Abstract
Octyl methoxycinnamate (OMC) is one of the most widely used sunscreen ingredients. To analyze biological effects of OMC, an in vitro approach was used implying ultraviolet (UV) exposure of two human cell lines, a primary skin fibroblast (GM00498) and a breast cancer (MCF-7) cell lines. End points include cell viability assessment, assay of cyclobutane pyrimidine dimers (CPDs) and oxidated DNA lesions using alkaline elution and lesion-specific enzymes, and gene expression analysis of a panel of 17 DNA damage–responsive genes. We observed that OMC provided protection against CPDs, and the degree of protection correlated with the OMC-mediated reduction in UV dose. No such protection was found with respect to oxidative DNA lesions. Upon UV exposure in the presence of OMC, the gene expression studies showed significant differential changes in some of the genes studied and the expression of p53 protein was also changed. For some genes, the change in expression seemed to be delayed in time by OMC. The experimental approach applied in this study, using a panel of 17 genes in an in vitro cellular system together with genotoxicity assays, may be useful in the initial screening of active ingredients in sunscreens.
Keywords: octyl methoxycinnamate, cyclobutane pyrimidine dimers, oxidative DNA lesions, UV, sunscreens, gene expression
Ultraviolet (UV) radiation from sunlight is considered one of the most important environmental factors affecting humans and has been implicated as the main cause for skin cancer (Ananthaswamy et al., 1997). It is generally thought that ultraviolet B (UVB) (280–320 nm) and to a lesser extent ultraviolet A (UVA) (320–400 nm) are responsible for the sunlight-induced cancers (Armstrong and Kricker, 2001; Cole et al., 1986). The UVA contribution to melanoma skin cancer has been discussed (Mitchell et al., 2007). The cellular effects of UV irradiation include DNA damage, cell cycle arrest, immunological depression, apoptosis, and transcriptional changes. UVB irradiation directly causes bulky DNA adducts such as cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts (6-4PPs), which are usually repaired by nucleotide excision repair (NER) generally removing bulky DNA adducts (Lehmann, 1995). UV light—in particular UVA but also UVB—mediates oxidative stress indirectly via reactive oxygen species (ROS) (Pelle et al., 2003). The UV-generated ROS can induce DNA single-strand breaks (SSBs), DNA-protein cross-links, and oxidized base derivatives, such as 7,8-dihydro-8-oxoguanine (8-oxoG) (Mitra et al., 1997). Oxidative DNA damage is preferentially removed by base excision repair (BER) (Helbock et al., 1999; Mitra et al., 1997).
Sunscreens have become the most popular choice of photoprotection and are recommended in addition to using protective clothing and avoiding the sun (International Agency for Research on Cancer, 2001). Ideally, sunscreens should protect not only against skin cancer but also against effects on the immune system and photoaging of the skin (Elmets and Anderson, 1996; Young and Walker, 2002). There is growing interest in the photostability of sunscreens due to a dramatic increase in their use. The photoinstability of active ingredients in sunscreen has been reported by several studies (Bredholt et al., 1998; Serpone et al., 2002; Tarras-Wahlberg et al., 1999). Upon UV exposure, sunscreens may be degraded to form photoproducts that are potentially toxic. It has been shown that some sunscreens change their spectral performance or may act as photooxidants via generation of free radicals and ROS upon UV exposure (Brezova et al., 2005; Gulston and Knowland, 1999). There are some reports of the protective effect of sunscreens against DNA damage formation in cell cultures (Reinhardt et al., 2003) and in the skin of volunteers (Al Mahroos et al., 2002). The decrease in the efficiency of sunscreens can be caused by different mechanisms: photoisomerization, photodecomposition, and interaction with the formulation or other sunscreen agents (Maier et al., 2001). Therefore, knowledge about sunscreen photostability on cellular processes is important.
Octyl methoxycinnamate (OMC) is the most commonly used UVB filter in sunscreens and cosmetics and is listed as a high production volume chemical in the European chemical Substances Information System database (http://ecb.jrc.ec.europa.eu/esis/). Topical application of OMC is well tolerated, with little or negligible skin irritation, allergic contact reaction, and phototoxic effects. However, we have previously reported increased toxicity as a result of breakdown of OMC following UV irradiation (Butt and Christensen, 2000). Such UV-induced molecular breakdown may interfere with cellular processes or induce oxidative damage in human skin. OMC has been shown to degrade into photoproducts when exposed to sunlight, which leads to a decrease in UV absorption efficiency (Butt and Christensen, 2000; Pangnakorn et al., 2007). These photoproducts may have a higher toxicity than OMC itself. Other adverse effects of sunscreens have been suggested, including formation of singlet oxygen and various estrogenic effects after in vitro and in vivo exposure to several UV filters (Allen et al., 1996; Schlumpf et al., 2001).
To address the need for nonanimal testing of ingredients in cosmetics, we established an in vitro cellular system based on two human cell lines, the primary skin fibroblast cell line (GM00498) and the breast cancer cell line (MCF-7). We investigated the effect of OMC with and without UV irradiation on the expression of a panel of 17 genes, selected because of their role in DNA damage response pathways, by quantitative real-time PCR (qRT-PCR) and the expression of p53 protein by Western blotting. The photoprotective capacities of OMC against UV-induced DNA damage, particularly CPDs and oxidative DNA damage, were evaluated by measuring DNA damage using the alkaline elution assay. Investigation of the expression of DNA damage response–related genes, along with genotoxicity assays, may provide sensitive biological end points that could be useful for screening and evaluation of agents to be used in sunscreens.
METHODS
Cell Culture and Chemical Treatment
The human breast carcinoma cell line MCF-7 was obtained from the American Type Culture Collection (ATCC no. HTB22) and was grown in Dulbecco's Modified Eagle's Medium supplemented with 5% fetal bovine serum and 1% penicillin-streptomycin (BioWhittaker, Lonza, Switzerland). The human primary skin fibroblast cell line GM00498 was obtained from Coriell Cell Repositories (http://ccr.coriell.org/) and was grown in Quantum medium with L-glutamine (PAA Laboratories GmbH, Austria) and 1% penicillin-streptomycin (BioWhittaker). The two cell lines were incubated at 37°C with 5% CO2 in air with saturated humidity.
OMC (Eusolex 2292) was purchased from Acros Organics (Geel, Belgium) and dissolved in ethyl alcohol and stored at 4°C. A working solution of OMC was prepared by dissolving the ethyl alcohol dissolved OMC in PBS (without Ca2+ and Mg2+).
This experimental design was used to mimic a situation in which an OMC-containing sunscreen lotion is applied to the skin, the individual is exposed to the sun, and then the direct exposure is terminated without removing the remaining sunscreen from the skin. To achieve such a scenario, exponentially growing cells are exposed to UV in the presence or absence of OMC, whereafter cells are grown without UV but still in presence of the UV-irradiated OMC. All cells were pretreated for 1 h with OMC (0, 5, 10, 16, 27, 43, or 60 ppm). Thereafter, half of the samples were irradiated at 4°C with UV (0–2000 s, corresponding to a maximum of 492 J/m2) at a distance of 28 cm from the light source, whereas the other half was kept in the dark. Thereafter, the PBS/OMC solutions were removed and replaced with culturing medium containing OMC (0–60 ppm), which had been treated with the same UV dose as the corresponding cell sample but without cells. The samples that had been kept in the dark were incubated in medium with OMC (0–60 ppm), which had not been UV treated. OMC was dissolved in PBS and not in medium during its UV exposure to avoid interactions with medium constituents and UV-filtering effects. For cell viability assays, cells were treated as described. Selected UV doses and OMC concentrations were used for downstream analysis. For alkaline elution, the UV dose range was 0–246 J/m2 (with or without 10 ppm OMC). For gene expression analyses, a single OMC concentration (27 ppm) was used and the UV exposure was 45 J/m2.
The light source used consisted of five Wolff Helarium B1-01-40W tubes (Germany) mounted inside the lid of a box fitted with an electrical extractor fan for cooling, and the emission spectrum contained wavelengths between 290 and 400 nm (Kinley et al., 1997), as measured with a UDT detector (UDT371; United Detector Technology, Hawthorne, CA) and probe 222UV (3.05 mW/cm2). UV doses in figures are indicated as exposure times (seconds) or as Commission Internationale de l'Eclairage (CIE)-weighted irradiance (J/m2), i.e., the efficient spectrum of the light source after spectral filtration through OMC at varying concentrations. During UV irradiation, cells were covered with PBS/OMC. The thickness of the PBS/OMC layer was 2.55 cm, selected to obtain a substantial filtration effect and at the same time reducing the toxicity of OMC. The degree of spectral filtration depends on the OMC concentration (Fig. 1, and Supplementary table 1). For example, OMC at 10 ppm reduced the irradiance by ∼50% at the level of attached cells, and OMC at 27 ppm reduced the irradiance by ∼70% at the level of attached cells.
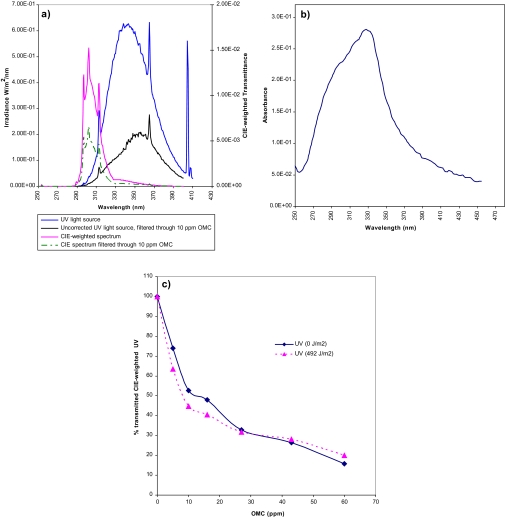
FIG. 1.
Light source and transmittance of OMC. (a) Characteristics of the light source, its CIE-weighted spectrum, and the spectrum transmitted through OMC. Spectral irradiance of the UV source (blue line, left axis), spectral irradiance after transmission through OMC (10 ppm, 2.55 cm) (black line, left axis), CIE-weighted spectrum calculated for the light source (purple line, right axis), or after transmission through OMC (10 ppm, 2.55 cm) (broken green line, right axis). The CIE-weighted spectrum was calculated by multiplying the spectral radiation of the lamp with the biological efficiency spectrum for each wavelength. (b) OMC spectral curve, as measured spectrophotometrically in a 27 ppm solution. (c) OMC concentration–dependent reduction in CIE-weighted UV dose (0 or 492 J/m2 [corresponding to 2000 s]). CIE-weighted light transmittance calculated from measurement of spectra of OMC (0–60 ppm) at a depth of 2.55 cm.
Genotoxicity Measurement by Alkaline Elution Assay
A semi-automated alkaline elution system was performed as previously described with some modifications (Brunborg et al., 1988) but using exogenous enzymes for removing modified bases. Briefly, MCF-7 cells were treated with 10 ppm OMC dissolved in PBS before irradiation with varying doses of UV. Cell samples were then analyzed as described (Brunborg et al., 1988) using crude extracts of Fpg and T4 endonuclease V (T4-endo V) enzymes. The Fpg enzyme extract was purified from Escherichia coli ER 2566 strain harboring the pFPG230 plasmid as previously described (Boiteux et al., 1990). The T4-endo V enzyme extract was purified from E. coli AB2480 (uvrA, recA, F'lac IQ1) plus ptac-den V(Apr) strain as previously described (Nakabeppu et al., 1982). The activity of the Fpg extract was measured by incubation with a substrate containing 3H-labeled FaPy residues and quantifying the number of 3H-labeled bases released from the substrate as previously described (Olsen et al., 2003). The enzymatic activity of the T4-endo V crude extracts was determined using a nicking assay with pure T4-endo V enzyme as a positive control. Various amounts of the extracts were used in initial experiments to establish first-order kinetics of the conversion of specific UV-induced lesions into DNA SSBs (data not shown).
Calibration and calculation of lesion frequencies were carried out using x-rays at the relevant elution pH and assuming an induction of 90 × 10−9/nucleotide/Gy (∼1000 SSB/Gy/diploid cell [= 3.6 × 1012 Da]). The normalized area above curve (NAAC) unit, calculated from elution profiles and used for lesion quantification in our alkaline elution assay (Brunborg et al., 1996), was determined at pH 12.25 ± 0.05: 1 NAAC = 22.8 lesions/1012 Da. All alkaline elution data (NAAC values) are reported as lesion frequencies (per 1012 Da) and presented as the average of five independent experiments.
Determination of Cell Viability
MCF-7 and GM00498 cells were seeded at a density of 5000 cells/100 μl in 96-well culture plates and grown for 24 h. The following day, cells were treated with combinations of OMC (0–60 ppm) and UV (0–2000 s) as described above. After 24-h incubation in medium containing (UV-irradiated) OMC, cell viability was measured using an 3-(4,5-dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT) assay (CellTiter 96 Non-Radioactive Cell Proliferation Assay; Promega, Madison, WI) according to the manufacturer's instructions. The absorbance was measured at 570 nm by Sunrise plate reader and analyzed with Magellan software, v 1.11 (Tecan, Switzerland). Cell viability was calculated as percentage of absorbance, relative to untreated (no UV and no OMC) control cell cultures; wells containing culturing medium only were used as blanks and were subtracted as background from each sample. The value of untreated control cells was defined as 100% viable, and all data were calculated accordingly. Results are from three independent experiments.
Gene Expression Analysis
RNA preparation.
Total RNA was isolated as previously described (Duale et al., 2007) using the GenElute kit (Sigma-Aldrich, Inc., Oslo, Norway). RNAs were isolated from three independent experiments. Its quantity and quality were determined using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA). RNA integrity was determined by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
qRT-PCR analysis.
qRT-PCR was used to analyze the transcriptional response of GM00498 and MCF-7 cell lines following the exposure of OMC (27 ppm) alone, co-exposure of OMC (27 ppm) plus UV (45 J/m2), and UV (45 J/m2) alone. The reverse transcription reaction and RT-PCR were carried out as previously described (Duale et al., 2007). The primer sequences used are listed at RTPrimerDB, real-time PCR primer and probe database (Pattyn et al., 2003), or purchased from Qiagen (Germany). The real-time PCR experimental layout of the plate was as follows: For each sample, there were three biological repeats, and for each of those, three technical replicates were run (3 × 3), and 10 samples (i.e., 3 × 3 × 10). We also included three control samples (cDNA from pooled RNA from control samples) in each 96-well plate. This layout allowed simultaneous measurements of all samples in one 96-well plate for each gene, reducing run-to-run variations. All PCR reactions were performed in triplicate, and data are expressed as an average of the triplicates. The average cycle threshold (Ct) measurements for the three independent experiments were used in calculations of relative expression. The endogenous reference genes (house keeping genes [HKGs]) for gene expression studies may depend on the applied treatments; we therefore evaluated the stability of the HKGs (18S rRNA, glyceraldehyde 3-phosphate dehydrogenase [GAPDH], and beta-actin) used in this study by the BestKeeper algorithm (Pfaffl et al., 2004). The three HKGs were correlated well with each other (Supplementary table 2 and Supplementary fig. 1). We therefore normalized target genes to the average of all three HKGs in the 2−ΔΔCt method (Livak and Schmittgen, 2001). The cDNA from pooled RNA from control samples was used as calibrators, and the same cDNA was included in each 96-well plate, allowing control of run-to-run variation. All gene expression data are reported as log2-transformed 2−ΔΔCt values and presented as the average of three independent experiments.
Western Blot Analysis
MCF-7 and GM00498 cells (∼2 × 106) were treated with OMC (27 ppm) alone, OMC (27 ppm) plus UV (45 J/m2), or UV alone (45 J/m2). Samples were washed with ice-cold PBS and incubated for 10 min on ice in cold lysis buffer with a protease inhibitor cocktail (60mM Tris-HCl [pH 6.8], 10% glycerol, 3% SDS, 1mM EDTA, 1mM sodium orthovanadate, 50mM sodium fluoride, 10mM glycerol 2-phosphate disodium salt hydrate [phosphatase inhibitors], and Complete Mini [Roche, Switzerland]). Whole-cell lysates were sonicated on ice. Protein concentration was determined by the Lowry method using Bio-Rad's detergent-compatible protein assay kit according to the manufacture's instruction (Bio-Rad Laboratories, Inc., Hercules, CA). Samples (20 μg per well) were heated in SDS sample buffer for 5 min at 95°C, separated by SDS-PAGE and electroblotted onto Immobilon-P membrane (Millipore Inc., Billerica, MA). After the proteins were transferred, the membrane was blocked in 5% blotting-grade nonfat dry milk (Bio-Rad Laboratories, Inc.). The following antibodies were used: anti-p53 (Cell Signaling Technology Inc., Danvers, MA) and anti-GAPDH (Biogenesis, Inc., Hackensack, NJ), used as internal control. Membranes were incubated with the appropriate peroxidase-coupled secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and subsequently detected by enhanced chemiluminescence using Amersham's ECL Western Blotting Detection Reagents (GE Healthcare, Norway).
Statistical Analysis
The alkaline elution assay data and the cell viability data were analyzed by a nonparametric Mann-Whitney U-test. The gene expression data were analyzed by one-way ANOVA, followed by a post hoc Dunnett's test to allow for multiple comparisons, i.e, comparison of the treatment groups [ΔCt exposed: (Ct exposed target gene − Ct exposed reference gene)] versus common untreated control groups [ΔCt control: (Ct control target gene − Ct control reference gene)], and two-sample t-test was used to check for cell type–specific gene expression differences between the two cell lines. All statistical analyses were performed using SPSS software (SPSS, Inc., Chicago, IL), and p < 0.05 was accepted as statistically significant.
RESULTS
Photoabsorption and Stability of OMC
The spectral irradiance of the light source in Figure 1 peaks at 350 nm but with a significant irradiance also in the UVB region (< 320 nm). The CIE-weighted spectrum for erythema induction was used to calculate the total efficiency of the light source with respect to CPD formation since the CIE-weighted spectrum is similar to the spectral efficiency for CPDs (Young et al., 1998). The resulting convolution curve is obtained by multiplying—for each wavelength—the irradiance of the light source with the CIE-weighted efficiency. The spectral absorbance of OMC dissolved in PBS was measured spectrophotometrically (Fig. 1b) and was used to calculate the CIE-weighted spectral irradiance without or with OMC (10 ppm) (Fig. 1a); the integrated radiant exposure at 10 ppm OMC was reduced to 52%. Also included in Figure 1a is the irradiance of the light source filtered through 10 ppm OMC (i.e., with no CIE correction). This curve is more relevant for oxidative lesions, which—unlike CPDs—are induced at highest efficiency in the UVA region. The integrated radiant exposure of the light source was reduced by OMC (10 ppm and 2.55 cm depth) to 32%, illustrating that OMC has some absorption also in the UVA region.
The photostability of OMC was investigated (Fig. 1c). OMC (0–60 ppm) dissolved in PBS was exposed for up to 2000 s (corresponding to 492 J/m2) at the depth (2.55 cm) used during cell exposure. The transmitted CIE-weighted spectral UV irradiance is shown as a function of OMC concentration; there was no major change during the exposure.
Photoprotective Efficacy of OMC Measured by Alkaline Elution Assay
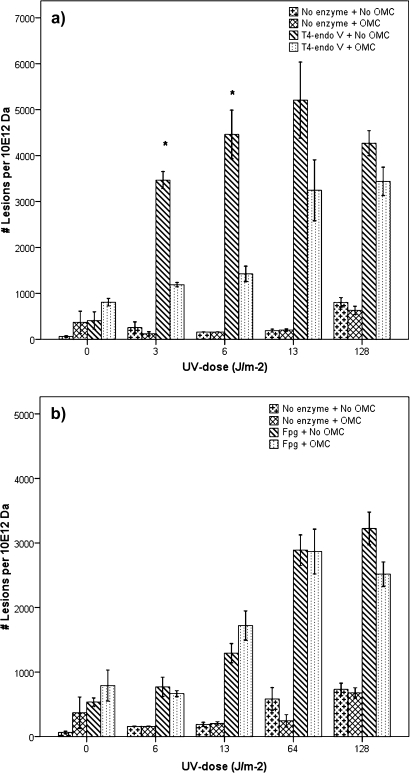
The semi-automated alkaline elution assay, in combination with Fpg or T4-endo V enzymes, was used to measure 8-oxoG and CPDs. The T4-endo V enzyme recognizes CPD, which is a representative lesion after UV exposure, whereas 6-4PPs—which are also formed—are not recognized by the enzyme. The main mutagenic lesion recognized by Fpg is 8-oxoG, but the enzyme has also affinity to other modified purines, such as ring-opened dG and dA.
Figure 2 shows UV dose–dependent increase in both CPDs and Fpg-sensitive lesions. OMC-protected cells had significantly lower levels of CPDs (p < 0.01) after exposure to UV (3 or 6 J/m2) compared to exposure without OMC. At higher UV exposure times (13 or 128 J/m2), the CPD levels for OMC-protected cells were also lower than those for unprotected cells; however, the observed OMC protection was not statistically significant (Fig. 2a). Figure 2b shows that, within the UV dose range used (0–128 J/m2), OMC provided no significant protection in the induction of the Fpg-sensitive lesions. There was no net increase in DNA SSBs (i.e., using no enzymes) at the lowest UV doses but a slight increase at 64 and 128 J/m2 of UV exposure (Fig. 2). We observed a very high response using T4-endo V at low UV doses approaching saturation (i.e., exceeding the dynamic range of the alkaline elution system) at 13 J/m2. Therefore, all high dose exposures were not included, and the 128 J/m2 dose illustrates that no further increase was found with this dose.
FIG. 2.
DNA lesions measured with the alkaline elution assay in cellular DNA after irradiation with UV, unfiltered or filtered through OMC. Exposure of MCF-7 cell line with UV (0–128 J/m2). (a) The number of CPDs (with T4-endoV enzyme). Asterisk represents significant reduction in net amount of induced CPDs (p < 0.01) in the presence of OMC (10 ppm, 2.55 cm) at low UV exposure times (3 and 6 J/m2). (b) The number of oxidated DNA lesions (with Fpg enzyme). No significant reduction in net amount of Fpg-sensitive DNA lesions in the presence of OMC at all UV exposure times. Bars represent means ± SE of five independent experiments.
In Table 1, these data are compared with the reduction in UV dose provided by OMC, calculated both with and without CIE correction. For CPDs, there were substantial relative reductions in lesion frequencies in the presence of OMC. For Fpg-sensitive sites, on the other hand, no such change was observed with OMC filtration. The results indicate that OMC-mediated UV light filtration reduced the formation of CPDs but not Fpg-sensitive sites. OMC by itself did not induce any significant genotoxic effects as measured by the alkaline elution assay.
TABLE 1.
CPDs and Fpg-Sensitive Sites Induced by UV Light, With and Without OMC Filtration
| UV exposure time (s) | UV dose (J/m2)a | OMC-filtered UV dose (J/m2)a | Mean lesions/1012 Da |
Relative (%) lesions with OMCb | |
| No OMC | OMC | ||||
| CPDs | |||||
| 25 | 6 | 3 | 4064 | 1451 | 36 |
| 50 | 12 | 6 | 4934 | 1370 | 28 |
| 100 | 25 | 13 | 6034 | 2628 | 44 |
| 1000 | 246 | 128 | 4852 | 3581 | 74 |
| Fpg-sensitive sites related to UVB irradiance | |||||
| 50 | 12 | 6 | 771 | 665 | 86 |
| 100 | 25 | 13 | 1292 | 1389 | 108 |
| 500 | 123 | 64 | 2919 | 2834 | 97 |
| 1000 | 246 | 128 | 2764 | 2503 | 91 |
Notes. UV dose–dependent CPD formation was observed, and there was a substantial relative reduction in lesion frequency with OMC (Column 6). (At the highest dose, there was less change due to saturation of alkaline elution lesion detection.) Fpg-sensitive sites were clearly induced in a dose-dependent manner, but no substantial relative change in their frequency with OMC was found (Column 6). The calculated UV dose was significantly reduced by OMC, irrespective of CIE correction (52% UV light remains after filtration of the CIE-corrected spectrum through OMC (10 ppm), and 32% UV light remains after filtration of the uncorrected spectrum through OMC).
CIE-corrected UV dose.
Relative (%) lesions remaining, with OMC [(Column 5)/Column 4 × 100]. OMC filtration: 10 ppm, 2.55 cm depth.
Cell Viability Measurements
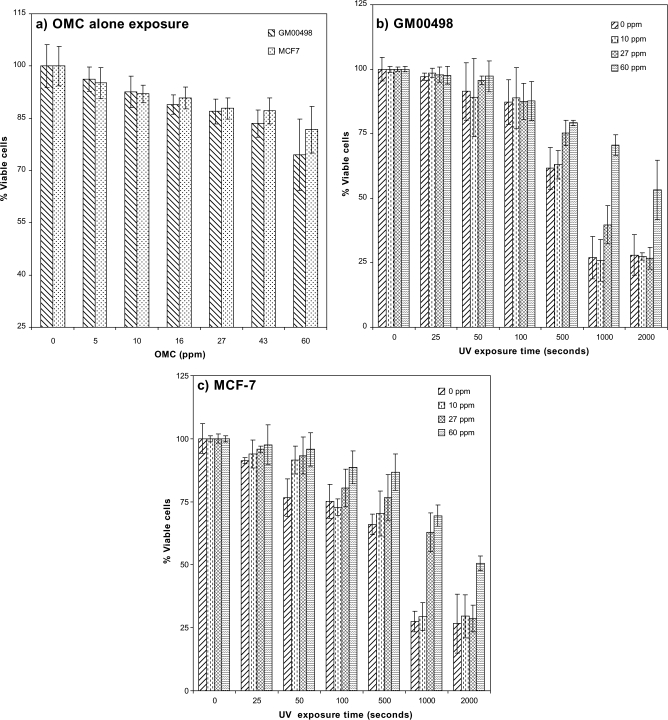
The MTT assay was used to measure cell viability of GM00498 and MCF-7 cells after UV exposure and in the presence or absence of OMC. Figure 3a shows that, for both cell types, there was around 10–25% reduction in cell viability from OMC-alone treatments at the two highest concentrations (43 and 60 ppm). Figures 3b and c show UV exposure–dependent reduction in cell viability. This reduction was highest at the lowest OMC concentrations after UV exposure time at and above 500 s (Figs. 3b and c and Supplementary table 2). This trend was the same for both cell types. In microscopic examination of propidium iodide (PI)/Hoechst-stained cells, changes in cell viability followed similar trends as with the MTT assay (data not shown). Based on these results, for the gene expression studies, we chose a low cytotoxic OMC (27 ppm) concentration, which resulted to ∼12% reduction in cell viability and when co-exposed with UV (500 s, corresponding to 45 J/m2 with 27 ppm OMC) resulted to ∼25% reduction in cell viability.
FIG. 3.
Cell viability measurements. Cell viability measured with the MTT assay. (a) GM00498 and MCF-7 cells exposed to OMC (0–60 ppm), with cell viabilities expressed as percentage of viable cells relative to untreated controls. (b) GM00498 cells exposed to various doses of UV in the presence of OMC (0–60 ppm), and cell viabilities were expressed as percentage of viable cells relative to UV unirradiated cells. (c) MCF-7 cells exposed to various doses of UV in the presence of OMC (0–60 ppm), and cell viabilities were expressed as percentage of viable cells relative to UV unirradiated cells. Bars represent means ± SE.
Gene Expression
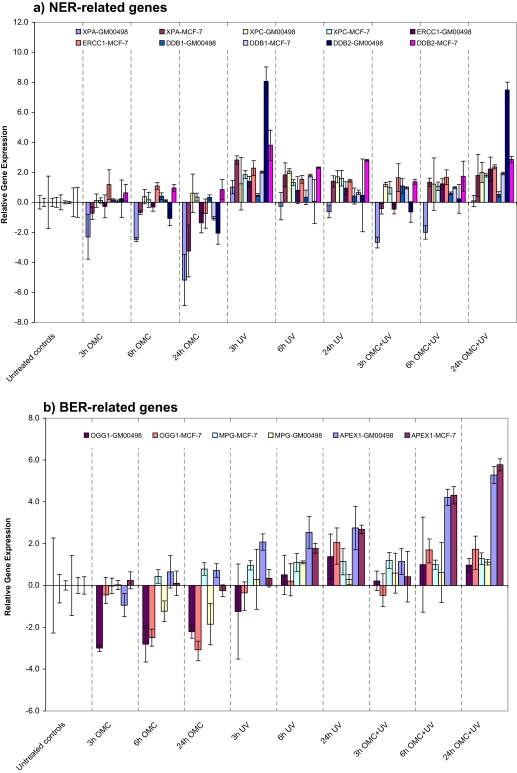
We were interested in identifying genes that are significantly differentially expressed among cells protected by OMC during UV irradiation compared to those exposed to UV alone and OMC alone in the dark. For this purpose, qRT-PCR was used to evaluate the transcriptional responses of a panel of 17 DNA damage response–related genes. These transcripts consist of five NER pathway–related genes, three BER pathway–related genes, five antioxidant-related enzymes, two immediate-early response genes, and GADD45A and TP53. By visual inspection of Figures 4a–d, we observe that there were clear differences between the gene expression profiles of the cells co-exposed with OMC and UV or UV-alone on the one hand and those exposed to OMC in the dark on the other hand. Overall, there were no significant differences in gene expression patterns between the GM00498 and the MCF-7 cell lines, and the observed gene expression changes therefore did not suggest cell type–specific differences. Furthermore, exposure of cells with OMC alone had no major effects on the transcription of several genes. It should be noted, however, that some genes showed downregulation upon the exposure to OMC alone (Figs. 4a–d and Supplementary table 3).
FIG. 4.
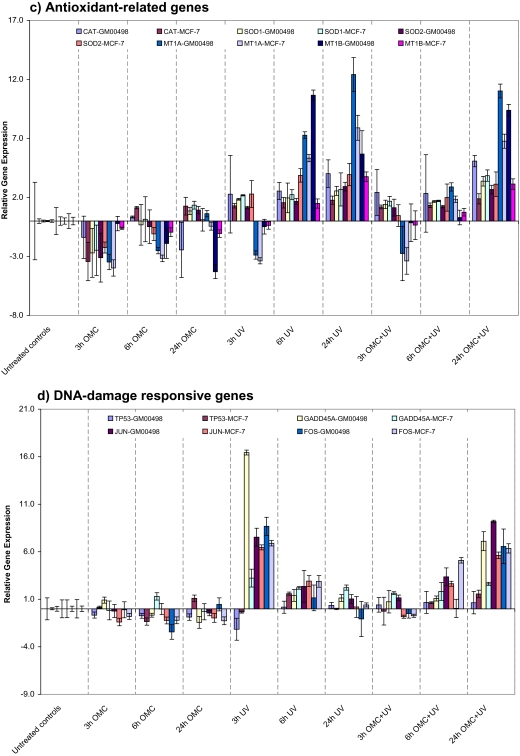
Time-dependent transcriptional response. The mRNA level of GM00498 and MCF-7 cells exposed to UV (45 J/m2) alone, OMC (27 ppm) plus UV (45 J/m2), or OMC (27 ppm) in the dark were analyzed by qRT-PCR. (a) The mRNA level of five NER pathway–related genes. (b) The mRNA level of three BER pathway genes. (c) The mRNA level of five antioxidant-related genes. (d) The mRNA level of two immediate-early response genes (FOS and JUN) and two DNA damage response–related genes (TP53 and GADD45A). The relative mRNA expression level data are presented as log2-transformed 2−ΔΔCt values. Bars represent means ± SE of three independent experiments.
Sunscreens (OMC) protect the cells from the UV-induced DNA lesions. Therefore, we expected that the expression of NER pathway genes would not be significantly affected in the OMC-protected cells. Figure 4a shows the expression pattern of the five NER pathway genes studied: DDB1, DDB2, ERCC1, XPA, and XPC. We observed a time-dependent induction of the messenger RNA (mRNA) levels of these genes (some of them were expressed at moderate levels and others at low levels; however, their transcription pattern was similar) in cells co-exposed to OMC and UV, with maximum induction at 24-h postirradiation (Fig. 4a). With UV alone, the mRNA level of these genes was increased. The induction was maximum at 3-h postirradiation, thereafter slowly declining to control levels at 24 h (Fig. 4a). The expression pattern of these genes was in somehow similar, even though some of them showed a moderate or a low expression level. Downregulation of xeroderma pigmentosum, complementation group A (XPA), a key enzyme in the NER pathway, was observed in both cells with OMC-alone treatment.
We observed a time-dependent upregulation of the mRNA level also of the BER pathway gene APEX1 following exposure to OMC plus UV or to UV alone (Fig. 4b). Two other BER pathway genes, MPG and OGG1, were also somewhat upregulated upon OMC plus UV at 24-h postirradiation, but their induction was not statistically significant. Downregulation of OGG1 was observed with OMC-alone treatment.
The gene expression patterns of the five antioxidant-related genes: CAT, SOD1, SOD2, MT1A, and MT1B, which protect cells against oxidative stress, are shown in Figure 4c. We observed time-dependent upregulation of the mRNA levels of these genes in both cell lines following UV-alone or OMC plus UV exposure; the net increase was less pronounced in the latter case (Fig. 4c). Some of these genes showed downregulation upon the treatment to OMC alone (Fig. 4c).
We also conducted gene expression analysis for some other selected DNA damage response–related genes: FOS, JUN, GADD45A, and TP53 (Fig. 4d). Time-dependent induction of the immediate-early genes, JUN and FOS, was observed following OMC plus UV exposure, with maximum upregulation at 24 h. Exposure to UV alone had an opposite effect, with mRNA levels increasing to maximum levels at 3-h postirradiation and then declining at longer times (Fig. 4d). In addition, the transcriptional level of GADD45A gene, which is involved in DNA damage response, cell cycle arrest, and apoptosis, was significantly induced both by UV alone and by OMC plus UV, although the temporal changes were somewhat different (Fig. 4d). The mRNA level of TP53 gene was not affected in either cell lines following the exposure.
The Involvement of p53 Protein in the UV-Induced DNA Damage Signaling Pathways
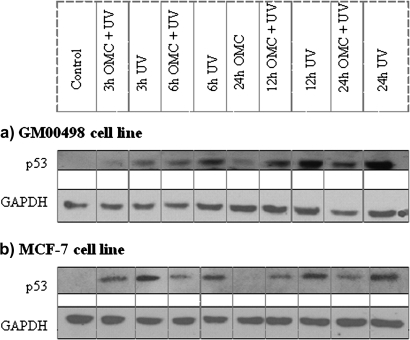
The tumor suppressor gene p53 is an important transcription factor activated by DNA damage. Some of the genes affected by UV alone or OMC plus UV are known p53 target genes. Using Western blot, we investigated the p53 protein expression levels following UV alone or OMC plus UV (Figs. 5a and b). The level of p53 protein was markedly increased by UV exposure; this increase was lower after OMC plus UV. However, the p53 expression level after OMC plus UV exposure was higher than in control cells and cells exposed to OMC alone (Fig. 5). The expression of GAPDH protein levels was not affected upon the exposures.
FIG. 5.
p53 protein expression analysis. Time-dependent induction of the p53 protein following the exposure of GM00498 or MCF-7 cells with OMC (27 ppm), UV (45 J/m2) alone, or OMC (27 ppm) plus UV (45 J/m2), as analyzed with Western blot. (a) GM00498 cells. (b) MCF-7 cells. GAPDH protein was used as internal control.
DISCUSSION
In this study, we evaluated photoprotection of OMC as well as the effects on UV-induced gene expression of this common sunscreen. With 10 ppm OMC in a buffer solution at a depth of 2.55 cm, we observed 52% reduction in the CIE-weighted dose (Fig. 1). This amount of OMC (25.5 μg/cm2) is comparable to the concentration found in the epidermis of human skin (5–25 μg/cm2) at normal use of OMC-containing sunscreen of protection factor 15 (Jiang et al., 1999). Furthermore, the UV doses used were in a range relevant to human solar light exposure.
The alkaline elution assay allows the sensitive measurement of DNA SSBs. To evaluate the photoprotection efficiency of OMC against UV-induced DNA damage, this assay was modified (Epe et al., 1993; Kielbassa et al., 1997) to include the T4-endo V enzyme, which has specific activity against UV-induced CPDs, or the Fpg enzyme, which recognizes oxidative purines, particularly 8-oxoG. With these modifications, it was possible to measure these lesions in a dose-dependent manner.
OMC significantly protected against UV-induced CDP formation (Fig. 2a). OMC is marketed as a UVB filter and is not claimed to provide protection against lesions induced by UVA. OMC absorbs also in the UVA region, and one would expect a reduced level of UVA-induced oxidative lesions. No such protection was observed (Fig. 2 and Table 1). Mouse lymphoma LY-R showed similar trends in DNA lesion-specific protection of OMC (data not shown).
Sunscreens are used as a major defense against solar UV, and one would anticipate major reductions in UV-induced transcriptional changes in DNA damage response genes when the cells are protected with OMC. A main finding from the gene expression analysis was the OMC-mediated characteristic delay of some genes known to be involved in DNA damage response. This information should be considered as relevant in systematic screening of sunscreen ingredients. We observed modulation of several genes that participate in DNA damage response pathways upon UV irradiation of OMC-protected cells (Fig. 4). There were clear differences in gene expression profiles between cells co-exposed with OMC plus UV compared to OMC-treated cells in the dark. Cells exposed with OMC plus UV showed similar expression patterns as the UV alone–treated cells. For some but not all genes studied, there was an apparent UV dose reductive effect of OMC at short times after exposure; furthermore, the change in expression of some genes seemed to be delayed in time (Fig. 4). Both cell lines responded similarly to the exposures, although further optimization would be expected to lead to more clearly distinguishable effects.
There are studies suggesting that the use of sunscreens can reduce DNA damage and p53 protein expression following UV exposure (Berne et al., 1998; Marrot et al., 2002). On the basis of these studies, we expected reduction in the p53 protein expression levels following UV irradiation in the OMC-protected cells. A slight induction of p53 protein expression was found in OMC-protected cells (Fig. 5). It is possible that the observed p53 protein induction in OMC-protected cells is a response to UV-induced oxidative damage rather than response to CPDs. OMC in the dark had no effect on p53 protein expression (Fig. 5). A similar accumulation of p53 protein in cells protected with photounstable sunscreen formulation has been reported (Fourtanier et al., 2006). Furthermore, p53 expression in MCF-7 cells was observed following UVB but not UVA (Wang et al., 1998).
One consequence of p53 induction is cell cycle arrest, giving the cells sufficient time to repair the damage in its genome. In our study, we observed an upregulation of the GADD45A gene both in OMC-protected and in UV alone–exposed cells (Fig. 4). It has been reported that GADD45A is induced by a wide spectrum of DNA-damaging agents, including UV (Brown, 2003; Hollander et al., 1993). It is also worth noting that the GADD45A gene is one of several known p53 target genes involved in a variety of DNA damage response–related pathways (Wang et al., 1999).
The decision whether the cells should undergo apoptosis or arrest in the cell cycle for DNA damage repair seems to be determined by the UV dose. We identified several important DNA repair genes related to different DNA repair pathways to be induced in response to OMC plus UV (Fig. 4). The product of DDB2 gene, which was clearly induced in our study (Fig. 4), forms—together with DDB1 gene—the UV-damaged DNA-binding protein complex. This complex is involved in several processes including repair, transcription, and cell cycle regulation and is essential for the initial recognition of CPDs during global genomic repair (Hwang et al., 1998; Tang et al., 2000). We found a downregulation of XPA by OMC alone, and the biological relevance of this change is unclear; however, moderate changes in the expression of XPA has limited effects on NER efficiency (Muotri et al., 2002).
Concerning BER pathway–related genes, we observed induction of APEX mRNA level following OMC plus UV exposure (Fig. 4). APEX1 is responsible for cleavage of apurinic/apyrimidinic (AP) sites via its 5′-endonuclease activity, and it also plays a role in p53 activation and redox-dependent activation of AP-1 (c-Jun/c-Fos) (Evans et al., 2000). The expression level of the highly relevant BER gene OGG1 was significantly downregulated by OMC alone. OGG1 gene is responsible for the excision of 8-oxoG and may be regulated at a posttranslational level and has been reported to be acetylated upon oxidative stress (Bhakat et al., 2006).
A notable observation in this study is a change in the temporal pattern of gene expression from OMC, suggesting a possible interference with normal progression of cell division. This change was found for the FOS and JUN genes (Fig. 4). UV belongs to the extracellular influences that activate the immediate-early mitogen-regulated genes, such as c-fos and c-jun (Holbrook and Fornace, 1991), and these genes may promote cell growth after the DNA lesions have been repaired. UV irradiation has been reported to upregulate the expression of FOS and JUN genes, via phosphorylation of JNK and p38, leading to the activation of AP-1 and NFκB and increasing damage tolerance (Xia et al., 1995).
Cells are also protected against oxidative stress by an interacting network of antioxidant enzymes, such as superoxide dismutases, catalases, metallothioneines, and various peroxidases (Packer and Valacchi, 2002; Vayalil et al., 2003). We observed time-dependent upregulation of the mRNA level of these genes following the exposure of OMC plus UV or UV alone (Fig. 4). Antioxidant genes are considered to be constitutively expressed; however, their mRNA levels can be regulated by various environmental stresses. The expression patterns of these genes did not follow the same temporal trend as was observed for the DNA damage response genes (FOS, JUN, GADD45, and DDB2) following OMC plus UV and UV-alone exposure.
In conclusion, our data support the notion that OMC provides protection against CPDs in cellular DNA and the degree of protection correlates with the reduced transmission of UV light in OMC. With regard to oxidative DNA lesions, protection did not seem to be provided. The gene expression results suggest that the overall cellular response to DNA damage was significantly altered by OMC, and the effects were in general similar in the two cell types used. Among the panel of 17 selected genes studied, several are considered as general biomarkers of genotoxic response. The systematic approach applied in this study—with appropriate standardization—may have a potential use in the initial screening and evaluation of sunscreen active agents.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
Supplementary Material
Acknowledgments
We thank Bjørn Johnsen at the Norwegian Radiation Protection Authority for spectral measurements of the UV light source. We are grateful to Magnar Bjørås (University of Oslo, Norway) for providing the E. coli ER 2566 strain harboring the pFPG230 plasmid and Bernd Epe (Institute of Pharmacy, University of Mainz, Germany) for providing the E. coli AB2480 (uvrA, recA, F'lac IQ1) plus ptac-den V(Apr) strain harboring T4-endonuclease V plasmid. We also thank Minh Hoang, Markus Brun Hustad, and Petter Hamborg for their excellent technical assistance, and Birgitte Lindeman for valuable help with cell viability measurements. The authors declare that they have no competing interests. All authors have read and approved the final version of the manuscript.
References
- Al Mahroos M, Yaar M, Phillips TJ, Bhawan J, Gilchrest BA. Effect of sunscreen application on UV-induced thymine dimers. Arch. Dermatol. 2002;138:1480–1485. doi: 10.1001/archderm.138.11.1480. [DOI] [PubMed] [Google Scholar]
- Allen JM, Gossett CJ, Allen SK. Photochemical formation of singlet molecular oxygen in illuminated aqueous solutions of several commercially available sunscreen active ingredients. Chem. Res. Toxicol. 1996;9:605–609. doi: 10.1021/tx950197m. [DOI] [PubMed] [Google Scholar]
- Ananthaswamy HN, Loughlin SM, Cox P, Evans RL, Ullrich SE, Kripke ML. Sunlight and skin cancer: inhibition of p53 mutations in UV-irradiated mouse skin by sunscreens. Nat. Med. 1997;3:510–514. doi: 10.1038/nm0597-510. [DOI] [PubMed] [Google Scholar]
- Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J. Photochem. Photobiol. B. 2001;63:8–18. doi: 10.1016/s1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- Berne B, Ponten J, Ponten F. Decreased p53 expression in chronically sun-exposed human skin after topical photoprotection. Photodermatol. Photoimmunol. Photomed. 1998;14:148–153. doi: 10.1111/j.1600-0781.1998.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, O’Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- Bredholt K, Christensen T, Hannevik M, Johnsen B, Seim J, Reitan JB. [Effects of sunscreening agents and reactions with ultraviolet radiation] Tidsskr. Nor. Laegeforen. 1998;118:2640–2645. [PubMed] [Google Scholar]
- Brezova V, Gabcova S, Dvoranova D, Stasko A. Reactive oxygen species produced upon photoexcitation of sunscreens containing titanium dioxide (an EPR study) J. Photochem. Photobiol. B. 2005;79:121–134. doi: 10.1016/j.jphotobiol.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Brown EJ. The ATR-independent DNA replication checkpoint. Cell Cycle. 2003;2:188–189. [PubMed] [Google Scholar]
- Brunborg G, Holme JA, Soderlund EJ, Omichinski JG, Dybing E. An automated alkaline elution system: DNA damage induced by 1,2-dibromo-3-chloropropane in vivo and in vitro. Anal. Biochem. 1988;174:522–536. doi: 10.1016/0003-2697(88)90052-8. [DOI] [PubMed] [Google Scholar]
- Brunborg G, Soderlund EJ, Holme JA, Dybing E. Organ-specific and transplacental DNA damage and its repair in rats treated with 1,2-dibromo-3-chloropropane. Chem. Biol. Interact. 1996;101:33–48. doi: 10.1016/0009-2797(96)03709-x. [DOI] [PubMed] [Google Scholar]
- Butt ST, Christensen T. Toxicity and phototoxicity of chemical sun filters. Radiat. Prot. Dosimetry. 2000;91:283–286. [Google Scholar]
- Cole CA, Forbes PD, Davies RE. An action spectrum for UV photocarcinogenesis. Photochem. Photobiol. 1986;43:275–284. doi: 10.1111/j.1751-1097.1986.tb05605.x. [DOI] [PubMed] [Google Scholar]
- Duale N, Lindeman B, Komada M, Olsen AK, Andreassen A, Soderlund EJ, Brunborg G. Molecular portrait of cisplatin induced response in human testis cancer cell lines based on gene expression profiles. Mol. Cancer. 2007;6:53. doi: 10.1186/1476-4598-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmets CA, Anderson CY. Sunscreens and photocarcinogenesis: an objective assessment. Photochem. Photobiol. 1996;63:435–440. doi: 10.1111/j.1751-1097.1996.tb03065.x. [DOI] [PubMed] [Google Scholar]
- Epe B, Pflaum M, Boiteux S. DNA damage induced by photosensitizers in cellular and cell-free systems. Mutat. Res. 1993;299:135–145. doi: 10.1016/0165-1218(93)90091-q. [DOI] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Fourtanier A, Bernerd F, Bouillon C, Marrot L, Moyal D, Seite S. Protection of skin biological targets by different types of sunscreens. Photodermatol. Photoimmunol. Photomed. 2006;22:22–32. doi: 10.1111/j.1600-0781.2006.00188.x. [DOI] [PubMed] [Google Scholar]
- Gulston M, Knowland J. Illumination of human keratinocytes in the presence of the sunscreen ingredient Padimate-O and through an SPF-15 sunscreen reduces direct photodamage to DNA but increases strand breaks. Mutat. Res. 1999;444:49–60. doi: 10.1016/s1383-5718(99)00091-1. [DOI] [PubMed] [Google Scholar]
- Helbock HJ, Beckman KB, Ames BN. 8-Hydroxydeoxyguanosine and 8-hydroxyguanine as biomarkers of oxidative DNA damage. Methods Enzymol. 1999;300:156–166. doi: 10.1016/s0076-6879(99)00123-8. [DOI] [PubMed] [Google Scholar]
- Holbrook NJ, Fornace AJ., Jr Response to adversity: molecular control of gene activation following genotoxic stress. New Biol. 1991;3:825–833. [PubMed] [Google Scholar]
- Hollander MC, Alamo I, Jackman J, Wang MG, McBride OW, Fornace AJ., Jr. Analysis of the mammalian gadd45 gene and its response to DNA damage. J. Biol. Chem. 1993;268:24385–24393. [PubMed] [Google Scholar]
- Hwang BJ, Toering S, Francke U, Chu G. p48 Activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. IARC Handbooks of Cancer Prevention Sunscreens. 2001. Vol. 5, International Agency for Research on Cancer, Lyon (France) [Google Scholar]
- Jiang R, Roberts MS, Collins DM, Benson HA. Absorption of sunscreens across human skin: an evaluation of commercial products for children and adults. Br. J. Clin. Pharmacol. 1999;48:635–637. doi: 10.1046/j.1365-2125.1999.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- Kinley JS, Brunborg G, Moan J, Young AR. Photoprotection by furocoumarin-induced melanogenesis against DNA photodamage in mouse epidermis in vivo. Photochem. Photobiol. 1997;65:486–491. doi: 10.1111/j.1751-1097.1997.tb08595.x. [DOI] [PubMed] [Google Scholar]
- Lehmann AR. The molecular biology of nucleotide excision repair and double-strand break repair in eukaryotes. Genet. Eng. (N Y) 1995;17:1–19. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maier H, Schauberger G, Brunnhofer K, Honigsmann H. Change of ultraviolet absorbance of sunscreens by exposure to solar-simulated radiation. J. Invest. Dermatol. 2001;117:256–262. doi: 10.1046/j.0022-202x.2001.01425.x. [DOI] [PubMed] [Google Scholar]
- Marrot L, Belaidi JP, Meunier JR. Comet assay combined with p53 detection as a sensitive approach for DNA photoprotection assessment in vitro. Exp. Dermatol. 2002;11(Suppl. 1):33–36. doi: 10.1034/j.1600-0625.11.s.1.8.x. [DOI] [PubMed] [Google Scholar]
- Mitchell D, Paniker L, Sanchez G, Trono D, Nairn R. The etiology of sunlight-induced melanoma in Xiphophorus hybrid fish. Mol. Carcinog. 2007;46:679–684. doi: 10.1002/mc.20341. [DOI] [PubMed] [Google Scholar]
- Mitra S, Hazra TK, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol. Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- Muotri AR, Marchetto MCN, Suzuki MF, Okazaki K, Lotfi CFP, Brumatti G, Amarante-Mendes GP, Menck CFM. Low amounts of the DNA repair XPA protein are sufficient to recover UV-resistance. Carcinogenesis. 2002;23:1039–1046. doi: 10.1093/carcin/23.6.1039. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Yamashita K, Sekiguchi M. Purification and characterization of normal and mutant forms of T4 endonuclease V. J. Biol. Chem. 1982;257:2556–2562. [PubMed] [Google Scholar]
- Olsen AK, Duale N, Bjoras M, Larsen CT, Wiger R, Holme JA, Seeberg EC, Brunborg G. Limited repair of 8-hydroxy-7,8-dihydroguanine residues in human testicular cells. Nucleic Acids Res. 2003;31:1351–1363. doi: 10.1093/nar/gkg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer L, Valacchi G. Antioxidants and the response of skin to oxidative stress: vitamin E as a key indicator. Skin Pharmacol. Appl. Skin Physiol. 2002;15:282–290. doi: 10.1159/000064531. [DOI] [PubMed] [Google Scholar]
- Pangnakorn P, Nonthabenjawan R, Ekgasit S, Thammacharoen C, Pattanaargson WS. Monitoring 2-ethylhexyl-4-methoxycinnamate photoisomerization on skin using attenuated total reflection fourier transform infrared spectroscopy. Appl. Spectrosc. 2007;61:193–198. doi: 10.1366/000370207779947648. [DOI] [PubMed] [Google Scholar]
- Pattyn F, Speleman F, De Paepe A, Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelle E, Huang X, Mammone T, Marenus K, Maes D, Frenkel K. Ultraviolet-B-induced oxidative DNA base damage in primary normal human epidermal keratinocytes and inhibition by a hydroxyl radical scavenger. J. Invest. Dermatol. 2003;121:177–183. doi: 10.1046/j.1523-1747.2003.12330.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Reinhardt P, Cybulski M, McNamee JP, Mclean JR, Gorman W, Deslauriers Y. Protection from solar simulated radiation-induced DNA damage in cultured human fibroblasts by three commercially available sunscreens. Can. J. Physiol. Pharmacol. 2003;81:690–695. doi: 10.1139/y03-062. [DOI] [PubMed] [Google Scholar]
- Schlumpf M, Cotton B, Conscience M, Haller V, Steinmann B, Lichtensteiger W. In vitro and in vivo estrogenicity of UV screens. Environ. Health Perspect. 2001;109:239–244. doi: 10.1289/ehp.01109239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpone N, Salinaro A, Emeline AV, Horikoshi S, Hidaka H, Zhao J. An in vitro systematic spectroscopic examination of the photostabilities of a random set of commercial sunscreen lotions and their chemical UVB/UVA active agents. Photochem. Photobiol. Sci. 2002;1:970–981. doi: 10.1039/b206338g. [DOI] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarras-Wahlberg N, Stenhagen G, Larko O, Rosen A, Wennberg AM, Wennerstrom O. Changes in ultraviolet absorption of sunscreens after ultraviolet irradiation. J. Invest. Dermatol. 1999;113:547–553. doi: 10.1046/j.1523-1747.1999.00721.x. [DOI] [PubMed] [Google Scholar]
- Vayalil PK, Elmets CA, Katiyar SK. Treatment of green tea polyphenols in hydrophilic cream prevents UVB-induced oxidation of lipids and proteins, depletion of antioxidant enzymes and phosphorylation of MAPK proteins in SKH-1 hairless mouse skin. Carcinogenesis. 2003;24:927–936. doi: 10.1093/carcin/bgg025. [DOI] [PubMed] [Google Scholar]
- Wang XW, Zhan Q, Coursen JD, Khan MA, Kontny HU, Yu L, Hollander MC, O’Connor PM, Fornace AJ, Jr, Harris CC. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rosenstein B, Goldwyn S, Zhang XS, Lebwohl M, Wei HC. Differential regulation of p53 and Bcl-2 expression by ultraviolet A and B. J. Invest. Dermatol. 1998;111:380–384. doi: 10.1046/j.1523-1747.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- Young AR, Chadwick CA, Harrison GI, Nikaido O, Ramsden J, Potten CS. The similarity of action spectra for thymine dimers in human epidermis and erythema suggests that DNA is the chromophore for erythema. J. Invest. Dermatol. 1998;111:982–988. doi: 10.1046/j.1523-1747.1998.00436.x. [DOI] [PubMed] [Google Scholar]
- Young AR, Walker SL. Effects of solar simulated radiation on the human immune system: influence of phototypes and wavebands. Exp. Dermatol. 2002;11(Suppl. 1):17–19. doi: 10.1034/j.1600-0625.11.s.1.5.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.