerase chain reaction (PCR) with a sense primer containing a BglII site and a reverse primer containing a SacI site. The mOrange2 gene was amplified by PCR with a sense primer containing a SacI site and the sequence for the MT1-MMP substrate peptide CPKESCNLFVLKD from proMMP-2 (24), and a reverse primer containing a SacII site, a stop codon, and a HindIII site. The PCR products were cloned into pRSETb (Invitrogen) using BglII/HindIII for bacterial expression and into pDisplay (Invitrogen) using BglII/SacII for mammalian cell expression, as shown in Fig. 1A. The AHLR MT1-MMP biosensor was constructed by replacing the original substrate peptide (CPKESCNLFVLKD) in the MT1-MMP biosensor with an optimized cleavage peptide sequence (CRPAHLRDSG) flanked by GGS linker peptides. For the Src biosensor, the mOrange2 or mCherry gene was amplified by PCR using a sense primer containing a BamHI or SacI site, and a reverse primer containing a SphI site or a stop codon together with an EcoRI site, respectively. The membrane-targeted Src biosensor was constructed by fusion of prenylation substrate sequence (KKKKKKSKTKCVIM) from KRas to the C-terminus of Citrine or mCherry. The human MT1-MMP plasmid is a gift from Stephen J. Weiss at University of Michigan at Ann Arbor (25).
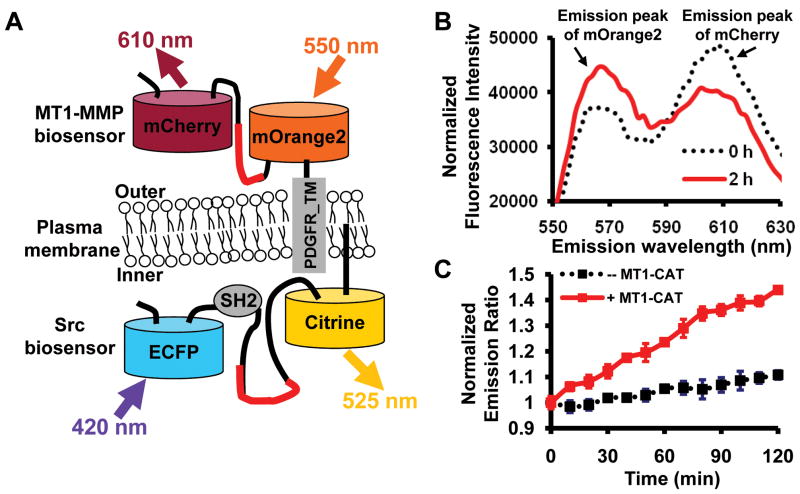
Figure 1.
The characterization of mOrange2 and mCherry as a FRET pair in vitro. (A) A scheme depicting the membrane-tethered MT1-MMP and Src biosensors. The MT1-MMP biosensor is fused to the transmembrane domain of PDGFR and located outside of the plasma membrane. Src biosensor is anchored at the inner face of the plasma membrane. (B) Emission spectra of purified NL MT1-MMP biosensor based on mOrange2 and mCherry (excited at 515 ± 10 nm) before (black dashed line) and after (red solid line) the addition of 2 μg/ml MT1-CAT for 2 hr at 37°C. Arrows point to the emission peaks of mOrange2 and mCherry. (C) The time courses of mOrange2/mCherry emission ratio (mean ± S.D.) of the MT1-MMP biosensor with (red line) or without (black line) the presence of MT1-CAT.
Expression of the biosensor proteins and in vitro assays
The biosensor proteins were expressed with N-terminal 6x His tags in Escherichia coli which were cultured for 16 hr at 37°C and purified by nickel chelation chromatography as previously described (23)(Supplementary Fig. 1A). Emission ratio of mOrange2/mCherry (564 nm/604 nm) with excitation wavelength at 515 ± 10 nm was measured by a fluorescence plate reader (TECAN, Sapphire II). The cleavage assay for MT1-MMP biosensor was conducted at 37°C by incubating the recombinant catalytic domain of human MT1-MMP (MT1-CAT) (2 μg/ml) with 1μM biosensor protein in a MT1-MMP proteolysis assay buffer (50 mM HEPES, pH 6.8, 10 mM CaCl2, 0.5 mM MgCl2, 50 μM ZnCl2, and 0.005% Brij-35)(26). To compare the cleavage efficiency of different substrates, MT1-CAT was incubated for 3 h at 37°C with the protein substrates at a 1:25–1:1600 molar ratio with Human plasma α1-antitrypsin (AAT, Calbiochem), MBP-J37, and the NL(ECFP/YPet) biosensor.
Cell culture and transfection
Cell culture reagents were obtained from GIBCO BRL. HeLa cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1 mM sodium pyruvate in a humidified 95% air, 5% CO2 incubator at 37°C before imaging experiments. The different DNA plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen).
Microscopy, image acquisition and analysis
Cells expressing various exogenous proteins were starved with 0.5% FBS for 36–48 hr and cultured in cover-glass-bottom dishes (Cell E&G) before EGF (50 ng/ml) stimulation. During the imaging process, the cells were maintained in CO2-independent medium (GIBCO BRL) without serum at 37°C. Imageswere collected by a Zeiss Axiovert inverted microscope equipped with a cooled charge-coupled device camera (Cascade 512B; Photometrics) using MetaFluor 6.2 software (Universal Imaging). The parameters of dichroic mirrors, excitation and emission filters for FRET and different fluorescent proteins are shown in Supplementary Table 1. The pixel-by-pixel ratio images of CFP/YFP or mOrange2/mCherry were directly calculated based on the background-subtracted fluorescence intensity images of FPs by the MetaFluor software to represent the FRET efficiency and the activation levels of biosensors. Emission ratios of CFP/YFP or mOrange2/mCherry were averaged on chosen regions of interest to allow the quantification and statistical analysis by Excel software (Microsoft). For the pre-treatment experiments with Src inhibitor, cells were incubated with 10 μM PP1 or 0.1% DMSO (v/v) as control for 1 hr before imaging.
Inhibitor washout assay
For the GM6001 washout assay, HeLa cells expressing MT1-MMP and indicated biosensors were pre-incubated with 5 μM GM6001 for 12 hr before being washed by HBSS (Hanks Balanced Salt Solution) for five time. The FRET responses of the biosensors were monitored in the CO2-independent medium afterwards. For the TIMP-2 washout assay, the cells were pretreated with 100 nM TIMP-2 (2 μg/ml) for 12 hr, washed twice with HBSS, exposed to acidic conditions for 1.5 min (50 mM Glycine, pH 4.0, and 100 mM NaCl) to remove the bound TIMP-2, and neutralized with 0.5 M HEPES, pH 7.5 and 100 mM NaCl. The cells were then washed by the CO2-independent medium for three times before the FRET responses of the biosensor were monitored.
Results
Characterization of mOrange2 and mCherry as a novel FRET pair in vitro
Among the recently developed FPs (18), mOrange2 and mCherry may form a second FRET pair that is spectrally distinguishable from CFP and YFP variants. We have previously developed specific FRET biosensors for monitoring the activities of Src and MT1-MMP separately (22, 23). To visualize Src and MT1-MMP activities concurrently in the same cells, we generated a novel MT1-MMP biosensor, by replacing the efficient ECFP and YPet pair (27) in the original biosensor with mOrange2 and mCherry (Fig. 1A). It is expected that active MT1-MMP would cleave the biosensor substrate peptide CPKESCNLFVLKD, thus separating mOrange2 and mCherry to result in a FRET decrease that could be tracked by an increase in the emission ratio of mOrange2/mCherry. Indeed, following the incubation of the purified biosensor (supplementary Fig. 1A) with the catalytic domain of MT1-MMP (MT1-CAT), a decrease in mCherry emission was detected along with a concomitant increase in mOrange2 emission, indicative of a FRET decrease (Fig. 1B). The mOrange2/mCherry emission ratio gradually and consistently increased upon the incubation of MT1-CAT (Fig. 1C), confirming the effect of MT1-CAT on the biosensor. Analysis by gel electrophoresis further verified that the biosensor was completely cleaved upon the MT1-CAT incubation for 8 hr (Supplementary Fig. 1B).
Characterization of mOrange2 and mCherry as a FRET pair in mammalian cells
The mOrange2/mCherry-based MT1-MMP biosensor was also targeted to anchor outside of the plasma membrane in mammalian cells by a PDGFR transmembrane domain (22). This membrane-targeted biosensor was transfected together with the wild-type MT1-MMP or its control vector into HeLa cells, which express minimal levels of endogenous MT1-MMP(26). Strong fluorescence signals can be observed in both mOrange2 and mCherry channels with the mOrange2 excitation (Supplementary Fig. 2), suggesting an efficient FRET between these two FPs. EGF induced an increase in the emission ratio of mOrange2/mCherry and hence a FRET decrease of the biosensor in HeLa cells transfected with the wild-type MT1-MMP (Fig. 2A and 2C), but not with the control vector (Fig. 2B and 2C). Hence, these results indicate that the MT1-MMP FRET biosensor based on the mOrange2/mCherry pair can report MT1-MMP activity in vitro and in mammalian cells.
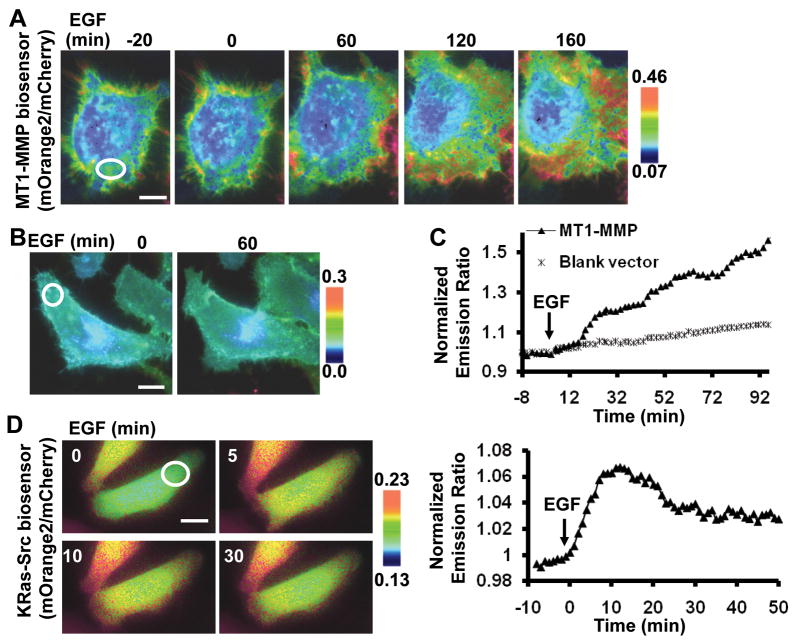
Figure 2.
The characterization of mOrange2 and mCherry as a FRET pair in mammalian cells. (A, B) Representative mOrange2/mCherry emission ratio images of the MT1-MMP biosensor before and after EGF stimulation in HeLa cells expressing (A) MT1-MMP or (B) a blank vector. (C) Representative time courses of normalized mOrange2/mCherry emission ratio of the MT1-MMP biosensor in HeLa cells expressing MT1-MMP (▲) or a blank vector (*). (D) Representative images (left panels) and time course of normalized mOrange2/mCherry emission ratio (right panel) in HeLa cells expressing the Src biosensor before and after EGF stimulation. All the scar bars represent the length of 30 μm.
To further examine whether mOrange2 and mCherry are an efficient FRET pair for other kinds of biosensors, a Src FRET biosensor developed previously (23) was modified by replacing its ECFP and Citrine with mOrange2 and mCherry, respectively. A prenylation substrate sequence (KKKKKKSKTKCVIM) from KRas was then fused to the C-terminus of mCherry to anchor the Src biosensor at the inner surface of the plasma membrane (KRas-Src biosensor) (Fig. 1A) (22). EGF induced an apparent increase in the mOrange2/mCherry emission ratio of this KRas-Src biosensor (Fig. 2D). This result confirmed that mOrange2 and mCherry can act as an efficient FRET pair for the development of different biosensors.
Visualization of Src and MT1-MMP activities concurrently in a single live cell
The Src activation is related to the activity of MT1-MMP in regulating cancer development (8, 9). However, it remains unclear on the dynamic interrelationship between Src and MT1-MMP at subcellular levels, which is particularly important given that the function of Src is largely dependent on its subcellular location/environment, e.g. Src inhibits RhoA at the focal adhesion sites (28) but activates RhoA at podosomes (29). We first investigated the spatiotemporal distribution of Src and MT1-MMP by co-transfection of EGFP-conjugated Src and mCherry-conjugated MT1-MMP. As shown in Supplementary Fig. 3, EGF induced a significant co-localization of Src-GFP and MT1-MMP-mCherry at the cell periphery, especially at the lamellipodium-like regions. This result is consistent with previous reports that active Src can be transported to the plasma membrane to act on downstream signaling molecules (30, 31). We then combined the membrane-targeted and CFP/YFP-based KRas-Src biosensor together with the mOrange2/mCherry-based MT1-MMP biosensor so that the MT1-MMP activity at the outer surface and the Src activity at the inner surface of the plasma membrane could be monitored concurrently with high spatiotemporal resolution (Fig. 1A). The KRas-Src biosensor, MT1-MMP biosensor and MT1-MMP were co-transfected into the same HeLa cells. EGF induced an immediate, fast, and relatively dispersed increase of Src activity which reached the peak within a few minutes. In contrast, the MT1-MMP activity increased slowly and was concentrated at the cell periphery upon EGF stimulation in the same cell (Fig. 3A). The quantification of the FRET response at a local region proximal to cell periphery further confirmed a fast and transient activation of Src but a slow and gradual activation of MT1-MMP in response to EGF (Fig. 3B). A similar phenomenon with a fast Src and slow MT1-MMP activation in response to EGF was also observed when the KRas-Src biosensor is switched to the mOrange2/mCherry pair and the MT1-MMP biosensor to the ECFP/YPet pair (Data not shown). These results suggest that the different spatiotemporal responses of Src and MT1-MMP observed are not due to the difference in FRET pairs.
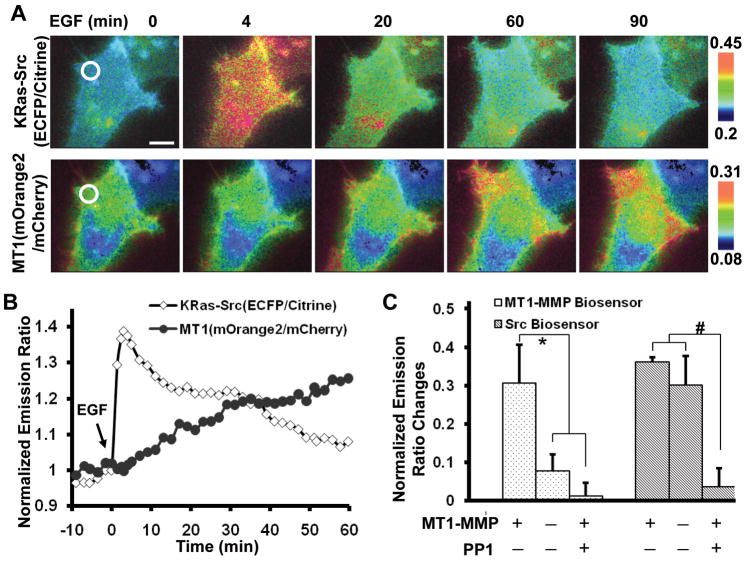
Figure 3.
Concurrently visualizing the MT1-MMP and Src activities. (A) Emission ratio images of HeLa cells expressing Src (ECFP/Citrine, upper panels) and MT1-MMP (mOrange2/mCherry, lower panels) biosensors before and after EGF stimulation. (B) The normalized emission ratio time courses of the KRas-Src and MT1-MMP biosensors averaged on the chosen region of interest as indicated in (A). (C) Bar graphs show the emission ratio changes of the Src (ECFP/Citrine) and MT1-MMP (mOrange2/mCherry) biosensors in response to EGF. ‘MT1-MMP− ’ or ‘MT1-MMP +’: HeLa cells co-transfected with a blank vector or MT1-MMP; ‘PP1 − ’ or ‘PP1 +’: cells pretreated with DMSO or 10 μM PP1 for 1 hr. The * and # signs represent statistically significant difference between the indicated groups.
Since the Src activity is related to the activity of MT1-MMP (8, 9), the different time courses of Src and MT1-MMP suggest that Src may play a role upstream of MT1-MMP in response to EGF. Indeed, the inhibition of Src by PP1 abolished the EGF-induced FRET response of the MT1-MMP biosensor in HeLa cells expressing exogenous MT1-MMP (Fig. 3C). In contrast, although the absence of exogenous MT1-MMP expression eliminated the response of the MT1-MMP biosensor in HeLa cells, EGF still induced a significant FRET response of the Src biosensor (Fig. 3C). Consistently, the inhibition of MT1-MMP by GM6001 did not block the EGF-induced FRET response of the Src biosensor (Data not shown). These results suggest that Src activity acts upstream of MT1-MMP activation in response to EGF.
The development of a MT1-MMP biosensor with a fast cleavage speed
It remains possible that the slow response of the MT1-MMP FRET biosensor upon EGF stimulation is attributed to the slow cleavage of the biosensor by active MT1-MMP, but not the slow activation of MT1-MMP itself. To address this question, we have first examined the cleavage efficiency of the MT1-MMP biosensor (labeled as ‘NL’ with the original substrate peptide sequence CPKESCNLFVLKD). The cleavage rate of this biosensor upon the incubation with MT1-CAT was compared to two typical MT1-MMP substrate molecules, AAT and MBP-J37. The purified substrate biosensors/proteins and MT1-CAT were incubated together for 3 hr with the enzyme/substrate ratios ranging 1:25–1:1600. As shown in Fig. 4A, a 50% cleavage of AAT and MBP-J37 can be accomplished at an enzyme/substrate ratio between 1:200 and 1:400 while the 50% cleavage of the NL biosensor was achieved at the ratio between 1:25 and 1:50. As a control, GM6001, a potent inhibitor of MMPs, inhibited MT1-CAT activity and blocked the cleavage of different substrates. Therefore, the NL biosensor is not an efficient substrate for MT1-MMP, suggesting a significant potential in improving the biosensor sensitivity by modifying the substrate peptide. A fast-cleavage biosensor can also allow the investigation of activation speed of MT1-MMP in response to EGF with high accuracy.
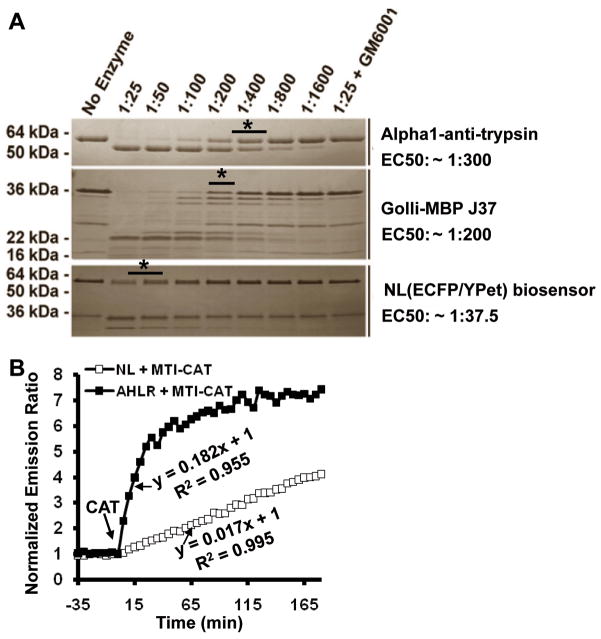
Figure 4.
The development of an optimized MT1-MMP biosensor. (A) The comparison of cleavage speeds of three MT1-MMP substrates (AAT, MBP-J37 and the NL ECFP/YPet biosensor) by MT1-CAT. MT1-CAT was incubated with the different substrate proteins at 1:25–1:1600 molar ratios for 3 h at 37°C. ‘*’ indicates the ratio condition that half of the substrate was cleaved. The corresponding EC50 value was shown at the right side. (B) Comparison of the cleavage speed of the purified NL (□) and AHLR (■) biosensors. The time courses show the normalized emission ratio (ECFP/YPet) change of biosensors (1 μM) before and after the addition of MT1-CAT (2 μg/ml). The equations and R-square values represent the initial slopes of the emission curves upon MT1-CAT incubation.
We have hence developed an optimized MT1-MMP biosensor containing a selective and efficient peptide sequence (CRPAHLRDSG) capable of being rapidly and specifically cleaved by MT1-MMP, but not by gelatinases MMP-2 or MMP-9. The selection of this sequence was the result of our extensive substrate phage cleavage studies performed at the Center for Proteolytic Pathways in the Burnham Institute for Medical Research to identify the cleavage preferences of the individual MMPs including MT1-MMP (manuscript in preparation).
This optimized MT1-MMP substrate peptide (CRPAHLRDSG) flanked by GGS on both sides (labeled as ‘AHLR’) was applied to replace the NL substrate peptide (CPKESCNLFVLKD) in the MT1-MMP biosensor. Following the incubation with MT1-CAT, a rapid and significant decrease in YPet of the new AHLR biosensor was detected along with a concomitant increase in ECFP, suggesting an efficient FRET loss (Supplementary Fig. 4A). Gel electrophoresis further revealed that the biosensor can be completely cleaved by MT1-CAT (Supplementary Fig. 4B), confirming the high sensitivity of the AHLR biosensor. When the two purified biosensors (1 μM) were incubated with MT1-CAT (2 μg/ml), the initial slopes of the emission ratio time courses for the NL and AHLR biosensors are 0.017 and 0.182, respectively (Fig. 4B). Therefore, the optimized AHLR biosensor is 10 fold more sensitive than the original NL biosensor. The FRET response of this AHLR biosensor in vitro is in fact faster than that of the Src biosensor measured by kinase assay as reported before (32).
The activation speed of the optimized AHLR biosensor in mammalian cells
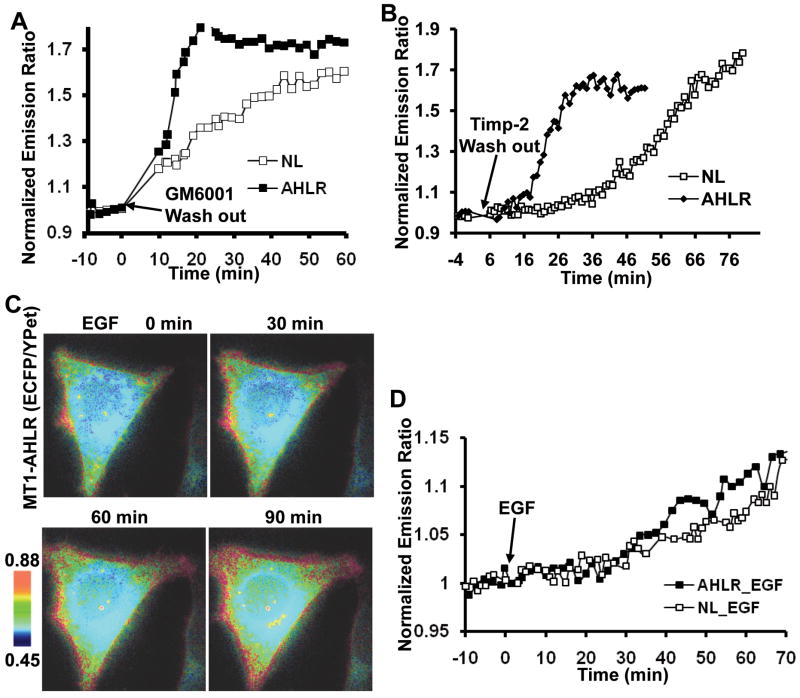
We further compared the cleavage speeds of the two NL and AHLR biosensors in HeLa cells transfected with MT1-MMP. The basal activity of MT1-MMP was pre-inhibited by GM6001 for 12 hr such that the newly cycled and membrane-integrated biosensors were kept intact to result in a low emission ratio (ECFP/YPet) at the plasma membrane. After GM6001 washout, the biosensor proteins were exposed to the restored basal MT1-MMP activity on the cell surface, which resulted in an increase of FRET ratio. As shown in Fig. 5A, the emission ratio of AHLR biosensor increased rapidly upon GM6001 washout, much faster than that of NL biosensor. A similar result can be observed when the MT1-MMP activity was pre-inhibited by a specific MT1-MMP inhibitor TIMP-2 (Fig. 5B). These results indicate that the AHLR biosensor can be cleaved by MT1-MMP with a much faster speed than the NL biosensor in mammalian cells.
Figure 5.
EGF induced a slow response of the optimized MT1-MMP biosensor. (A and B) The ECFP/YPet emission ratio changes of the NL (□) and AHLR (■) biosensors in response to (A) GM6001 or (B) TIMP-2 washout. HeLa cells expressing MT1-MMP and biosensors were pre-incubated with (A) GM6001 (5 μM) or (B) TIMP-2 (100 nM) for 12 hr, followed by the washout with HBSS or acid/neutralization solutions, respectively. (C) Representative ECFP/YPet emission ratio images of the optimized AHLR FRET biosensor upon EGF stimulation in HeLa cells expressing MT1-MMP. (D) The ECFP/YPet emission ratio time courses of the NL (□) and AHLR (■) biosensors before and after EGF stimulation in HeLa cells expressing MT1-MMP.
We then applied EGF to stimulate the AHLR biosensor in HeLa cells expressing MT1-MMP. As shown in Fig. 5C, the FRET response of the AHLR biosensor is still slow and comparable to that of the NL biosensor upon EGF stimulation. This EGF-induced response of the AHLR biosensor is significantly slower and weaker than that to the inhibitor washout, possibly suggesting a relatively high basal level of MT-MMP activity which is further inducible by EGF with moderate magnitude. The pretreatment of cells with the MT1-MMP inhibitor TIMP-2 completely blocked this EGF-induced response of the AHLR biosensor, suggesting the specificity of the AHLR biosensor (Data not shown). These results confirmed that, upon EGF stimulation, the slow response of the biosensor is due to the slow activation of MT1-MMP, but not to a slow cleavage rate of the biosensor by MT1-MMP. Therefore, the distinct spatiotemporal responses of the Src and MT1-MMP biosensors upon EGF stimulation indeed reflect the different activation patterns of Src and MT1-MMP upon EGF stimulation.
Discussion
Both Src and MT1-MMP have been shown to play pivotal roles in regulating cancer development and metastatic invasion. However, the coordination between these two critical molecules in space and time remains unclear. With mOrange2 and mCherry serving as a new FRET pair besides the popular CFP/YFP pair, we have combined two FRET biosensors with distinct colors for the concurrent visualization of Src and MT1-MMP activities in a single live cell. The results revealed distinctive spatiotemporal activation patterns of Src and MT1-MMP in response to EGF, with the Src activation occurring fast and dispersed whereas that of MT1-MMP being slow and concentrated at the cell periphery.
Cancer invasion is largely dependent on integrin-mediated cell adhesive interactions with the surrounding ECM (33, 34). These integrin-associated activities can be linked to MMP expression, activation and proteolytic activity (25), in part via Src kinase-dependent processes (35, 36). Upon integrin activation, focal adhesion kinase (FAK) becomes tyrosine phosphorylated at site 397 and recruits Src to phosphorylate various signaling molecules, including FAK itself, p130Cas, caveolin, and endophilin A2. These events further promote cell surface MT1-MMP activity and invasion (8, 9). Consistently, Src has been shown to be related to MT1-MMP in a variety of cell systems (8–10), which is also in line with our results that the inhibition of Src blocked the FRET response of the MT1-MMP biosensor (Fig. 3C). However, it remains unclear on how, in live cells, Src activity controls the MT1-MMP activity at subcellular levels. Our dual FRET imaging results indicate that the activation of Src and MT1-MMP differ both in space and time (Fig. 3A-B). Our optimized MT1-MMP biosensor capable of fast cleavage further confirmed that the EGF-induced MT1-MMP activation occurs slowly (Fig. 5C–D), in contrast to the fast activation of Src. This immediate activation of Src and the significant delay of MT1-MMP activation in the same subcellular location suggest that the effect of Src on MT1-MMP may require other molecular-intermediates/mechanisms in addition to the direct interactions, e.g. the direct phosphorylation of MT1-MMP cytoplasmic tail by Src as previously reported (10). The direct phosphorylation of MT1-MMP by Src is expected to occur much faster than the observed delay between these two signals. Indeed, this view of indirect interaction between Src and MT1-MMP is consistent with other reports that Src may indirectly regulate MT1-MMP through FAK/endophilin A2 (8) or caveolin (9). Src also regulates podosomes, in which MT1-MMP resides and plays an active role (37–39). It is hence possible that Src affects the podosome formation in modulating the MT1-MMP activation (40). Further studies are warranted to elucidate the molecular mechanism by which Src regulates MT1-MMP activity in space and time in response to EGF.
It has become increasingly clear that signaling molecules inside cells are not functional in isolation. Instead, different signaling pathways interact with each other in a non-linear fashion and form a coordinated network. Recent evidence further suggests that signaling networks or molecular hierarchies are largely dependent on subcellular localization, possibly due to the different molecular mediators at different subcellular locations. For example, Src induces the p190RhoGAP activation and subsequently inhibits RhoA at the focal adhesion sites (28), whereas Src activates RhoA at podosomes (29). RhoA also couples with its downstream molecule ROCK at the cell rear and a contractile region behind lamellipodium, but co-localizes with another substrate molecule mDia at the leading edge of a migrating cell (41, 42). Hence, the development and integration of multiple FRET biosensors, e.g. using mOrange2/mCherry and CFP/YFP pairs, can provide powerful tools to visualize molecular hierarchies at subcellular levels in live cells. The information obtained should significantly advance our in-depth and systematic understanding of spatiotemporal molecular network coordinating cellular functions, including those crucial for cancer invasion and metastasis.
Recently, multiple new FPs have been developed with different colors(17, 18). This has provided opportunities for new FRET pairs which are spectrally distinguishable from the popular CFP/YFP or BFP/GFP pairs. As we demonstrated here, mOrange2 and mCherry can act as a new FRET pair to monitor active signaling events in vitro and in mammalian cells. However, there is a certain overlap between the excitation spectra of mOrange2 and mCherry (17, 18), i.e. the excitation of donor mOrange2 can directly excite the acceptor mCherry and cause non-specific cross-talk. This artifact may have contributed to the lower dynamic range of the mOrange2/mCherry-based MT1-MMP biosensor (40% in vitro as shown in Fig. 1C) in comparison to the ECFP/YPet-based biosensor (570% in vitro) (22). FLIM technology could help to solve this problem because the emission lifetime measurement of the donor mOrange2 alone is sufficient to deduce the FRET efficiency, without the need to measure the emission lifetime of the acceptor mCherry (43). It is envisioned that the integration of FLIM and FRET will become increasingly accepted as a platform for FRET imaging, which is also an ongoing project in our laboratory.
Supplementary Material
Acknowledgments
We are indebted to Dr. Patrick Daugherty at University of California, Santa Barbara, for the YPet construct. This work is supported by grants from NIH CA139272, NS063405, NSF CBET0846429, CMMI0800870, and the Wallace H. Coulter Foundation.
References
- 1.Seiki M. Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 2003;194:1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]
- 2.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J Cell Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 3.Nakahara H, Howard L, Thompson EW, et al. Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proc Natl Acad Sci U S A. 1997;94:7959–64. doi: 10.1073/pnas.94.15.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh Y, Seiki M. MT1-MMP: a potent modifier of pericellular microenvironment. J Cell Physiol. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- 6.Lo HW, Hsu SC, Hung MC. EGFR signaling pathway in breast cancers: from traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006;95:211–8. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- 7.Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6:209–14. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Wu X, Simone J, Hewgill D, et al. Measurement of two caspase activities simultaneously in living cells by a novel dual FRET fluorescent indicator probe. Cytometry A. 2006;69:477–86. doi: 10.1002/cyto.a.20300. [DOI] [PubMed] [Google Scholar]
- 9.Labrecque L, Nyalendo C, Langlois S, et al. Src-mediated tyrosine phosphorylation of caveolin-1 induces its association with membrane type 1 matrix metalloproteinase. J Biol Chem. 2004;279:52132–40. doi: 10.1074/jbc.M409617200. [DOI] [PubMed] [Google Scholar]
- 10.Nyalendo C, Michaud M, Beaulieu E, et al. Src-dependent phosphorylation of membrane type I matrix metalloproteinase on cytoplasmic tyrosine 573: role in endothelial and tumor cell migration. J Biol Chem. 2007;282:15690–9. doi: 10.1074/jbc.M608045200. [DOI] [PubMed] [Google Scholar]
- 11.Kiyokawa E, Hara S, Nakamura T, Matsuda M. Fluorescence (Forster) resonance energy transfer imaging of oncogene activity in living cells. Cancer Sci. 2006;97:8–15. doi: 10.1111/j.1349-7006.2006.00141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Shyy JY, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 14.Kawai H, Suzuki T, Kobayashi T, et al. Simultaneous real-time detection of initiator-and effector-caspase activation by double fluorescence resonance energy transfer analysis. J Pharmacol Sci. 2005;97:361–8. doi: 10.1254/jphs.fp0040592. [DOI] [PubMed] [Google Scholar]
- 15.Peyker A, Rocks O, Bastiaens PI. Imaging activation of two Ras isoforms simultaneously in a single cell. Chembiochem. 2005;6:78–85. doi: 10.1002/cbic.200400280. [DOI] [PubMed] [Google Scholar]
- 16.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–3. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 17.Shaner NC, Campbell RE, Steinbach PA, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 18.Shaner NC, Lin MZ, McKeown MR, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS ONE. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant DM, Zhang W, McGhee EJ, et al. Multiplexed FRET to image multiple signaling events in live cells. Biophys J. 2008;95:L69–71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niino Y, Hotta K, Oka K. Simultaneous live cell imaging using dual FRET sensors with a single excitation light. PLoS One. 2009;4:e6036. doi: 10.1371/journal.pone.0006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouyang M, Lu S, Li XY, et al. Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J Biol Chem. 2008;283:17740–8. doi: 10.1074/jbc.M709872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Botvinick EL, Zhao Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita TSH, Takino T, Itoh M, Akizawa T, Seiki M. Processing of a precursor of 72-kilodalton type IV collagenase/gelatinase A by a recombinant membrane-type 1 matrix metalloproteinase. Cancer Res. 1996;56:2535–8. [PubMed] [Google Scholar]
- 25.Hotary KB, Allen ED, Brooks PC, et al. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhai Y, Hotary KB, Nan B, et al. Expression of membrane type 1 matrix metalloproteinase is associated with cervical carcinoma progression and invasion. Cancer Res. 2005;65:6543–50. doi: 10.1158/0008-5472.CAN-05-0231. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–60. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 28.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 29.Berdeaux RL, Diaz B, Kim L, Martin GS. Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J Cell Biol. 2004;166:317–23. doi: 10.1083/jcb.200312168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandilands E, Cans C, Fincham VJ, et al. RhoB and actin polymerization coordinate Src activation with endosome-mediated delivery to the membrane. Dev Cell. 2004;7:855–69. doi: 10.1016/j.devcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Sandilands E, Brunton VG, Frame MC. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J Cell Sci. 2007;120:2555–64. doi: 10.1242/jcs.003657. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang M, Sun J, Chien S, Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc Natl Acad Sci U S A. 2008;105:14353–8. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quaranta V. Cell migration through extracellular matrix: membrane-type metalloproteinases make the way. J Cell Biol. 2000;149:1167–70. doi: 10.1083/jcb.149.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carman CV, Springer TA. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr Opin Cell Biol. 2003;15:547–56. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Hauck CR, Hsia DA, Puente XS, Cheresh DA, Schlaepfer DD. FRNK blocks v-Src-stimulated invasion and experimental metastases without effects on cell motility or growth. Embo J. 2002;21:6289–302. doi: 10.1093/emboj/cdf631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauck CR, Hsia DA, Schlaepfer DD. The focal adhesion kinase--a regulator of cell migration and invasion. IUBMB Life. 2002;53:115–9. doi: 10.1080/15216540211470. [DOI] [PubMed] [Google Scholar]
- 37.Guegan F, Tatin F, Leste-Lasserre T, et al. p190B RhoGAP regulates endothelial-cell-associated proteolysis through MT1-MMP and MMP2. J Cell Sci. 2008;121:2054–61. doi: 10.1242/jcs.025817. [DOI] [PubMed] [Google Scholar]
- 38.Varon C, Tatin F, Moreau V, et al. Transforming growth factor beta induces rosettes of podosomes in primary aortic endothelial cells. Mol Cell Biol. 2006;26:3582–94. doi: 10.1128/MCB.26.9.3582-3594.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatin F, Varon C, Genot E, Moreau V. A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J Cell Sci. 2006;119:769–81. doi: 10.1242/jcs.02787. [DOI] [PubMed] [Google Scholar]
- 40.Buschman MD, Bromann PA, Cejudo-Martin P, et al. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol Biol Cell. 2009;20:1302–11. doi: 10.1091/mbc.E08-09-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moissoglu K, Schwartz MA. Integrin signalling in directed cell migration. Biol Cell. 2006;98:547–55. doi: 10.1042/BC20060025. [DOI] [PubMed] [Google Scholar]
- 42.Pertz O, Hahn KM. Designing biosensors for Rho family proteins--deciphering the dynamics of Rho family GTPase activation in living cells. J Cell Sci. 2004;117:1313–8. doi: 10.1242/jcs.01117. [DOI] [PubMed] [Google Scholar]
- 43.Buranachai C, Kamiyama D, Chiba A, Williams BD, Clegg RM. Rapid frequency-domain FLIM spinning disk confocal microscope: lifetime resolution, image improvement and wavelet analysis. J Fluoresc. 2008;18:929–42. doi: 10.1007/s10895-008-0332-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.