Abstract
Macrophages are major HIV target cells. They support both productive and latent HIV-1 infection. Susceptibility of primary macrophages to HIV depends on the anatomical location and activation state of the cells. We demonstrate that peritoneal macrophages (PMs) are abundant in ascitic fluid of patients with liver cirrhosis and are susceptible to HIV-1 infection. PMs expressed CD68, a differentiation marker, exhibited phagocytic activity, and survived in culture for 2 months without additional growth factors. Freshly-isolated PMs were susceptible to HIV-1 R5 strains but not to X4-T cell line adapted (TCLA) strains. Interestingly, after 7 days in culture, PMs acquired susceptibility to X4-TCLA strains. HIV entry inhibitors, TAK779 and AMD3100, blocked HIV infection of PMs, indicating that infection by R5 and X4 strains was mediated by CCR5 and CXCR4, respectively. Although PMs did not express detectable cell surface levels of CXCR4 and CCR5, they did express mRNAs of these HIV co-receptors and responded to stimulation by their natural ligands, SDF-1α and RANTES. PMs were susceptible to HIV-1 X4, R5, and X4R5 primary isolates. PMs after 7 days in culture produced greater amounts of X4 and X4R5 HIV than freshly-isolated PMs. The day-7 PMs were more susceptible to R5 infection in a single-cycle infection assay, but there was no increase in viral production in a multiple-round infection assay. The level of CXCR4 mRNA and production of CC-chemokines (MIP-1α, MIP-1β and RANTES) increased significantly during 7 days in culture. Our results indicate that PMs are susceptible to receptor-mediated infection by a broad range of HIV strains. These primary macrophages could provide a valuable system for investigating the role of primary macrophages in HIV pathogenesis.
Keywords: Human immunodeficiency virus (HIV-1), peritoneal macrophages, CCR5, CXCR4
Introduction
Macrophages are one of the major cell types in which HIV can both productively replicate and persist in a latent state 1. Macrophages play an important role in HIV-1 pathogenesis because of their ability to generate progeny virions, resistance to HIV-induced killing, and long lifespan 1–4. In some settings, tissue macrophages contribute significantly to viral load. High levels of HIV-1 RNA are present in macrophages of rhesus macaques infected by a pathogenic simian immunodeficiency/HIV type 1 chimeric virus (SHIV-1) that eliminates CD4+ T cells 5. Macrophages in liver and brain are thought to be the source of the high levels of HIV-1 DNA that are present in certain AIDS patients with very low CD4+ T cell counts 6, 7. Opportunistic infections, which occur more frequently during the advanced stages of AIDS, increase the amount of HIV produced by macrophage, suggesting that activated macrophages may be a major source of the X4-utilizing variants that often emerge in patients with advanced disease 8, 9. Further studies regarding HIV-infected macrophages are needed, as this information may lead to more effective strategies for HIV eradication 1–4.
The susceptibility of macrophages to HIV-1 infection varies depending on their location in the body and on the strain of the virus. HIV-infected Kupffer cells (hepatic macrophages) are present in patients with AIDS 6, 7, 10, and these cells are susceptible to HIV infection in vitro 11. Macrophages in human tonsils can be infected ex vivo by primary HIV-1 X4 and X4R5 dual tropic viruses but not X4-T cell-line adapted (TCLA) strains 12. Alveolar macrophages are susceptible to R5 and X4R5 primary isolates 13, but intestinal macrophages are not susceptible to HIV-1 infection in vitro 14.
Experimental investigation of the interactions between HIV-1 and macrophages has been impeded by the difficulty of isolating human primary macrophages. Monocyte-derived macrophages (MDMs) are often used to study HIV-1 infection; however, the functional properties of MDMs vary depending on the methods used for isolation and cultivation 15–17, and it is unclear to what extent MDMs model the characteristics of tissue macrophages. MDMs typically express abundant CCR5 and minimal CXCR4 18, 19–21. They can be infected by both primary X4 and X4R5 dual tropic viruses, but not by X4-TCLA strains.
Primary cultures of peritoneal macrophages (PMs) are a potential system for investigating interactions between HIV-1 and primary macrophages. These cells, which reside in the peritoneal cavity, have distinctive properties, including the ability to suppress T-lymphocyte activation 22. PMs from peritoneal fluid of women undergoing diagnostic laparoscopy are known to be susceptible to R5 HIV-1BaL 23; however, the susceptibility of PMs to other HIV strains is unknown and merits investigation. In this study, we developed methods for preparing, culturing, and cryopreserving large numbers of PMs from ascitic fluid of patients with liver cirrhosis. We demonstrate that PMs supported receptor-mediated HIV-1 infection, including that of X4-TCLA strains. HIV production in PMs with X4 and X4R5 primary isolates was enhanced in PMs during 7 days in culture. The level of CXCR4 mRNA was significantly increased in these cultures compared to that in day-1 PMs. Our study indicates that HIV-1 infection of PMs from ascitic fluid provides a novel and useful model for identifying the factors that modulate macrophage susceptibility to HIV-1 infection.
Materials and Methods
Sources of peritoneal macrophages (PMs) and peripheral blood mononuclear cells (PBMCs)
Ascitic fluid (AF) was collected under sterile conditions from patients with liver cirrhosis and refractory ascites who were undergoing therapeutic large volume paracentesis. With approval from the Mount Sinai School of Medicine (MSSM) IRB, subjects gave written consent for medical record review and AF collection. The etiology of cirrhosis was alcoholic liver disease or hepatitis C virus (HCV) infection; subjects did not have bacterial peritonitis. Ascitic mononuclear cells (AMCs) were prepared by centrifugation at 250 xg for 15 min followed by Ficoll-Hypaque gradient centrifugation. AMCs at 10×106/ml were cryopreserved by slow freezing cells in media containing 10% (v/v) DMSO and 90% (v/v) FBS at −80°C overnight, followed by storage in liquid nitrogen. AMCs were recovered from frozen stocks by quick thawing at 37°C and transferring in warm RPMI media with 10% FBS followed by centrifugation to remove DMSO. Cells were plated immediately, as described below.
PMs were prepared by plating AMCs in a 48-well plate at 1×105 cells per well, culturing for 16 h to allow adherence, and vigorously washing four times with PBS to remove non-adherent cells. Cultures were maintained in RPMI with 10% FBS. To deplete CD3 cells from AMCs prior to plating, freshly-isolated AMCs were treated with 10% human serum for 10 min at room temperature, incubated with CD3 magnetic beads (Miltenyi Biotec, Auburn, CA), and passed over a column. The efficiency of CD3 depletion was confirmed by FACS analysis.
PBMCs from normal healthy blood donors were isolated by Ficoll-Hypaque gradient centrifugation. Monocytes were isolated from PBMCs using CD14 isolation kits (Miltenyi Biotec), placed in dishes coated with human serum. Cells were cultured in RPMI with 20% FBS for 10 days and allowed to differentiate into monocyte-derived macrophages (MDMs).
FACS analysis
Adherent cells were incubated at 4°C in ice-cold PBS for 15 min and then detached using cell scrapers. For surface molecule staining, cells were stained with mAbs conjugated with phycoerythrin (PE), allophycocyanin (APC), or fluorescein isothiocyanate (FITC) (BD Pharmingen). For the intracellular staining of CD68, macrophages were fixed with 2% paraformaldehyde and permeablized with 0.2 % saponin before staining. Appropriate isotype controls were included in all assays. Stained samples were analyzed on FACSCalibur flow cytometer using CellQuest (BD). Results were analyzed with FlowJo software (BD).
Immunofluorescence microscopy
AMCs were plated in dishes containing cover slips or in 8-well chamber slides and cultured in RPMI 1640 with 10% FBS overnight followed by extensive washing. Cells were fixed with 2% paraformaldehyde for 20 min. After washing with PBS, cells were permeabilized with 0.5% Triton X-100 for 10 min, blocked with 3% BSA/0.3% Triton X-100 in PBS for 30 min, incubated with appropriate primary antibodies for 1 h, and washed four times with washing solution (0.3% Triton X-100 in PBS) for 10 min per wash. Cells were then incubated with appropriate secondary antibodies for 1 h, washed four times with washing buffer and then mounted with VECTASHIELD® HardSet™ mounting medium with DAPI. For CD68 detection, anti-CD68 antibody (KP1 at 1:300, DAKO) was used followed by a secondary goat anti-mouse antibody conjugated with Texas-Red. For double staining with HIV p24 and CD68, mouse anti-HIV p24 mAb (clone #24-2) from AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, NIH at a dilution of 1:200 and rabbit anti-human CD68 antibody (Santa Cruz Biotech, Santa Cruz, CA) at a dilution of 1:100 were used followed by donkey anti-mouse Alexa-594 and goat anti-rabbit Alexa-488 at a dilution of 1:400. Images were captured using Zeiss Axiophot2 and analyzed with Image J (http://rsb.info.nih.gov/ij).
Phagocytosis
Cells were cultured with FluoSphere carboxylate-modified microspheres (1.0 mm in diameter; Invitrogen, Carlsbad, CA) in RPMI with 10% FBS at 37°C for 2 h, washed with cold PBS, fixed, and permeablized.
HIV-1 infection
For use in single-cycle infection assays, replication-defective HIV-1HxB2 and HIV-1JR-FL Env-pseudotyped reporter viruses expressing luciferase were produced in HEK293T cells, as described previously 24–26. Virus particles at approximately 8 ng of HIV p24 per sample in a 48-well plate were used. Macrophages were exposed to HIV-1JR-FL or HIV-1HxB2 pseudotyped luciferase reporter viruses for 2 h at 37°C. Unbound virus was then removed by washing. Cells were incubated for 48 h, and lysed with Passive Lysis Buffer (Promega Inc, Madision, WI). Luciferase activity was measured using Promega’s luciferase assay systems. In multiple-round infection assays, HIV-1 primary isolates (provided by ARRRP and The UNAIDS Network for HIV-1 Isolation and Characterization, and the DAIDS, NIAID) prepared from PHA-activated PBMCs were incubated with cells for 2 h. Cells were then washed with media and maintained in RPMI containing 10% FBS. HIV-1 p24 levels in the media were measured using the HIV-1 p24 ELISA kit (SAIC Frederick).
Immunoblotting analysis
PMs were pre-treated with or without AMD3100 or TAK 779, specific inhibitors for CXCR4 or CCR5, respectively for 1 h. Cells were then exposed to a CXCR4 ligand, SDF-1α (Peprotech, Rocky Hill, NJ) at 300 ng/ml, or a CCR5 ligand, RANTES (R&D Systems, Minneapolis, MN) at 200 ng/ml in the presence or absence of inhibitors for 15 min. Whole-cell extracts (WCE) were prepared by lysis of cells in 20 mM HEPES buffer (pH 7.9) containing 0.2% NP-40, 10% glycerol, 200 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The protein concentration was determined using the Bio-Rad Protein Assay. MAPK (p44/p42) activity was analyzed by immunoblotting on PDVF membranes, as described previously 27. Blots were probed with rabbit polyclonal antibodies against phospho-p44/p42 MAPK (Cell Signaling Technology), washed, and reacted with horseradish peroxidase-linked goat anti-rabbit antibody (KPL, Inc. Gaithersburg, MD). Bands were visualized using the Amersham ECL kit. Blots were incubated with Restore™ Western blot stripping buffer (Thermo Fisher Scientific, Rockford, IL), and reprobed with antibodies against p44/p42 MAPK proteins.
Real-time PCR analysis of CCR5, CXCR4, and CD4 mRNAs
Total RNA was isolated from cells using Qiagen RNeasy®Total RNA Mini Kit (Qiagen, Valencia CA) and treated with RNase-free DNase I. To synthesize first-strand cDNA, 500 ng of total RNA, oligo d(T)16 (Invitrogen) at 25 μg/ml and dNTP at 0.5 mM were incubated at 65°C for 5 min and quick-chilled on ice. Reverse transcription (RT) was performed at 42°C for 50 min using SuperScript™ II (Invitrogen). The PCR reaction contained cDNA equivalent to 25 ng of RNA input, 200 nM of primer sets and SYBR Green Master Mix (QIAGEN) in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Each reaction was performed in triplicate. The primer sequences were: CD4 forward (5′-AAGGGGATACAGTGGAACTGAC-3′), CD4 reverse (5′-GGACCTTTAGTTAAGAAG GAGCC-3′); CXCR4 forward (5′-TACACCGAGGAA ATGGGCTCA-3′), CXCR4 reverse (5′-TTCTTCACGGAAACAGGGTTC-3′); CCR5 forward (5′AGGGCTGTGAGGCTTATC TTC-3′), CCR5 reverse (5′-CACCTGCATAGCTTGGTCCA-3′); β-actin forward (5′-GTGGACTTGGGAGAGGACTG-3′); β-actin reverse (5′-ACTGGAACGGTGAAGGT GAC-3′). PCR conditions included denaturation at 95°C for 10 minutes followed by 40 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. Quantification of PCR products was normalized using the housekeeping β-actin gene. Relative quantification of gene expression was calculated by using a ΔCt (Ct, threshold cycle of real-time PCR) method according to the following formula: 2−ΔCT = [2−(sample Ct − β-actin Ct)].
Luminex fluorescent-bead assay
CC-chemokines measurement was performed by using a Human Chemokine Five-Plex Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using the two-tailed Student’s t test, with significance set at a p-value of <0.05.
Results
Macrophages are the most abundant cell type in AMCs
AMCs were prepared from the ascitic fluid of patients with liver cirrhosis. The yield was approximately 3–9×109 cells for each 3–9 liter specimen. As determined by FACS analysis, over 80% of the AMCs expressed CD68, a marker of differentiated macrophages (Table 1), indicating that ascitic fluid is an abundant source of mature macrophages. In contrast, less than 30% of the PBMCs were CD68+ macrophages. The macrophages remained viable and functional following a freeze-thaw cycle of the AMCs (not shown). Comparable results were obtained whenever frozen or freshly-isolated AMCs were used for preparation of peritoneal macrophages (PMs). Experiments, such as those using CD3-depleted cell cultures, were only performed on freshly-isolated cells.
Table 1. Cell populations in AMCs.
Freshly isolated AMCs were subjected for cell surface staining for CD3. CD14, CD19. For CD68 staining, cells were fixed and permeabilized. Results of FACS analysis of AMCs from two donors with liver diseases (LD1 and LD2) and PBMCs from two healthy donors (HD1 and HD2) were shown.
| AMCs LD1 | AMCs LD2 | PBMCs HD1 | PBMCs HD2 | |
|---|---|---|---|---|
| CD3 | 12% | 10% | 36.5% | 36% |
| CD14 | 70% | 74% | 15% | 4.23% |
| CD19 | 0.03% | 0.07% | 2.38% | 4.52% |
| CD68 | 82% | 85% | 26% | 12% |
PMs were isolated from AMCs by selecting for adherent cells after 16 h attachment. The majority of the adherent cells had classical macrophage morphology and expressed CD68 (Fig. 1A). FACS analysis revealed that more than 95% of the adherent cells were CD68+ (Fig 1B). The macrophages remained viable for more than two months (data not shown) and efficiently phagocytosed fluorescent beads (Fig. 1C). They produced high levels of TNFα in response to LPS treatment, as did monocyte-derivded macrophages (MDMs) (data not shown).
Figure 1. Peritoneal macrophages (PMs) are highly abundant in ascitic fluid.
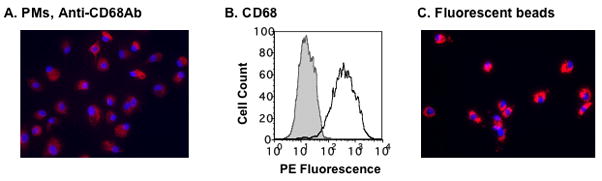
(A) PMs were prepared by plating AMCs (1–2 × 106 per well) in 6-well plates overnight, followed by washing the cultures four times with PBS. Adherent cells, PMs, were cultured in RPMI with 10% FBS and stained with a mouse monoclonal antibody against CD68, a marker of differentiated macrophages. (B) The expression of CD68 in day-1 PMs was determined by FACS analysis. The gray area represents the signal from cells stained with isotype control antibody, whereas the solid line indicates the signal from cells stained with CD68 antibody. (C) Day-1 PMs phagocytosed Fluoresbrite particles (6.0 mm in diameter). Panels A, and C are 20x magnification. The results represent experiments carried out on cells from three or more patients.
Expression of cell surface CD4, CXCR4 and CCR5 on peritoneal macrophages
PMs were obtained by plating AMCs for 16 h and removing non-adherent cells. FACS analysis revealed that these cells expressed cell surface CD4, albeit at levels far below those of MDMs or freshly-isolated PBMCs (Fig. 2A). Similar to intestinal macrophages and alveolar macrophages 13, 14, PMs expressed little, if any, cell surface CXCR4 or CCR5. In contrast, MDMs expressed both cell surface CXCR4 and CCR5 and PBMCs expressed a high level of CXCR4, but undetectable CCR5 (Fig. 2A).
Figure 2. Cell surface expression of CD4, CXCR4 and CCR5 on day-1 PMs.
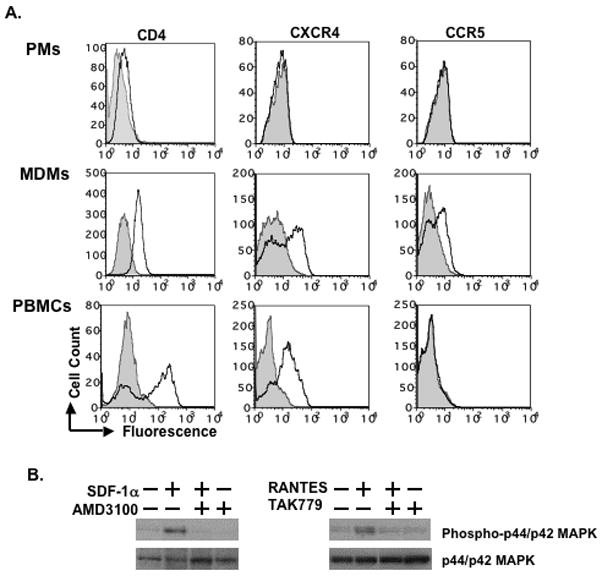
(A) Cell surface expression of CD4, CXCR4 and CCR5 was determined by FACS analysis. MDMs and PBMCs were included as a comparison. The gray area represents the signal from cells stained with isotype control antibody, whereas the solid line indicates the signal from cells stained with CD4, CXCR4 or CCR5 antibody. (B) To examine whether PMs expressed functional CXCR4 or CCR5, cells were exposed to SDF-1α at 300 ng/ml or RANTES at 200 ng/ml for 15 min, respectively. PMs were also pre-treated with or without specific inhibitors of CXCR4 and CCR5 (AMD3100 or TAK779 at 10 μM, respectively) for 1 h followed by stimulation with SDF-1α or RANTES in the presence of inhibitors. Unstimulated PMs were included as controls. Whole cell extracts were prepared and phosphorylation of MAPK (p44/p42) was determined by Western blotting. Blots were then stripped and re-probed with antibodies against p44/p42 MAPK. The results represent three independent experiments using PMs from different patients. Similar data were obtained when day-1 PMs from frozen or freshly-isolated AMCs were used.
To determine whether PMs expressed functional CXCR4 and CCR5, PMs were exposed to SDF-1α or RANTES, high affinity ligands for CXCR4 and CCR5, respectively, followed by assessment of MAPK phosphorylation (Fig. 2B). Cells were also treated with small molecule inhibitors of CXCR4 and CCR5, AMD3100 and TAK779, respectively. SDF-1α and RANTES induced phosphorylation of p44/p42 MAPK in PMs. MAPK activation was blocked by AMD3100 or TAK779 (Fig. 2B). This result demonstrates that PMs express functional CXCR4 and CCR5, even though these receptors could not be detected by FACS analysis.
Freshly-isolated PMs are susceptible to the R5 strain, HIV-1BaL, but not to the X4 T cell-line adapted strain (TCLA), HIV-1IIIB
Freshly-isolated PMs were exposed to the R5 strain, HIV-1BaL, and to the X4 strain, HIV-1IIIB, at multiplicities of infection (MOIs) of 0.01 and 0.1 for 2 h. Unbound virus was removed by washing, and the cells were maintained in RPMI containing 10% FBS. HIV-1 virus particles released into media were measured by HIV p24 ELISA. Freshly-isolated PMs supported replication of an R5 strain, HIV-1BaL, but were resistant to an X4-TCLA strain, HIV-1IIIB (Fig. 3A).
Figure 3. Day-1 PMs are susceptible to R5 HIV-1BaL but not to X4 HIV-1IIIB.
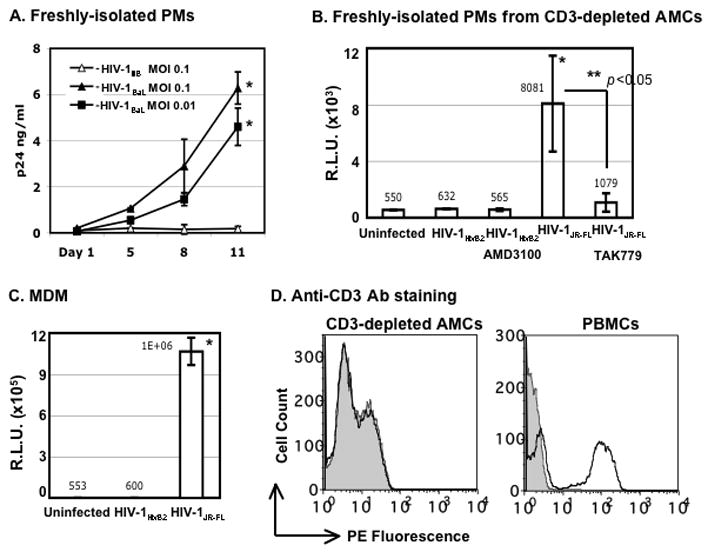
(A) Freshly-isolated (day-1) PMs were exposed to HIV-1BaL or HIV-1IIIB for 2 h. Unbound virus was removed by washing with PBS, and cells were cultured in RPMI with 10% FBS without additional growth factors. HIV-1 infection was monitored by measuring virus particles in media using an HIV-1 p24 ELISA. The difference in the HIV p24 level from HIV-1BaL-infected cells at day 1 vs day 11 after viral infection was significant (* p< 0.05). (B) Freshly-isolated PMs from CD3-depleted AMCs were exposed to pseudotyped X4 HIV-1HxB2 or R5 HIV-1JR-FL luciferase reporter viruses for 2 h. Cells were washed and cultured for 48 h before measurement of luciferase activity. To determine sensitivity to inhibitors, PMs were treated with entry inhibitors (AMD3100 or TAK-799 at 10μM) for 1 h before viral exposure. Inhibitors were added back to the culture during and after viral exposure. The mean value of luciferase activity is shown. *p< 0.05, uninfected control vs HIV-1JR-FL-exposed cells; **p< 0.05 for HIV-1JR-FL-exposed cells in the presence or absence of TAK-799. (C) MDMs were exposed to pseudotyped X4 HIV-1HxB2 or R5 HIV-1JR-FL luciferase reporter virus and HIV infection was determined at 48h after infection. *p< 0.05, uninfected control vs HIV-1JR-FL-exposed cells. (D) The CD3+ population in AMCs after CD3 depletion was determined by FACS analysis. Freshly-isolated PBMCs were included as a comparison. For experiments in panels A to C, data are mean ± SD of triplicate samples and represent three independent experiments from different donors.
To confirm that PMs support receptor-mediated HIV infection, AMCs were depleted of CD3+ T cells prior to plating and the HIV susceptibility of the purified PM cultures was examined in a single-cycle infection assay. After overnight plating and selection of adherent cells, PMs were exposed to HIV luciferase reporter viruses pseudotyped with envelopes derived from either the R5 strain HIV-1JR-FL or the X4-TCLA strain HIV-1HxB2. Unbound viruses were removed by washing. Cells were treated with entry inhibitors before and during HIV infection. After cells were cultured for 48 h at 37°C, luciferase activity was measured. PM cultures lacking CD3+ T cells were susceptible to the R5 HIV-1 and were resistant to the X4-TCLA virus (Fig. 3B). Infection by the R5 virus was blocked by TAK779, indicating that viral entry was mediated by CCR5. The PMs produced a much lower signal from the pseudotyped R5 virus than MDMs (Fig. 3C). FACS analysis confirmed that the CD3+-depleted PM population did not contain CD3+ T cells (Fig. 3D).
Peritoneal macrophages acquire susceptibility to the HIV-1 laboratory X4-TCLA strain, HIV-1IIIB
Because PMs survive for several weeks in vitro, it is possible to evaluate changes in HIV susceptibility over time. To seek evidence of altered susceptibility, we cultured PMs in RPMI containing 10% FBS for 7 days and then exposed them to the R5 strain, HIV-1BaL, or the X4-TCLA strain, HIV-1IIIB, at an MOI 0.1, as described above. Day-7 PMs were susceptible to HIV-1BaL (Fig. 4A). Interestingly, these PMs had acquired susceptibility to HIV-1IIIB. Comparable amounts of the R5 strain, HIV-1BaL, and the X4-TCLA strain, HIV-1IIIB HIV were produced from PM cultures, regardless of whether they were derived from AMCs with or without CD3 depletion (Figs 4A and 4B). These results establish that PMs developed susceptibility to the X4-TCLA strain during 7 days in culture. As expected, MDMs were susceptible to HIV-1BaL and released large quantities of virus particles; however, they were not susceptible to HIV-1IIIB (Fig. 4C).
Figure 4. PMs acquire susceptibility to the HIV-1 laboratory X4-TCLA strain.
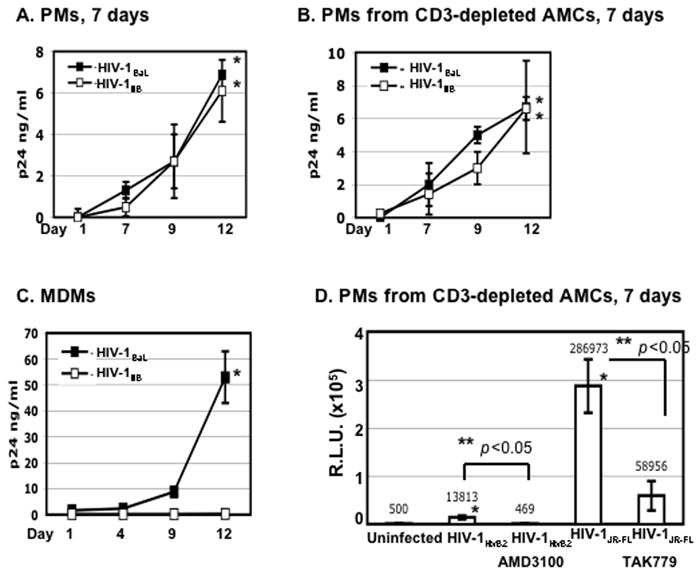
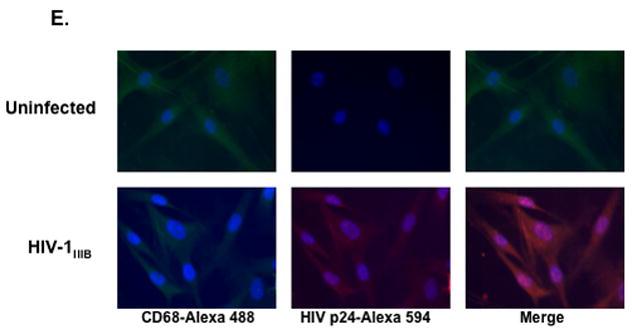
(A and B) PMs from AMCs with or without CD3 depletion were cultured for 7 days before exposure to HIV-1BaL or HIV-1IIIB for 2 h. Unbound virus was removed by washing with PBS, and cells were cultured in RPMI with 10% FBS. HIV-1 infection was determined by HIV-1 p24 ELISA. The difference in the HIV p24 level at day 1 vs day 12 after infection was significant for both strains (*p< 0.05). (C) As a comparison, MDMs were exposed to HIV-1BaL or HIV-1IIIB and HIV production was determined by ELISA. *p<0.05, HIV-1BaL at day 1 vs day 12 after infection. (D) PMs from CD3-depleted AMCs were cultured for 7 days before exposure to pseudotyped X4 HIV-1HxB2 or R5 HIV-1JR-FL luciferase reporter viruses for 2 h. Cells were washed and cultured for 48 h before measurement of luciferase activity. To determine sensitivity to inhibitors, PMs were treated with entry inhibitors (AMD3100 at 10 μM and TAK-799 at 10μM) for 1 h before viral exposure. Inhibitors were added back to the culture during and after viral exposure. The difference in the luciferase activity between uninfected cells and HIV-1HxB2 PMs (*p< 0.05) as well as HIV-infected PMs with or without inhibitors (**p< 0.05) was significant as determined by two-tailed student t test. (E) PMs were cultured for 7 days before exposure to HIV-1IIIB at an MOI 0.1. Unbound virus was removed by washing; cells were cultured for 10 days and stained with antibodies against HIV-1 p24 and CD68 and appropriate secondary antibodies (20x). For experiments in panels A to D, data are the mean±SD of triplicate samples and represent at least three independent experiments using PMs from different patients.
To further confirm these results, single-cycle infection assays were preformed on day-7 PMs prepared from CD3-depleted AMCs (Fig. 4D). Similar to the results in the multiple-round infection assays, these PMs were susceptible to infection with both R5 and X4-TCLA strains (Fig. 4D). The signal from the X4 virus was much lower than that from the R5 virus even though similar amounts of HIV p24 were used during viral exposure. The susceptibility of day-7 PM cultures to the X4-TCLA strain was also shown by immunostaining. In these co-localization experiments, PMs infected by HIV-1IIIB reacted with antibodies against HIV p24 and CD68 (Fig. 4E) and PMs infected by a pseudotyped HIV-1HxB2 GFP-expressing virus expressed GFP and reacted with anti-CD68 antibodies (data not shown).
HIV infection of day-7 PMs by HIV-1 X4 or R5 virus was blocked by AMD3100 or TAK779, respectively, indicating that HIV infection was mediated through HIV co-receptors. Interestingly, in the single-cycle infection assay, the R5 signal from day-7 PMs was significantly higher than that of day-1 cultures (Figs. 3B and 4D); however, in the multiple-cycle infection assay, there was no significant difference in R5 HIV-1BaL production between the two (Figs. 3A, 4A and 4B).
Peritoneal macrophages are susceptible to primary isolates of X4, R5 and X4R5 HIV-1
Day-1 and day-7 PMs were exposed to HIV-1 X4 (92HT599), R5 (92IN905), or dual tropic X4R5 (92RW009) primary isolates for 2 h, washed and cultured in complete media. HIV production was determined by HIV p24 ELISA. Productive HIV infection could be detected in both day-1 and day-7 PMs (Fig 5). Significant enhancement of viral production occurred in day-7 PMs infected with X4 and dual-tropic X4R5 viruses, but not with R5 viruses (Fig. 5).
Figure 5. PMs are susceptible to HIV-1 X4, R5, X4R5 primary isolates.
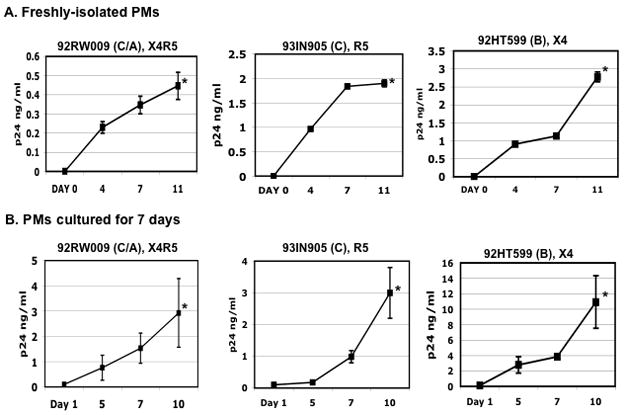
Freshly-isolated (day-1 PMs) (Panel A) or day-7 cultures of PMs (Panel B) were exposed to HIV-1 X4, R5 and X4R5 primary isolates for 2 h at 37°C and then unbound virus was removed by washing. The names and tropisms of virus isolates are indicated and viral genotypes are shown in parentheses. HIV-1 production was determined by HIV p24 ELISA. Data are the mean±SD of triplicate samples and represent two independent experiments using peritoneal macrophages from different patients. The difference in the HIV p24 level at day 1 vs day 10 or 11 after infection was significant (*p< 0.05).
CCR5, CXCR4, and CD4 gene expression is up-regulated in PMs during 7 days in culture
It has been shown that over-expression of CXCR4 but not CD4 renders MDMs susceptible to X4-TCLA strains, indicating that high levels of CXCR4 can overcome the block to infection 20. Additionally, over-expression of CD4 enhances susceptibility to primary isolates of X4 viruses. These results suggest that higher expression of HIV-1 receptors, and/or enhancement of signal transduction pathways linked to these receptors may underlie the increased susceptibility to X4-TCLA strains that occurred during 7 days in culture. Because PMs become firmly attached to their support surface over time, it was not possible to detach the day-7 PMs for FACS analysis of HIV-1 receptors without causing cell damage. As an alternative, we analyzed mRNA levels of CD4, CCR5 and CXCR4 by real-time PCR in day-1 and day-7 PMs. Although expression of cell surface CCR5 and CXC4 on the day-1 PMs was undetectable by FACS analysis (Fig. 2), these cells contained detectable levels of CD4, CCR5 and CXCR4 mRNAs in day-1 PMs (Fig. 6). CXCR4 mRNA appeared to be less abundant than CCR5 and CD4 mRNAs. Importantly, the level of CXCR4 mRNA was significantly increased, by approximately 13-fold, in the day-7 cells compared to the day-1 cells. This induction might account for the acquired susceptibility to X4-TCLA HIV-1 observed in the day-7 cells.
Figure 6. CD4, CCR5 and CXCR4 gene expression is up-regulated in PMs after 7 days in culture.
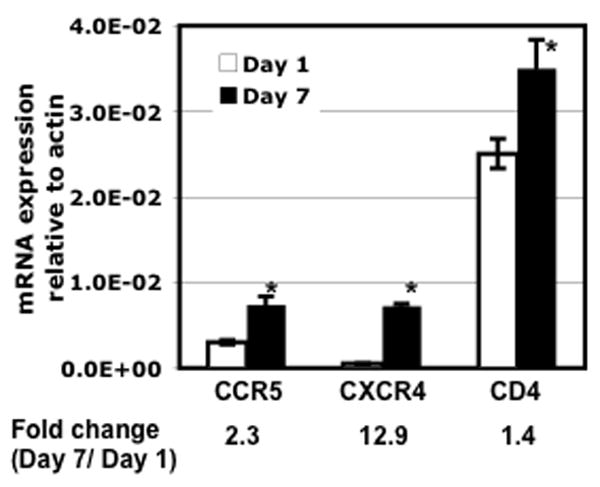
Total RNA was prepared from PMs after culturing for 1 or 7 days. The levels of CD4, CCR5, and CXCR4 mRNA were determined by real-time PCR analysis. The results represent three independent experiments using PMs from different patients. The difference in the level of gene expression from cells after culturing for 1 day vs 7 days was significant (*p<0.05).
PMs produce CC-chemokines during 7 days in culture
While day-7 PMs were more susceptible to HIV R5 infection in a single-cycle infection assay than day-1 PMs, there was no increase in HIV production in the day-7 PMs in a multiple-round infection assay. It is possible that CC-chemokines, natural HIV R5 entry inhibitors, might be induced in cultured PMs. CC-chemokines could block HIV infection in a multiple round infection but enhance viral infection by activation of macrophages in a single-cycle infection assay. To investigate CC-chemokine accumulation in the cell culture media, Luminex assays were used to measure MIP-1α, MIP-1β, and RANTES in the supernatants of PMs cultured for 1 or 7 days without a change of the media. Levels of MIP-1α and MIP-1β were significantly increased, by 5- to 30-fold, in the day-7 supernatants (Fig. 7A). RANTES was also increased, but to a lesser extent.
Figure 7. CC-chemokines are induced in PMs after culture.
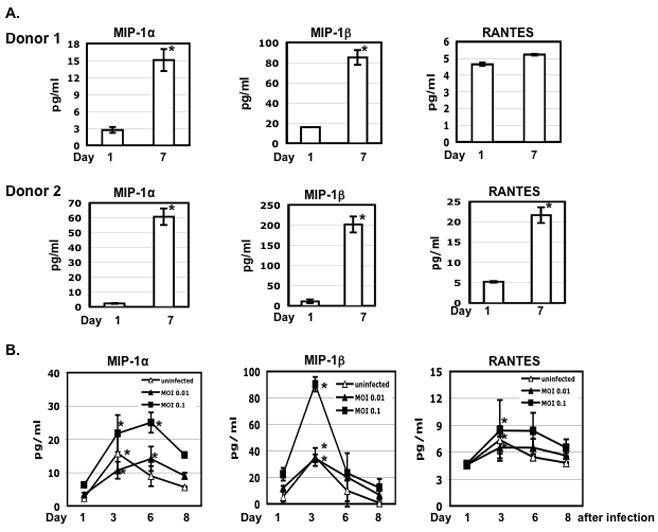
(A) AMCs (1×106 per well) were plated in a 6-well plate, cultured for 16 h to allow adherence, and washed four times with PBS to remove non-adherent cells. Media from PMs after 1 or 7 days in culture were collected and the level of chemokines were determined by Luminex assays. (B) PMs were prepared by plating AMCs (1×105 cells per well) in a 48-well plate for overnight following by washing. Cells were exposed to R5 HIV-1BaL at an MOI 0.01 or 0.1 for 2 h, washed and cultured in RPMI with 10% FBS. Uninfected cells were included as a comparison. One-half volume of media was collected at the indicated time points to determine the levels of chemokines and HIV p24. Fresh media were added back at each time point. Significant differences between the level of chemokines at day 1 vs day 4 or 7 after culture (panel A) or infection (panel B) are noted (*p< 0.05). The results represented four independent experiments from different patients.
HIV infection enhances production of CC-chemokines (reviewed in 28, 29). To determine whether HIV infection further increased CC-chemokine production, cells were exposed to HIV-1BaL at MOIs 0.01 and 0.1, washed, and then cultured in RPMI with 10% FBS. One-half volume of the supernatants was collected at various times and the levels of HIV p24 and CC-chemokines were measured. Fresh complete media were added back at each collection. Similar to the results obtained in the study above, which measured the cumulative amounts of these factors, the levels of MIP-1α, MIP-1β, and RANTES were increased in the supernatants of day-4 cultures. While HIV at an MOI 0.01 did not affect CC-chemokines production, HIV at an MOI of 0.1 promoted CC-chemokine production (Fig. 7B). An increase in the production of CC-chemokines in PMs after several days in culture may have contributed to the lack of increased HIV production in the day-7 PMs with exposure to replication competent R5 virus.
Discussion
We demonstrated that ascitic fluid from cirrhotic patients contains a large number of primary, differentiated macrophages. These macrophages were phagocytic, produced TNFα in response to LPS, and could be maintained in culture for over two months with only FBS and no additional growth factors or cytokines. Importantly, they are susceptible to infection by a broad range of HIV-1 isolates in vitro. It is likely that they differ physiologically from the PMs present in healthy individuals, but they have the advantage of being available in large quantities in a waste material (ascitic fluid) that is collected without surgery. While patient-to-patient variability is an inherent property of human material, we obtained consistent results when using PMs from different patients. Our data indicate that cultured PMs are a valuable system for investigating interactions between HIV-1 and primary macrophages. These cells also provide a relevant model for investigating HIV-1 infection of macrophages in patients with end stage liver disease due to HCV infection. In the United States, about 30% of HIV-infected patients are also infected with HCV. End stage liver disease is an increasingly important cause of death in the post-HAART era, but few experimental systems are available for investigating HIV/HCV co-infection. Because macrophages are reported to support HCV replication 30, PMs could serve as a potential in vitro model for HIV/HCV co-infection of primary macrophages.
The ease of maintaining PMs in cell culture allowed us to examine HIV-1 receptor and co-receptor expression over time. Freshly-isolated (day-1) PMs contained mRNAs for CD4 and HIV co-receptors. Stimulation of these PMs with ligands for CXCR4 and CCR5 induced MAPK activation. These cells were susceptible to X4, R5 and X4R5 primary isolates although they did not express detectable cell surface CCR5 or CXCR4. Intestinal macrophages are resistant to HIV infection and alveolar macrophages are susceptible to R5 and X4R5 primary isolates, although neither of these tissue macrophages expresses detectable levels of cell surface CCR5 or CXCR4 14, 31, 32. These results show that susceptibility to HIV infection does not correlate with expression of cell surface CCR5 and CXCR4, as determined by FACS analysis 32. HIV infection of PMs was mediated by CCR5 and CXCR4 as TAK779 and AMD3100, specific inhibitors for CCR5 and CXCR4, respectively, blocked viral infection. Interestingly, day-7 PMs were susceptible to X4-TCLA strains. Five-day old cultures of Kupffer cells are the only primary tissue macrophages previously reported to be susceptible to X4-TCLA strains 11.
The restriction of the X4-TCLA strain has been attributed to the low level of CXCR4 combined with the unfavorable status of cell signaling pathways (reviewed in 18). An increase in the level of cell surface CXCR4 on MDMs by transduction of CXCR4 or treatment with TGF-β increases susceptibility to X4-TCLA or X4-using dual tropic virus, respectively 20, 33, suggesting that the levels of CXCR4 and CXCR4-mediated cell signaling modulate the restriction of X4-TCLA strains and X4-using dual tropic viruses. Post-entry restriction of X4-TCLA strains in MDMs or alveolar macrophages occurs at the step of nuclear import 32, 34. Here we demonstrated that day-1 PMs were resistant to the X4-TCLA virus but day-7 PM cultures were sensitive. Analysis of the mRNA level of CCR5, CXCR4, and CD4 revealed that expression of CXCR4 was significantly increased in PMs after culturing for 7 days, which could contribute to the acquired susceptibility to the X4-TCLA strain.
R5 viruses are preferentially transmitted during infection of a new host and they dominate during the initial stages of infection 35, 36–38. Over time, X4 and dual-tropic viruses arise in ~50% individuals infected with subtype B 39, 40. Interestingly, our results demonstrated that day-7 PM cultures produced greater amounts of primary isolates of X4 and X4R5 viruses than day-1 cultures, while production of the R5 viruses was unaltered. Contact with the plastic substrate, oxidative stress, and exposure to soluble factors produced by PMs may have induced changes in permissiveness to the X4 and dual tropic strains. Human PMs produce fibronectin, cytokines, CC-chemokines (MIP-1α, MIP-1β and RANTES), and growth factors without exogenous stimulation 41, 42. In MDMs, CD40 ligands increase susceptibility to X4-TCLA strains and decrease R5 HIV infection43. Induction of CC-chemokines, MIP-1α, MIP-1β and RANTES, contributes to the decrease in R5 HIV infection of MDMs in response to CD40 ligands or LPS 44, 43, 45–47. CC-chemokines can activate cell signaling pathways and enhance HIV-1 infection 48; however, they can also block entry of R5 HIV-1 and decrease susceptibility 49. Indeed, we found that CC-chemokines were induced in PMs during culture, which could explain why HIV infection was increased in 7 day-old PM cultures in a single-cycle infection assay but the HIV p24 level was not elevated in a multiple-round infection assay.
The intensity of macrophage infection is strongly influenced by clinical status 8, 50. Inflammation, neoplasia, and opportunistic infections increase the viral load of HIV-1-infected tissue macrophages. Macrophage infection is often associated with advanced disease. PMs provide an opportunity to rigorously investigate whether susceptibility to HIV-1 infection is heightened by exposure to inflammatory cytokines and/or by microbial infection. Our unpublished data indicated that HIV DNA was detectable in PMs of patients with undetectable HIV-1 viral load. Analysis of the HIV-1 envelope sequences in these cells could shed light on the potential of macrophages to provide an HIV-1 reservoir.
In summary, large quantities of PMs can be obtained from ascitic fluid. These cells are susceptible to receptor-mediated HIV-1 infection. PMs provide a new in vitro model for molecular studies of interactions between HIV-1 and primary macrophages. Understanding the cellular determinants of HIV-1 susceptibility may provide insights into the increased permissiveness of human tissue macrophages to X4-using strains that has been reported during late-stage HIV-1 disease, establishing the foundation for developing new interventions.
Acknowledgments
Supported by NIH grants DA016156 and DK066939 to ADB and AI073205 to TLC.
We thank Mary Klotman and members of the Branch laboratory for helpful discussions. This work was supported by NIH grants DA016156 and DK066939 to ADB, AI073205 to TLC, and an innovation award from the MSSM Department of Medicine to TLC, TDS, Stephanie Factor, and ADB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cassol E, Alfano M, Biswas P, Poli G. Monocyte-derived macrophages and myeloid cell lines as targets of HIV-1 replication and persistence. J Leukoc Biol Nov. 2006;80(5):1018–1030. doi: 10.1189/jlb.0306150. [DOI] [PubMed] [Google Scholar]
- 2.Gorry PR, Churchill M, Crowe SM, Cunningham AL, Gabuzda D. Pathogenesis of macrophage tropic HIV-1. Curr HIV Res Jan. 2005;3(1):53–60. doi: 10.2174/1570162052772951. [DOI] [PubMed] [Google Scholar]
- 3.Kedzierska K, Crowe SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr Med Chem Nov. 2002;9(21):1893–1903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- 4.Clarke JR, White NC, Weber JN. HIV Compartmentalization: Pathogenesis and Clinical Implications. AIDS Rev. 2000;2:15–22. [Google Scholar]
- 5.Igarashi T, Brown CR, Endo Y, et al. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A. 2001 Jan 16;98(2):658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao YZ, Dieterich D, Thomas PA, Huang YX, Mirabile M, Ho DD. Identification and quantitation of HIV-1 in the liver of patients with AIDS. Aids Jan. 1992;6(1):65–70. doi: 10.1097/00002030-199201000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Donaldson YK, Bell JE, Ironside JW, et al. Redistribution of HIV outside the lymphoid system with onset of AIDS. Lancet. 1994 Feb 12;343(8894):383–385. doi: 10.1016/s0140-6736(94)91222-x. [DOI] [PubMed] [Google Scholar]
- 8.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997 Jun 20;276(5320):1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 9.Moriuchi M, Moriuchi H, Turner W, Fauci AS. Exposure to bacterial products renders macrophages highly susceptible to T-tropic HIV-1. J Clin Invest. 1998 Oct 15;102(8):1540–1550. doi: 10.1172/JCI4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hufert FT, Schmitz J, Schreiber M, Schmitz H, Racz P, von Laer DD. Human Kupffer cells infected with HIV-1 in vivo. J Acquir Immune Defic Syndr Jul. 1993;6(7):772–777. [PubMed] [Google Scholar]
- 11.Schmitt MP, Steffan AM, Gendrault JL, et al. Multiplication of human immunodeficiency virus in primary cultures of human Kupffer cells--possible role of liver macrophage infection in the physiopathology of AIDS. Res Virol Mar-Apr. 1990;141(2):143–152. doi: 10.1016/0923-2516(90)90016-c. [DOI] [PubMed] [Google Scholar]
- 12.Jayakumar P, Berger I, Autschbach F, et al. Tissue-resident macrophages are productively infected ex vivo by primary X4 isolates of human immunodeficiency virus type 1. J Virol Apr. 2005;79(8):5220–5226. doi: 10.1128/JVI.79.8.5220-5226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worgall S, Connor R, Kaner RJ, et al. Expression and use of human immunodeficiency virus type 1 coreceptors by human alveolar macrophages. J Virol Jul. 1999;73(7):5865–5874. doi: 10.1128/jvi.73.7.5865-5874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng G, Sellers MT, Mosteller-Barnum M, Rogers TS, Shaw GM, Smith PD. Lamina propria lymphocytes, not macrophages, express CCR5 and CXCR4 and are the likely target cell for human immunodeficiency virus type 1 in the intestinal mucosa. J Infect Dis Sep. 2000;182(3):785–791. doi: 10.1086/315790. [DOI] [PubMed] [Google Scholar]
- 15.Verreck FA, de Boer T, Langenberg DM, van der Zanden L, Ottenhoff TH. Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-gamma- and CD40L-mediated costimulation. J Leukoc Biol Feb. 2006;79(2):285–293. doi: 10.1189/jlb.0105015. [DOI] [PubMed] [Google Scholar]
- 16.Kedzierska K, Crowe SM, Turville S, Cunningham AL. The influence of cytokines, chemokines and their receptors on HIV-1 replication in monocytes and macrophages. Rev Med Virol Jan-Feb. 2003;13(1):39–56. doi: 10.1002/rmv.369. [DOI] [PubMed] [Google Scholar]
- 17.Amzazi S, Ylisastigui L, Bakri Y, et al. The inhibitory effect of RANTES on the infection of primary macrophages by R5 human immunodeficiency virus type-1 depends on the macrophage activation state. Virology. 1998 Dec 5;252(1):96–105. doi: 10.1006/viro.1998.9452. [DOI] [PubMed] [Google Scholar]
- 18.Khati M, James W, Gordon S. HIV-macrophage interactions at the cellular and molecular level. Arch Immunol Ther Exp (Warsz) 2001;49(5):367–378. [PubMed] [Google Scholar]
- 19.Collman RG, Yi Y. Cofactors for human immunodeficiency virus entry into primary macrophages. J Infect Dis May. 1999;179 (Suppl 3):S422–426. doi: 10.1086/314797. [DOI] [PubMed] [Google Scholar]
- 20.Tokunaga K, Greenberg ML, Morse MA, Cumming RI, Lyerly HK, Cullen BR. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J Virol Aug. 2001;75(15):6776–6785. doi: 10.1128/JVI.75.15.6776-6785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi Y, Rana S, Turner JD, Gaddis N, Collman RG. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol Jan. 1998;72(1):772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matlack R, Yeh K, Rosini L, et al. Peritoneal macrophages suppress T-cell activation by amino acid catabolism. Immunology Mar. 2006;117(3):386–395. doi: 10.1111/j.1365-2567.2005.02312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shadduck PP, Weinberg JB, Haney AF, et al. Lack of enhancing effect of human anti-human immunodeficiency virus type 1 (HIV-1) antibody on HIV-1 infection of human blood monocytes and peritoneal macrophages. J Virol Aug. 1991;65(8):4309–4316. doi: 10.1128/jvi.65.8.4309-4316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen BK, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol Feb. 1994;68(2):654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1--infected individuals. J Exp Med. 1997 Feb 17;185(4):621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Husain M, Gusella GL, Klotman ME, et al. HIV-1 Nef induces proliferation and anchorage-independent growth in podocytes. J Am Soc Nephrol Jul. 2002;13(7):1806–1815. doi: 10.1097/01.asn.0000019642.55998.69. [DOI] [PubMed] [Google Scholar]
- 27.Chang TL, Gordon CJ, Roscic-Mrkic B, et al. Interaction of the CC-chemokine RANTES with glycosaminoglycans activates a p44/p42 mitogen-activated protein kinase-dependent signaling pathway and enhances human immunodeficiency virus type 1 infectivity. J Virol Mar. 2002;76(5):2245–2254. doi: 10.1128/jvi.76.5.2245-2254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fantuzzi L, Belardelli F, Gessani S. Monocyte/macrophage-derived CC chemokines and their modulation by HIV-1 and cytokines: a complex network of interactions influencing viral replication and AIDS pathogenesis. J Leukoc Biol Nov. 2003;74(5):719–725. doi: 10.1189/jlb.0403175. [DOI] [PubMed] [Google Scholar]
- 29.Yi Y, Lee C, Liu QH, Freedman BD, Collman RG. Chemokine receptor utilization and macrophage signaling by human immunodeficiency virus type 1 gp120: Implications for neuropathogenesis. J Neurovirol. 2004;10 (Suppl 1):91–96. doi: 10.1080/753312758. [DOI] [PubMed] [Google Scholar]
- 30.Radkowski M, Bednarska A, Horban A, et al. Infection of primary human macrophages with hepatitis C virus in vitro: induction of tumour necrosis factor-alpha and interleukin 8. J Gen Virol Jan. 2004;85(Pt 1):47–59. doi: 10.1099/vir.0.19491-0. [DOI] [PubMed] [Google Scholar]
- 31.Opalek JM, Ali NA, Lobb JM, Hunter MG, Marsh CB. Alveolar macrophages lack CCR2 expression and do not migrate to CCL2. J Inflamm (Lond) 2007;4:19. doi: 10.1186/1476-9255-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang ZB, Potash MJ, Simm M, et al. Infection of macrophages with lymphotropic human immunodeficiency virus type 1 can be arrested after viral DNA synthesis. J Virol Nov. 1993;67(11):6893–6896. doi: 10.1128/jvi.67.11.6893-6896.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Tuttle DL, Oshier JT, et al. Transforming growth factor-beta1 increases CXCR4 expression, stromal-derived factor-1alpha-stimulated signalling and human immunodeficiency virus-1 entry in human monocyte-derived macrophages. Immunology Apr. 2005;114(4):565–574. doi: 10.1111/j.1365-2567.2004.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verani A, Pesenti E, Polo S, et al. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998 Sep 1;161(5):2084–2088. [PubMed] [Google Scholar]
- 35.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J Exp Med. 1997 Feb 17;185(4):621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarlatti G, Tresoldi E, Bjorndal A, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med Nov. 1997;3(11):1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 37.Schuitemaker H, Koot M, Kootstra NA, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol Mar. 1992;66(3):1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaul R, Thottingal P, Kimani J, et al. Quantitative ex vivo analysis of functional virus-specific CD8 T lymphocytes in the blood and genital tract of HIV-infected women. Aids. 2003 May 23;17(8):1139–1144. doi: 10.1097/00002030-200305230-00004. [DOI] [PubMed] [Google Scholar]
- 39.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 40.Koot M, Keet IP, Vos AH, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993 May 1;118(9):681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 41.Eischen A, Duclos B, Schmitt-Goguel M, et al. Human resident peritoneal macrophages: phenotype and biology. Br J Haematol Dec. 1994;88(4):712–722. doi: 10.1111/j.1365-2141.1994.tb05109.x. [DOI] [PubMed] [Google Scholar]
- 42.Kauma S, Clark MR, White C, Halme J. Production of fibronectin by peritoneal macrophages and concentration of fibronectin in peritoneal fluid from patients with or without endometriosis. Obstet Gynecol Jul. 1988;72(1):13–18. [PubMed] [Google Scholar]
- 43.Bakri Y, Mannioui A, Ylisastigui L, Sanchez F, Gluckman JC, Benjouad A. CD40-activated macrophages become highly susceptible to X4 strains of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2002 Jan 20;18(2):103–113. doi: 10.1089/08892220252779647. [DOI] [PubMed] [Google Scholar]
- 44.di Marzio P, Mariani R, Lui R, Thomas EK, Landau NR. Soluble CD40 ligand induces beta-chemokine production by macrophages and resistance to HIV-1 entry. Cytokine Oct. 2000;12(10):1489–1495. doi: 10.1006/cyto.1999.0594. [DOI] [PubMed] [Google Scholar]
- 45.Cotter RL, Zheng J, Che M, et al. Regulation of human immunodeficiency virus type 1 infection, beta-chemokine production, and CCR5 expression in CD40L-stimulated macrophages: immune control of viral entry. J Virol May. 2001;75(9):4308–4320. doi: 10.1128/JVI.75.9.4308-4320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee C, Liu QH, Tomkowicz B, Yi Y, Freedman BD, Collman RG. Macrophage activation through CCR5- and CXCR4-mediated gp120-elicited signaling pathways. J Leukoc Biol Nov. 2003;74(5):676–682. doi: 10.1189/jlb.0503206. [DOI] [PubMed] [Google Scholar]
- 47.Verani A, Scarlatti G, Comar M, et al. C-C chemokines released by lipopolysaccharide (LPS)-stimulated human macrophages suppress HIV-1 infection in both macrophages and T cells. J Exp Med. 1997 Mar 3;185(5):805–816. doi: 10.1084/jem.185.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trkola A, Gordon C, Matthews J, et al. The CC-chemokine RANTES increases the attachment of human immunodeficiency virus type 1 to target cells via glycosaminoglycans and also activates a signal transduction pathway that enhances viral infectivity. J Virol Aug. 1999;73(8):6370–6379. doi: 10.1128/jvi.73.8.6370-6379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995 Dec 15;270(5243):1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 50.Orenstein JM. HIV expression in surgical specimens. AIDS Res Hum Retroviruses Jul. 2008;24(7):947–955. doi: 10.1089/aid.2008.0265. [DOI] [PubMed] [Google Scholar]


