Abstract
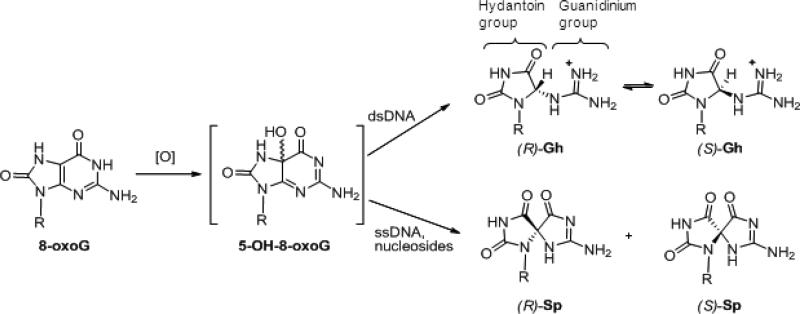
The oxidation of guanine generates one of the most common DNA lesions, 8-oxo-7,8-dihydroguanine (8-oxoG). The further oxidation of 8-oxoG can produce either guanidinohydantoin (Gh) in duplex DNA or spiroiminodihydantoin (Sp) in nucleosides and ssDNA. Although Gh can be a strong block for replicative DNA polymerases such as RB69 DNA polymerase this lesion is also mutagenic: DNA polymerases bypass Gh by preferentially incorporating a purine with a slight preference for adenine, which results in G•C → T•A or G•C → C•G transversions. The 2.15 Å crystal structure of the replicative RB69 DNA polymerase in complex with DNA containing Gh reveals that Gh is extrahelical and rotated toward the major groove. In this conformation Gh is no longer in position to serve as a templating base for the incorporation of an incoming nucleotide. This work also constitutes the first crystallographic structure of Gh, which is stabilized in the R configuration in the two polymerase/DNA complexes present in the crystal asymmetric unit. In contrast to 8-oxoG, Gh is found in a high syn conformation in the DNA duplex and therefore presents the same hydrogen bond donor and acceptor pattern as thymine, which explains the propensity of DNA polymerases to incorporate a purine opposite Gh when bypass occurs.
One of the most prevalent and common lesions in DNA is the oxidation product of guanine, 8-oxo-7,8-dihydroguanine (8-oxoG). This lesion is mutagenic due to its propensity to form a Hoogsteen base pair with adenine (8-oxoG•A), which leads to a G•C to T•A transversion after replication. Because of its lower oxidation potential, 8-oxoG is much more prone to oxidation than guanine (1). The further oxidation products of 8-oxoG are guanidinohydantoin (Gh) and the two stable stereoisomers of spiroiminodihydantoin (Sp1 and Sp2) (Figure 1) (2-4). Gh is the principal hydantoin product of 8-oxoG oxidation in duplex DNA, while Sp is more common in nucleoside oxidation (5). Sp has been detected in genomic DNA in Escherichia coli cells exposed to chromate, which supports the biological relevance of the further oxidation products of 8-oxoG (6). The biological effects of Gh and Sp have been extensively studied in vitro and in vivo (7-10). Gh-containing DNA can be bypassed by E. coli DNA pol I, which inserts dAMP or dGMP opposite the lesion (7). In contrast, Gh and Sp constitute strong replication blocks for Pol α (calf thymus) and human pol β (7).
Figure 1.
Oxidation products of guanine. 8-oxoguanine is the oxidation product of guanine, and its further oxidation in vitro can yield diastereomers of guanidinohydantoin (Gh) or spiroiminodihydantoin (Sp) (8). (R) and (S)-Gh diastereomers are slowly interconvertible, whereas Sp diastereomers have stable configurations.
In vivo mutagenesis assays have shown that Gh and Sp are both highly mutagenic, leading to a mixture of G•C to T•A and G•C to C•G transversions (8, 11). In the first of these studies, the Gh lesion in the sequence context 5’-TXG-3’ (X = Gh) showed high lesion bypass (~75%) in E. coli and the resulting mutation was 98% G•C to C•G. Furthermore, studies with E. coli DNA pol I (Klenow exo-) showed that dGMP incorporation opposite Gh was the most efficient compared to any other hydantoin:nucleotide pair. Taken together, these initial studies suggested that a base pair could be forming between the Gh lesion and an incoming dGMP during replication. This prompted us to pursue crystallographic studies of the Gh lesion in a template DNA strand bound to a replicative DNA polymerase in order to examine the orientation of the Gh lesion during nucleotide insertion opposite the lesion.
Here we describe the first crystal structure of a replicative DNA polymerase bound to Gh. The structure of RB69 DNA polymerase in complex with Gh reveals why this oxidized DNA lesion is a strong block for most DNA polymerases. Moreover, this 2.15 Å structure is also the first structure of Gh, which was found to be in the R configuration.
EXPERIMENTAL PROCEDURES
Enzymes and oligonucleotides
The exonuclease deficient variant (D222A and D327A) of RB69 gp43 (RB69 exo-) was expressed and purified as previously described (12). The K279A RB69 exo- variant was obtained by creating a point mutation using the QuikChange XL kit (Stratagene). The resulting construct was sequenced in its entirety prior to expression and purification (12).
The 13-mer oligonucleotide primer (5’-GCGGACTGCTTAA) was purchased from The Midland Certified Reagent Co. (Midland, TX) and was purified on a 16% (w/v) polyacrylamide gel containing 8 M urea and de-salted on a Sep-Pak (Millipore) cartridge. The 18-mer DNA template (5’-ACT(Gh)TTAAGCAGTCCGCG with Gh=guanidinohydantoin) was synthesized as described previously (13). The 18-mer template was annealed to the complementary 13-mer primer by mixing the two strands in a buffer containing 10 mM Tris-HCl (pH 7.5) and 50 mM NaCl, heating to 90°C for 5 min, and slowly cooling to room temperature. The primer used in the primer extension assays was 5’-labeled with tetrachlorofluorescein.
Primer extension assays
In a volume of 10 μL, 200 nM enzyme (either RB69 exo- or K279A RB69 exo-) and 100 nM duplex oligonucleotide were pre-incubated in a buffer containing 35 mM NaCl, 10 mM Hepes pH 7.5 and 2.0 mM DTT. The reaction was started by adding 10 μl of solution containing 20 mM MgCl2 and 250 μM of either individual dNTPs or a dNTP mixture. The final concentrations in the reaction mixture were: 100 nM enzyme, 50 nM oligonucleotides, 125 μM dNTP, and 10 mM of MgCl2. At the indicated time, 10 μl aliquots were quenched with 10 μL of formamide. Extended primers were separated from the template on 16% polyacrylamide gels containing 8 M urea. DNA bands were visualized on a Bio-Rad (Hercules, CA) Molecular Imager FX at the Alexis 532 setting to excite the tetrachlorofluorescein label.
Crystallization
The exonuclease deficient DNA polymerase (RB69 exo-) was mixed in an equimolar ratio (0.1 mM) with annealed primer template DNA and 10 mM dideoxyATP (Sigma-Aldrich). Hanging drops were made by mixing 0.5 μL of reaction mixture and 0.5 μL of reservoir solution [12% (v/v) PEG 2000 MME, 100 mM NaCl, 150 mM MgSO4, 100 mM Hepes (pH 7.0), 2 mM β-mercapto-ethanol, and 6% (v/v) glycerol] and equilibrated against 1 mL of reservoir solution at 20°C. Crystals (160×40×40 μm3) grew within two weeks and were found to belong to space group P21 with unit cell parameters a = 78.73 Å, b = 120.25 Å, c = 139.96 Å, and β = 95.96°. The cell parameters are substantially smaller than those of the P21 binary complexes previously published (12, 14, 15) especially for a and b, which are usually ~130 Å and ~165 Å, respectively. As a result, the crystal comprises two molecules per asymmetric unit compared to the four observed previously, with an estimated solvent content of 65%. The crystals were cryoprotected by increasing the concentration of PEG 2000 MME and glycerol to 15% and 18%, respectively and flash-cooled into liquid nitrogen.
Data collection
X-ray data were collected at beamline 23-ID-B of the Advanced Photon Source at the Argonne National Laboratory on a MAR m300 CCD detector. One complete 2.15 Å data set was collected at 100 K at λ = 1.03321 Å. The data set was processed and scaled using HKL 2000 (16). Data collection statistics are reported in Table I.
Table 1.
Data collection and refinement statistics.
| Gh Complex | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 78.25, 120.16, 139.91 |
| β (°) | 95.95 |
| Resolution (Å) | 50-2.15 (2.23-2.15)* |
| R merge | 12.6 (70.7)* |
| I / σI | 9.09 (2.0)* |
| Completeness (%) | 99.9 (99.5)* |
| Redundancy | 4.3 (4.0)* |
| Refinement | |
| Resolution (Å) | 50-2.15 |
| No. reflections | 583223 (137162)** |
| Rwork / Rfree (%) | 21.90 / 25.45 |
| No. atoms | 16717 |
| Protein | 14527 |
| DNA | 1182 |
| Water | 998 |
| Average B-factors (Å2) | |
| Protein (A, B) | 26.7, 29.8 |
| DNA (A, B) | 34.7, 36.9 |
| Water | 32.2 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.0062 |
| Bond angles (°) | 1.208 |
Highest resolution shell is shown in parentheses.
Number of all reflections. Number of unique reflections is in parentheses.
Structure determination and refinement
The previously determined structure of RB69 gp43 in a ternary complex with undamaged DNA (17) (PDB ID code 1IG9) devoid of all non-protein atoms was used as a model for molecular replacement using Phaser (18). Rigid body refinement was performed on the solution obtained (RFZ = 36.2, TFZ = 76.6 and LLG = 10182). The structure refinement consisted of cycles of model building with COOT (19), followed by positional refinement and individual B factor refinement with CNS 1.21 (20, 21). The Gh parameter and topology files used for refinement in CNS were generated by the Dundee PRODRG2 server (22). The Rfree value was calculated using 10% of the reflections that were set aside during refinement. Water molecules were added with COOT during the last refinement cycles.
The final model includes residues 1 to 903 for molecules A and B. Residues 254 to 260 (molecule A) and 253 to 259 (molecule B) were disordered in the electron density map and were therefore not built into the model. These residues belong to a β-hairpin located in the exonuclease domain, which was shown to be very flexible (23). The first two bases at the 5’-end of the DNA template are missing in both protein/DNA complexes. The quality of the model was assessed with PROCHECK (24): All non-glycine residues are found in the allowed regions of the Ramachandran plot except Thr622 (in both molecules A and B), a residue that adopts a distorted geometry because of its proximity to the active site aspartates Asp621 and Asp623 (12, 14, 15, 17, 23). No density was observed for the C-terminal tri-histidine tag in either molecule. Refinement statistics are found in Table 1. Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank (www.pdb.org) and are available under the 3L8B accession code. Figures 3-6 were drawn using PyMOL (25).
Figure 3.
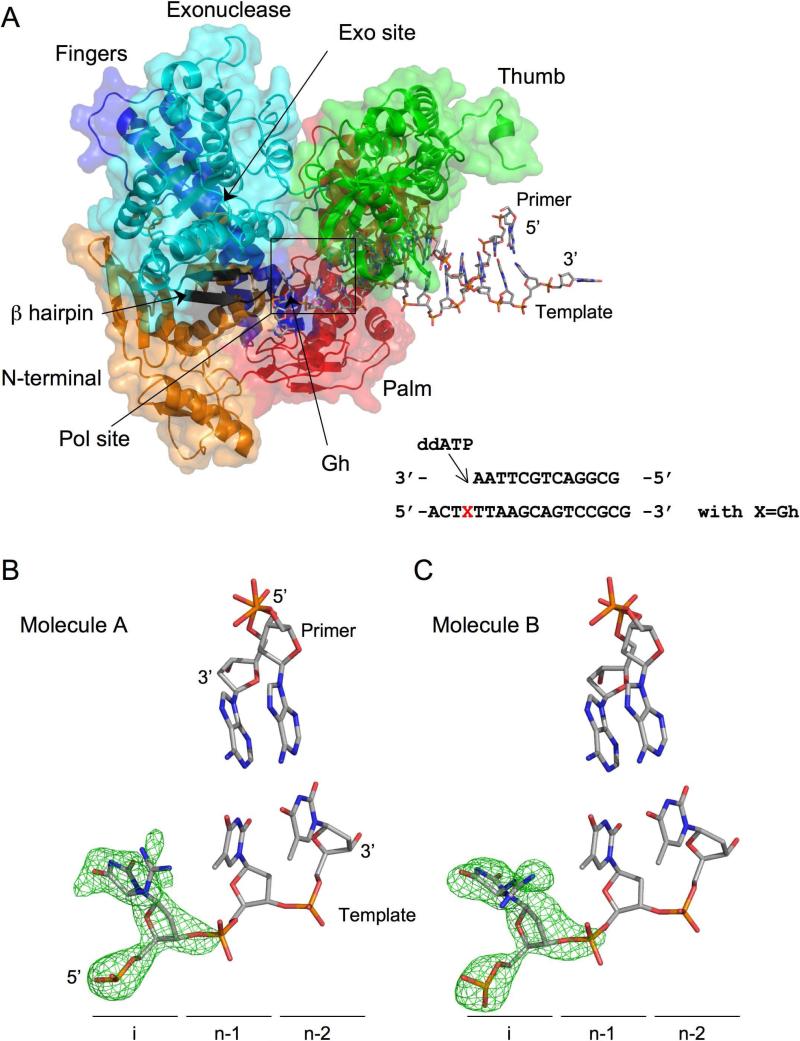
A) Overall view of RB69 gp43 exo- in complex with Gh containing DNA (Only one of the two complexes per asymmetric unit is shown). The DNA (shown as a stick model) lies in the polymerase active site (pol). The polymerase domains are colored in red for the palm, green for the thumb, blue for the fingers, cyan for the exo domain with the β hairpin in black and orange for the N-terminal domain. The sequence of the DNA oligonucleotide is shown.
Close up view of Gh in the polymerase active site in the same orientation as A). Simulated annealing omit maps centered on the Gh lesion and contoured at 4.0 σ (B) and 3.0 σ (C) are shown (green). Although Gh is observed in the R configuration in both cases, the position of the guanidinium group above the hydantoin group differs in molecules A and B.
Figure 6.
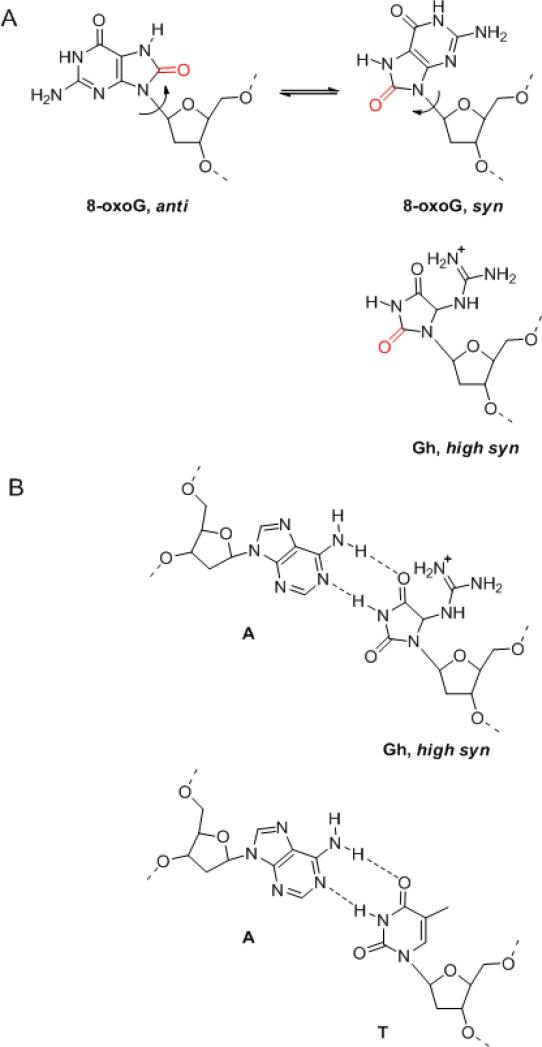
A) Diagram of 8-oxoguanine in the anti and syn conformations and Gh in high syn conformation. The oxygen at the 8 position is highlighted in red. B) Diagram of a regular Watson Crick T•A base pair, and possible Gh•A base pair, where Gh is in a high syn conformation. The hydrogen bonds are shown as black dashed lines.
RESULTS
Gh is a strong block to the RB69 DNA polymerase in vitro
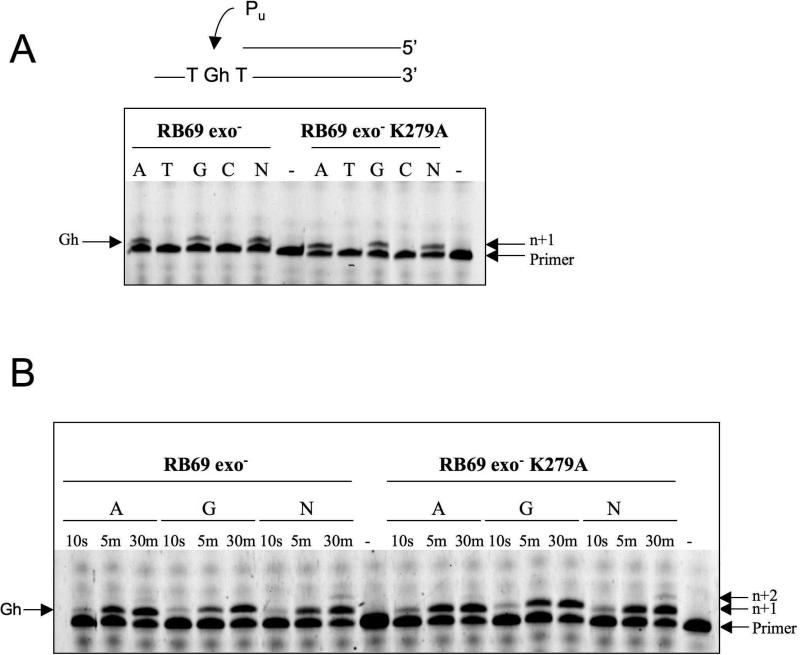
Earlier work from other groups has established that DNA polymerases differ in their propensity to bypass Gh: It constitutes a strong block in vitro for both calf thymus DNA pol α (a B family polymerase) and human DNA pol β (X family) but not for Klenow fragment (A family) (7, 9, 10). In vivo, Escherichia coli DNA polymerase can efficiently bypass the Gh lesion in the sequence context 5’-TXG-3’ (8). However, a later study, also by the Essigmann laboratory, examined Gh in the 5’-GXA-3’ sequence context, and the extent of lesion bypass was only ~20% (11). In addition, the mutation type changed from 98% GC in the first study to a 57:40 mixture of G•C and G•T in the second. We performed a primer extension assay of the replicative DNA polymerase from bacteriophage RB69 (RB69 gp43), a member of the B family, with DNA containing Gh at the templating position. Because RB69 gp43 has a potent proofreading activity, an exonuclease deficient variant was used (D222A, D327A). The results indicate that Gh constitutes a very strong pause site for RB69 DNA pol exo-: The polymerase was able to incorporate a modest amount of purine opposite Gh after 5 min (Figure 2a; left panel, lanes A and G). Moreover, we observed a difference between adenine and guanine incorporation: RB69 gp43 exo- can incorporate dATP opposite Gh slightly faster than dGTP (Figure 2b; left panel, lanes A and G at 5 minutes). Incorporation of pyrimidines was undetectable in our assay. When a mixture of dNTPs is added, the primer is not fully extended and the polymerase stops after adding 1 or 2 bases (Figure 2a,b; left panel, lane N). These results are consistent with the results previously reported for DNA pol α, also a polymerase of the B family (7).
Figure 2.
Gh is a block to replication but when bypass occurs a purine (Pu) is incorporated opposite Gh. A) Primer extension assay with each nucleotide dATP (A), dTTP (T), dGTP (G), dCTP (C) or a mix of the four nucleotides (N), all with an incubation time of 5 min. RB69 exo- and RB69 exo- K279A stall after incorporation of A or G opposite Gh. B) In this primer extension assay the incubation time was varied from 10 seconds to 30 minutes with dATP, dGTP or the four nucleotide mix (N).
Overall structure description of RB69 gp43 exo- in complex with Gh-containing DNA
An 18-mer oligodeoxynucleotide containing a single Gh lesion was synthesized and purified by ion-exchange HPLC according to previous protocols (9). Synthesis produces the Gh lesion as a mixture of two diastereoisomers with configurations differing only at the guanidinium-bearing carbon of the hydantoin ring. In nucleoside studies, we found that the Gh diastereomers interconverted, presumably via enolization, in a matter of hours after initial isolation (4, 26). Therefore, the oligomers used in crystallographic studies contained a diastereomeric mixture.
We obtained a stable complex of DNA pol RB69 gp43 exo- with Gh-containing DNA by mixing the DNA polymerase with annealed primer/template and ddATP in the presence of magnesium. Crystallization trays were also set up with dATP, but in the resulting crystals the DNA was found in the exonuclease active site. RB69 gp43 has two active sites, a polymerase and exonuclease active site where proofreading takes place in the event of a mispair. Different factors can affect the equilibrium of the primer DNA between the two sites, such as the concentration of the incoming nucleotide or the presence of a DNA lesion. Crystals with DNA in the exonuclease site revealed that one dAMP had been incorporated prior to the DNA switching to the proofreading site. The structure was not further analyzed because the Gh lesion on the template strand was disordered and not visible in the electron density map.
Diffraction data on the complex with ddATP were collected to 2.15 Å resolution at the Advanced Photon Source National Laboratory (Argonne, IL). The structure was solved by molecular replacement using the previously solved ternary complex (PDB ID: 1IG9) of RB69 gp43 exo- with undamaged DNA devoid of all non-protein atoms (17). The structure was refined to Rwork and Rfree values of 22.05% and 25.54%, respectively (Table 1). The crystals grew in the same space group (P21) as previously reported for other binary complexes (12, 14, 15, 23). The unit cell dimensions, however, are smaller and the crystal asymmetric unit contains two polymerase-DNA complexes instead of four (Figure 3a). Our work and that of others has shown that the DNA can be found in the polymerase or exonuclease active site depending on the nature of the lesion (12, 15, 27). In the two protein/DNA complexes in the crystal asymmetric unit (A and B), the DNA duplex is located in the polymerase site (Figure 3a). The two complexes are very similar with an overall rms deviation of ~ 0.7 Å (on Cα). In both polymerase molecules the fingers domain adopts an open conformation, as expected for a binary complex (12, 14, 15, 28). We compared the Gh complex with two other binary complexes with DNA containing either tetrahydrofuran (THF) (PDB ID code 2P5O, monomer C) (12) or thymine glycol (Tg) (PDB ID code 2DY4) (12, 14) and found that the overall orientation of the subdomains in the Gh complex is very similar to that described for the THF and Tg complexes.
Structure of Gh in the polymerase active site
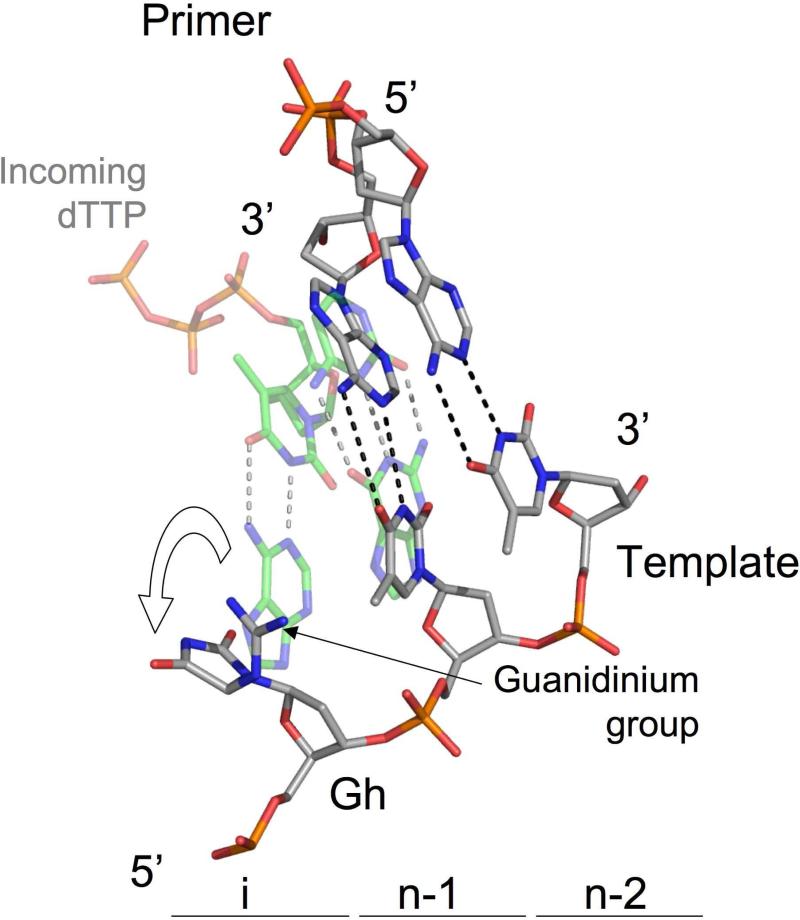
The Gh complex crystals comprise two independent RB69 exo-/DNA copies per asymmetric unit. In the two molecules we observe only the (R) isomer of Gh. Gh comprises two moieties, a hydantoin and a guanidinium group (Figure 1). The hydantoin moiety is well defined in the residual Fo-Fc electron density map (Figure 3b,c). The guanidinium group, on the other hand, is more flexible and can adopt two different conformations (Figure 3b,c), as seen in molecules A and B. This flexibility is illustrated by the fact that the residual map around the guanidinium group is not as well-defined as around the hydantoin group. Gh is non-planar and the guanidinium group is almost perpendicular to the plane of hydantoin group, preventing the adjacent 3’-thymine from stacking with the oxidized base. The guanidium group of Gh makes a van der Waals contact with the 5-methyl group of the 3’-thymine instead. In both molecules, Gh is extrahelical and rotated toward the major groove and is thus no longer in position to serve as a templating base for the incorporation of an incoming nucleotide (Figure 4). In fact, in the Gh complex, there was no visible electron density for either an incoming ddATP or incorporated ddAMP opposite Gh despite the high concentration of nucleotide (10 mM) used in the crystallization buffer (Figure 3b,c).
Figure 4.
Close up view of Gh with surrounding bases. Superposition of the Gh complex (light gray) with an incipient base pair (A•dTTP) from an RB69 gp43 complex with normal DNA (17) (green; PDB ID code 1IG9). The adenine (green) overlaid on the Gh structure (light gray) illustrates that Gh is rotated out of the DNA helix and the displacement is such that Gh is no longer in position to serve as templating base for incoming dNTP. The adjacent T•A base pair in the post insertion site (n-1) shows a high buckle angle. The hydrogen bonds are show as black dashed line in the Gh complex and gray dashed lines in the complex with normal DNA.
As mentioned above, the guanidinium group induces a displacement of the adjacent 3’ thymine, which affects its interaction with the opposite adenine in the primer strand (Figure 4, black dashed lines). The hydrogen bond on the major groove side of the base pair between O4 of thymine (template) and N6 of adenine (primer) is significantly longer than a typical H bond in a Watson Crick base pair (distance of 3.5Å and 3.4Å for molecules A and B, respectively, vs. 2.8-3.0 Å). The T•A base pair also displays a buckle angle (κ) with a high value (29) (-21.3° and -16.5° for molecules A and B, respectively) compared to the small buckling angle (a few degrees) usually seen in DNA (30), indicating that the presence of Gh in the insertion site perturbs the adjacent base pair.
Interaction between Gh and Lys 279
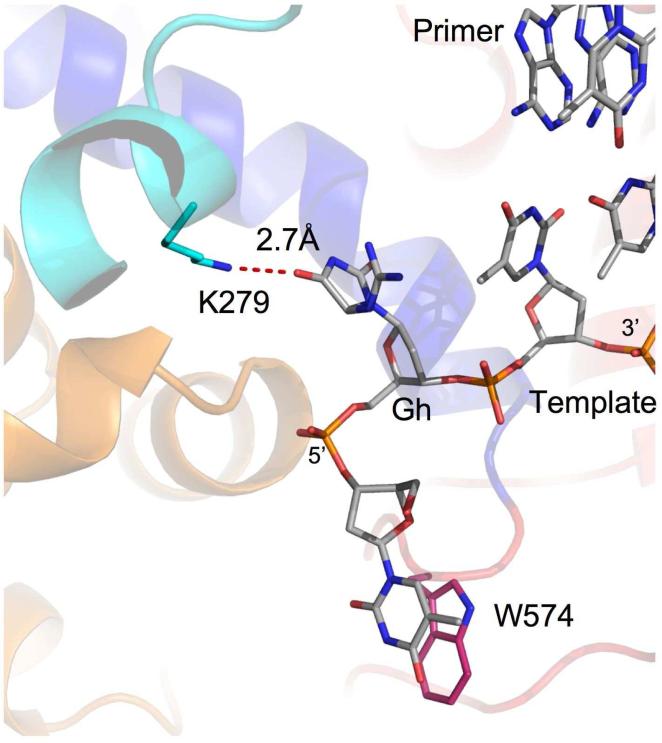
In the two polymerase/DNA complexes in the asymmetric unit, we observe only one direct interaction of the polymerase with Gh, a hydrogen bond between the amino group of lysine 279 and a carbonyl group (CO5) of Gh (Figure 5a,b), with a distance of 2.7 Å and 2.6 Å for molecules A and B, respectively. Lysine 279, which is always either a lysine or an arginine in B family DNA polymerases, is located in the exonuclease domain.
Figure 5.
Interactions of Gh with RB69 exo-. Gh (light gray) forms a hydrogen bond (red dashed line) with the side chain of lysine 279 (cyan, exo domain). The thymine 5’ of Gh is stabilized via a stacking interaction with Trp574, as observed previously (14, 15, 23).
In order to assess the possible role of lysine 279 in stabilizing Gh in the observed conformation we mutated lysine 279 to an alanine (K279A). As a control, we tested the activity of the RB69 exo- K279A variant with undamaged DNA and found that the mutation does not affect primer elongation with normal DNA (data not shown). We then compared the incorporation of all four dNTPs opposite Gh with the RB69 gp43 exo- and RB69 gp43 exo- K279A (Figure 2a). The K279A variant preferentially incorporated a purine opposite Gh, just like RB69 gp43 exo- and elongation stopped after one purine addition. The kinetics of purine incorporation (Figure 2b) does not differ from RB69 gp43 exo-. Our results indicate that the K279A mutation does not affect the enzyme's ability to elongate past Gh, and thus the hydrogen bond between Gh and lysine 279 is unlikely to play a key role in stabilizing Gh in a conformation that is incompatible with bypass.
DISCUSSION
Our work describes the first crystal structure of a replicative DNA polymerase (RB69 gp43) in complex with DNA containing guanidinohydantoin. In fact, it is the first crystal structure for guanidinohydantoin itself. Gh is the oxidation product of 8-oxo-7,8-dihydroguanine (4) and it seems relevant to compare the two lesions in the context of the RB69 gp43 polymerase active site. The structure of a ternary complex of RB69 gp43 exo- with dCTP opposite 8-oxoG was solved by Kisker and collaborators (28). This structure shows that 8-oxoG is in an anti conformation when cytosine is incorporated. RB69 and T4 polymerases can easily bypass this lesion (28) and preferentially add a cytosine. When 8-oxoG is in syn conformation, it can form a Hoogsteen base pair with adenine (31, 32). A major difference between Gh and 8-oxoG is that Gh is non-planar and adopts preferentially a high syn conformation in DNA (Figure 6a) (33). The O4’-C1’-N9-C4 torsion angles are 105° and 118° for molecules A and B, respectively (19). A decrease in the torsion angle from 105°/118° (high syn) to around -100° (anti) would result in a steric clash with Ser565 and Pro361.
Gh is poorly bypassed by RB69 DNA polymerase and the crystal structure presented here provides a rationale: Gh is extrahelical and no longer in a templating position. When bypass does occur the lesion is highly mutagenic (5): RB69 gp43 preferentially incorporates a purine opposite Gh, like pol η (34), albeit with a slight preference for adenine (Figure 2a,b), which would result in G•C → T•A or G•C → C•G transversions. Since Gh is positioned in the crystal in such a way it can no longer act as a template for incorporation of an incoming nucleotide, how can we explain the incorporation of dATP and dGTP opposite the lesion (Figure 2)? As mentioned earlier, it is not possible to study the two stereoisomers of Gh separately in solution because of slow isomerization (4). In the crystal, on the other hand, only the R configuration was captured. The R configuration is not compatible with nucleotide addition, and that happens to be the one that crystallized. In solution, the two configurations coexist and the S configuration might be better positioned for incorporation. In contrast to the R isomer, the S isomer of Gh in high syn conformation would fit well within the DNA duplex and could serve as a templating base. When Gh adopts a high syn conformation, the hydantoin group mimics a pyrimidine ring and displays the same hydrogen bond donor/acceptor pattern as a thymine (Figure 6a,b), which explains why purines are preferentially incorporated opposite this lesion (35). Only one addition would occur though because the protruding guanidinium group would prevent the stacking of the next 5’ template base. Significant differences have also been observed between the two isomers of the other hydantoin lesion, Sp: In this case the R and S stereoisomers are stable and can be separated, and differences in enzyme activities have been reported for polymerases, DNA glycosylases, and exonucleases (9, 36, 37).
RB69 gp43 harbors two activities, polymerase and exonuclease, and when the DNA polymerase senses a mispair, the DNA primer switches from the polymerase site to the exonuclease site, where the primer 3’-end is degraded (38). In the Gh complex structure, there are two molecules per asymmetric unit and both have the DNA in the pol site. The presence of the lesion in the insertion site (Figure 3) does not disturb the minor groove interactions of the base pair in the n-1 post-insertion site with Tyr567 and Lys706, two conserved residues which have been shown to contact the minor groove side of the base pairs at the n-1 and n-2 post-insertion positions and have been posited to detect mispairs (12, 17). The minor groove interactions are maintained, in spite of displacement of the adjacent 3’-thymine (n-1 post-insertion) towards the major groove and the DNA therefore remains in the polymerase site. In the complex obtained with dATP, on the other hand, the primer strand found in the exonuclease site included an additional dAMP at its 3’-end, indicating that the DNA polymerase had incorporated one dAMP prior to switching the primer strand from the polymerase to the exonuclease active site. This finding implies that the Gh•dAMP had been sensed as a mispair. This is consistent with our primer extension assays where we observed at most one base extension past Gh (Figure 2).
Since the presence of the Gh lesion perturbs the adjacent T•A base pair (high buckle angle, longer H-bonds between the two bases) we investigated the conformation of the β hairpin loop (residues 248-265, from the exonuclease domain, depicted in black in Figure 3A). This loop was shown to play a role in maintaining the stability of polymerase/DNA interactions as the DNA primer migrates to the exonuclease active site for proofreading (23) and also facilitates strand separation (39). The β hairpin has been captured in two conformations in crystals (SI Figure 1), “up” and “down”. The “up” conformation is generally observed in closed, ternary complexes with the proper incoming nucleotide and undamaged DNA (17, 28) whereas the “down” conformation is routinely observed in binary complexes with DNA containing a lesion (12, 14). In the latter conformation β hairpin residues make close contact with the single stranded 5’-end of the template (12) and these contacts are lost in the “up” conformation. We superimposed the structure of the Gh-containing DNA complex with those of a closed ternary complex with normal DNA (PDB ID: 1IG9) (17), a binary complex with DNA containing a THF•A pair (12) and a binary complex with DNA containing a Tg•A pair (14) using SSM (40). In all these structures the DNA is located in the polymerase active site. Even though the tip of the β hairpin is disordered in the electron density map the loop appears to be in the “up” conformation in the Gh complex. The T•A base pair in the n-1 post-insertion site maintains minor groove contacts with the polymerase, and is therefore sensed as a normal base pair despite the perturbations induced by the neighboring Gh.
In summary, the binary complex of RB69 DNA polymerase with Gh-containing DNA sheds light on the nature of the strong block to replication induced by the presence of Gh in the template strand. Our crystal structure revealed that Gh is extrahelical and rotated towards the major groove and, in that conformation, the incorporation of the incoming nucleotide opposite Gh is hindered. This work also represents the first crystallographic structure of Gh, which was found in the R configuration. Although Gh is a strong block for RB69 gp43, bypass does happen. In this case, it is probable that the isomerization of Gh to the S configuration would allow nucleotide incorporation opposite the lesion. In the high syn conformation, the hydantoin group mimics a pyrimidine ring and displays the same hydrogen bond donor/acceptor pattern as a thymine, which explains why purines are preferentially incorporated opposite this lesion.
Supplementary Material
Acknowledgements
We thank Wendy Cooper for help with protein purification, Dr. Matt Hogg for the K279A variant, Dr. Stéphanie Duclos for assistance with the primer extension assays, and Dr. Matt Hogg and Karl Zahn for critically reading the manuscript. We thank Stephen Corcoran, Dr. Ward Smith, and Dr. Ruslan Sanishvili for help with data collection at the GM/CA CAT beamline at the APS synchrotron.
The beamline GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advance Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, under contract No. DE-AC02-06CH11357. This work was supported by NIH grants CA52040 to SD and CA90689 to CJB.
ABBREVIATIONS
- Gh
guanidinohydantoin
- PEG 2,000 MME
Polyethylene glycol 2,000 monomethylether
- RB69 gp43 exo-
exonuclease deficient bacteriophage RB69 DNA polymerase (D222A/D327A)
- Sp
spiroiminodihydantoin
- Tg
thymine glycol
- THF
tetrahydrofuran
Footnotes
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank (www.pdb.org) and are available under the 3L8B accession code.
Supporting Information: A superposition of the β hairpin loop from four different RB69 gp43 DNA complexes is shown in Supporting Figure 1. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Steenken S, Jovanovic SV. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 2.Luo W, Muller JG, Burrows CJ. The pH-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2001;3:2801–2804. doi: 10.1021/ol0161763. [DOI] [PubMed] [Google Scholar]
- 3.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-Oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 4.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 5.Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals. Environ Health Perspect. 2002;110(Suppl 5):713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 9.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 10.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 11.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 12.Hogg M, Wallace SS, Doublié S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Muller JG, Burrows CJ. Synthesis and characterization of the oxidized dGTP lesions spiroiminodihydantoin-2'-deoxynucleoside-5'- triphosphate and guanidinohydantoin-2'-deoxynucleoside-5'- triphosphate. J Org Chem. 2006;71:2181–2184. doi: 10.1021/jo052484t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aller P, Rould MA, Hogg M, Wallace SS, Doublié S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci U S A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahn KE, Belrhali H, Wallace SS, Doublié S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and Development of Coot. Acta Crystallogr D Biol Crystallogr. 2010 doi: 10.1107/S0907444910007493. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 23.Hogg M, Aller P, Konigsberg W, Wallace SS, Doublié S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. Delano Scientific, Palo Alto, California, USA. 2008 http://www.pymol.org.
- 26.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of 13C, 15N, and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J. Am. Chem. Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Arnaiz P, Lazaro JM, Salas M, de Vega M. Involvement of phi29 DNA polymerase thumb subdomain in the proper coordination of synthesis and degradation during DNA replication. Nucleic Acids Res. 2006;34:3107–3115. doi: 10.1093/nar/gkl402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids: Structures, Properties, and Functions. University Science Books; Sausalito, CA: 2000. [Google Scholar]
- 31.Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 33.Saenger W. Principles of nucleic acid structure. Springer-Verlag; New York, NY: 1995. [Google Scholar]
- 34.Kino K, Ito N, Sugasawa K, Sugiyama H, Hanaoka F. Translesion synthesis by human DNA polymerase eta across oxidative products of guanine. Nucleic Acids Symp Ser (Oxf) 2004:171–172. doi: 10.1093/nass/48.1.171. [DOI] [PubMed] [Google Scholar]
- 35.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 36.Chen X. Ph. D. dissertation. University of Utah; 2008. DNA nuclease activity towards oxidized guanine lesions. [Google Scholar]
- 37.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steitz TA, Yin YW. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2004;359:17–23. doi: 10.1098/rstb.2003.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trzemecka A, Plochocka D, Bebenek A. Different behaviors in vivo of mutations in the beta hairpin loop of the DNA polymerases of the closely related phages T4 and RB69. J Mol Biol. 2009;389:797–807. doi: 10.1016/j.jmb.2009.04.055. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.