Abstract
Engineering the antibody Fc region to enhance the cytotoxic activity of therapeutic antibodies is currently an active area of investigation. The contribution of complement to the mechanism of action of some antibodies that target cancers and pathogens makes a compelling case for its optimization. Here we describe the generation of a series of Fc variants with enhanced ability to recruit complement. Variants enhanced the cytotoxic potency of an anti-CD20 antibody up to 23-fold against tumor cells in CDC assays, and demonstrated a correlated increase in C1q binding affinity. Complementenhancing substitutions combined additively, and in one case synergistically, with substitutions previously engineered for improved binding to Fc gamma receptors. The engineered combinations provided a range of effector function activities, including simultaneously enhanced CDC, ADCC, and phagocytosis. Variants were also effective at boosting the effector function of antibodies targeting the antigens CD40 and CD19, in the former case enhancing CDC over 600-fold, and in the latter case imparting complement-mediated activity onto an IgG1 antibody that was otherwise incapable of it. This work expands the toolkit of modifications for generating monoclonal antibodies with improved therapeutic potential and enables the exploration of optimized synergy between Fc gamma receptors and complement pathways for the destruction of tumors and infectious pathogens.
Key words: antibody, Fc, complement, CDC, C1q, ADCC, phagocytosis, CD20, CD19, CD40
Introduction
Monoclonal antibodies (mAbs) are successful as therapeutics due in part to their ability to bring to bear the destructive capabilities of the immune system against specific target cells. In a variety of in vivo and in vitro settings, antibody coating of targets has been shown to mediate potent killing mechanisms such as complement- dependent cytotoxicity (CDC), antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP). All of these effector functions are mediated by the antibody Fc region and over the past decade engineering of this region to enhance the cytotoxic activity of therapeutic antibodies has been a subject of intense investigation.1,2
Modification of antibodies to boost the response of the complement system is appealing due to its contribution to the mechanism of action of one of the most successful anti-cancer mAbs, anti-CD20 rituximab,3 as well as its important role in destroying invading pathogens. For antibody therapeutics, the relevant arm of the complement system is the classical (antibody-dependent) complement cascade, which consists of over twenty tightlyregulated proteins, C1 through C9. The trigger for classical complement activation is the initial binding to antibody-coated target by complement protein C1q, a bundle of six heterotrimeric subunits composed of globular heads and collagen-like tails. For human IgG1, CH2 domain residues D270, K322, P329 and P331 have been implicated as essential to the interaction between human IgG antibodies and C1q.4,5 Efforts have been made to increase the affinity of this interaction, with the goal of concomitantly increasing progression through the classical complement cascade to enhance lysis of the target cells. Several groups have taken advantage of the differences in CDC activity among human IgG isotypes (IgG3 > IgG1 ≫ IgG2 ≈ IgG46) by swapping segments between isotypes to generate various chimeric IgG molecules with enhanced complement recruitment.7–10 Others have engineered specific amino acid substitutions into either the hinge region11–13 or the CH2 domain.14
The most widely recognized mechanism of complement-mediated target destruction is lysis by the membrane-attack complex (MAC), a transmembrane channel created by complexation of C5b, C6, C7, C8 and C9 proteins. This non-cellular process, commonly referred to as CDC, is thought to be relevant to the clinical activity of some anti-tumor antibodies. 15 Less established for antibody drugs, although potentially no less relevant, are cellular-based complement mechanisms that are mediated by interaction between opsonic C3 and C4 components and complement receptors (CR1, CR3 and CR4) expressed on effector cells. Particularly intriguing from the standpoint of improving mAb clinical activity is the synergy between complement and Fc gamma receptor (FcγR) effector pathways. These include enhancement of FcγR-mediated effector functions by the CR/opsonin interaction, and the capacity of complement protein C5a as a potent chemoattractant and positive regulator of activating FcγR expression relative to inhibitory FcγRIIb.16,17 Although such crosstalk may typically rely on activation by microbial danger signals, tapping into the cooperation between complement and FcγRs is an enticing approach to improving the cytotoxic potency of mAbs.
Our long-term goal has been the engineering of therapeutic antibodies with improved clinical performance. Here we have built on previous Fc work18,19 to generate variants that provide enhanced complement- and FcγR-mediated activities. We have engineered a series of Fc variants with increased ability to recruit complement in the context of the humanized anti-CD20 monoclonal IgG1 antibody ocrelizumab.20 Variants demonstrated greatly enhanced potency in a cell-based CDC assay and improved binding affinity to C1q, and were transferable to antibodies targeting other antigens. By combining CDC-enhancing variants with previous variants that improve binding to FcγRs, we have generated antibodies simultaneously enhanced for multiple cellular and non-cellular immune effector arms.
Results
Engineering of CDC-enhanced anti-CD20 antibodies.
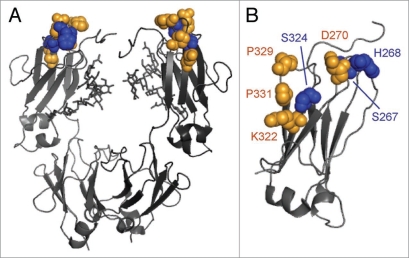
We have previously screened a large set of variants for binding to all FcγRs, C1q and the neonatal receptor FcRn (data not shown) and selected variants have been described previously.19,21 Based on the C1q data from these studies in conjunction with the reported location of the C1q binding center,4,5 we screened 38 single substitution variants for CDC activity (Suppl. Fig. 1). From these data, we identified three substitutions (S267E, H268F, S324T) in the human IgG1 CH2 domain (Fig.1) for further study. Variant Fc domains including one to three of these substitutions were constructed, expressed, and screened in the context of ocrelizumab,20 a humanized anti-CD20 IgG1 antibody.
Figure 1.
Cartoon representation of human IgG1 antibody Fc from Protein Data Bank record 1E4K65 with positions at which substitution modulates C1q binding affinity highlighted as space-filling spheres. The putative C1q binding center (D270, K322, P329 and P331) is colored orange. Residues S267, H268 and S324 are indicated in blue. Oligosaccharides are represented as sticks. (A) Full Fc. (B) CH2 domain only.
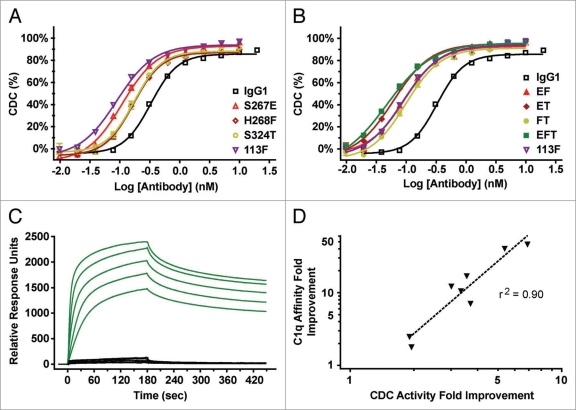
We examined the relative CDC activity of the anti-CD20 Fc variants against Burkitt’s lymphoma Ramos cells (Fig. 2A and B). Human serum complement was added to opsonized target cells, cell viability was determined from Alamar Blue fluorescence and half-maximal effective concentration (EC50) values of the antibody-dependent cell lysis were calculated (Table 1). The three single substitutions resulted in potency increases of 1.9- to 3.0-fold relative to native IgG1 ocrelizumab. When the single substitutions were combined, potencies further increased, ranging from 3.3-fold to 5.4-fold for double substitution variants and 6.9-fold for the triple. Several variants surpassed the potency of 113F, an IgG1/IgG3 chimera7 included as a positive control for CDC enhancement. Similar results were observed when targeting the Burkitt’s lymphoma Raji cell line (data not shown).
Figure 2.
Fc engineering generates variant anti-CD20 antibodies with enhanced binding affinity for C1q and enhanced cytotoxicity of CD20+ Ramos cells. (A and B) CDC activity of Fc variant anti-CD20 mAbs against opsonized Ramos cells using human complement. Antibody-dependent % lysis was measured at multiple antibody concentrations by Alamar Blue-based detection (mean ± SE of duplicate wells). EC50s are listed in Table 1. (C) SPR sensorgrams for native IgG1 (black) and variant EFT (green) are shown. C1q concentrations range from 100 nM to 6.25 nM by 2-fold serial dilution. (D) Correlation between the fold improvements in C1q affinity as determined by SPR (Table 1) and CDC activity (Table 1, A and B).
Table 1.
CDC activities and human C1q binding affinities of Fc variant antibodies
| CDC assay | Clq SPR binding | ||||
| ID | Variant | EC50 (nM)a | Foldb | Kd (nM)c | Foldd |
| IgG1 | Native IgG1e | 0.33 | 1 | 48 | 1 |
| — | S324T | 0.17 | 1.9 | 19 | 2.5 |
| — | H268F | 0.17 | 2.0 | 26 | 1.8 |
| — | S267E | 0.11 | 3.0 | 3.9 | 12 |
| FT | H268F/S324T | 0.098 | 3.3 | 4.6 | 11 |
| EF | S267E/H268F | 0.092 | 3.6 | 2.8 | 17 |
| ET | S267E/S324T | 0.061 | 5.4 | 1.2 | 41 |
| EFT | S267E/H268F/S324T | 0.048 | 6.9 | 1.0 | 47 |
| — | 113F | 0.089 | 3.7 | 6.9 | 7.1 |
aEC50s were from four-parameter sigmoidal dose-response fits (n = 2). bFold = EC50 (Native IgG1)/EC50 (variant). cKd = Kd1/(1 + 1/Kd2) from a global two-state binding fit of SPR data. dFold = Kd (Native IgG1)/Kd (variant). eAntibodies had ocrelizumab variable regions.
We next examined the binding of the Fc variants to human C1q using surface plasmon resonance (SPR). Sensorgrams (Fig.2C) were fit with a two-state binding model (Table 1). Although the fitted Kd values do not represent the actual Kd between C1q globular head and Fc, they nonetheless reflect the relative affinity of the C1q multimeric bundle for an opsonized surface. The C1q affinities of the variants showed similar rank order to their CDC potencies, with a correlation coefficient of r2 = 0.90 (p < 0.0005) (Fig. 2D). Consistent with its 6.9-fold increase in CDC potency, the S267E/H268F/S324T (EFT) variant had the tightest C1q affinity as well, an increase of 47-fold over native IgG1 and 6.5-fold over the 113F positive control.
Engineering of anti-CD20 antibodies with augmented CDC, ADCC and ADCP activity.
In an effort to develop variants with broadly enhanced effector function, we combined our CDC-enhancing substitutions with variants previously characterized for improved FcγR-mediated activity.18,19 These include two double substitution variants, one with broad affinity enhancement to all FcγRs (S239D/I332E, referred to here as DE), and the other with selective affinity enhancement to FcγRIIa and FcγRIIIa relative to FcγRIIb (G236A/I332E, referred to as AE). These substitutions were added via site-directed mutagenesis to H268F/S324T and S267E/H268F/S324T, resulting in a set of four variants, each with four or five substitutions. The EFT triple was chosen as the variant with the greatest CDC activity, while the FT double was of interest as the most potent CDC variant lacking S267E, which was expected from previous observations22 to impair FcγR-mediated effector function due to decreased FcγRIIIa and increased FcγRIIb affinity. For simplicity we refer to these variant combinations by adding the FcγR substitutions as a suffix to the CDC variants, i.e., FT + DE, FT + AE, EFT + DE and EFT + AE. Combination variants were constructed and expressed in the context of anti-CD20 IgG1 ocrelizumab.
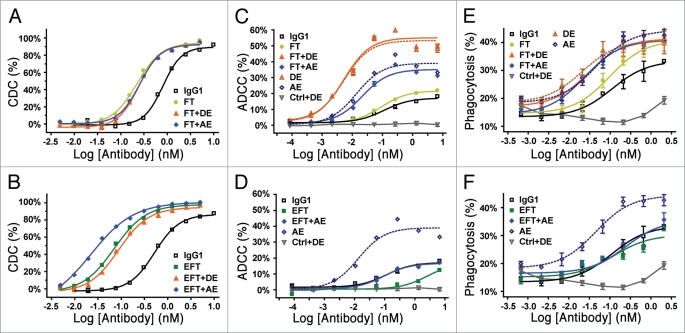
CDC assays of the combination variants confirmed that the enhanced potency conferred by the initial engineering remained upon adding the substitutions (Fig. 3A and B). These results were supported by correlated increases in C1q affinity (data not shown). Unexpectedly, we found that the EFT + AE variant gained an additional 3.3-fold in CDC activity (and, similarly, C1q affinity, data not shown) relative to the EFT triple variant (Table 2). However, this synergy was not observed with the EFT + DE combination (Fig. 3B). Subsequent work identified I332E and S267E as the synergistic pair, but that their synergy was absent in the presence of S239D (data not shown). The five-substitution EFT + AE variant was the most potent complement mediator, providing enhancement to CDC EC50 by 23-fold in the context of the anti-CD20 (Table 2). A transform of the data based on cell-surface binding indicated that the variants lowered the CD20 saturation level needed for CDC activity (Suppl. Fig. 2); remarkably, the CDC EC50 of the EFT + AE variant corresponded to less than 1% CD20 saturation level versus ∼15% for native IgG1.
Figure 3.
Fc variants enhance CDC, ADCC and ADCP. (A and B) CDC activity of Fc variant anti-CD20 mAbs against opsonized Ramos cells using human complement. Antibody-dependent % lysis was measured at multiple antibody concentrations by Alamar Blue-based detection (mean ± SE of duplicate wells). (C and D) ADCC activity of Fc variant anti-CD20 mAbs against Ramos cells using human PBMCs (FcγRIIa genotype was H131/R131, FcγRIIIa genotype was V158/F158). Antibody-dependent % cytotoxicity was measured at multiple antibody concentrations by lactate dehydrogenase release (mean ± SE of triplicate wells). (E and F) Phagocytosis activity of Fc variant anti-CD20 mAbs against Ramos cells using purified human monocytederived macrophages. Antibody-dependent % phagocytosis was measured at multiple antibody concentrations by flow cytometry (mean ± SE of triplicate wells). Macrophages for this experiment were H131/R131 FcγRIIa and V158/F158 FcγRIIIa genotype. Ctrl + DE for ADCC (C and D) and ADCP (E and F) experiments represents an isotype control anti-RSV antibody with DE substitutions. For all of the data shown, fold improvements in EC50 and maximal lysis relative to native IgG1 are listed in Table 2.
Table 2.
Summary of fold improvements for multiple cellular and non-cellular effector arms
| CDC | ADCC | ADCP | ||||
| IDa | Potencyb | Efficacyc | Potencyb | Efficacyc | Potencyb | Efficacyc |
| FT | 3.3 | 1.1 | 1.2 | 1.2 | 1.2 | 1.2 |
| EFT | 6.9 | 1.1 | 0.045 | 0.68 | 0.91 | 0.94 |
| FT + DE | 3.3 | 1.1 | 22 | 3.4 | 3.7 | 1.2 |
| FT + AE | 3.2 | 1.1 | 5.3 | 2.1 | 4.7 | 1.2 |
| EFT + AE | 23 | 1.1 | 1.2 | 1.0 | 0.46 | 1.0 |
| DE | 1.0 | 0.94 | 22 | 3.3 | 5.4 | 1.2 |
| AE | 1.2 | 1.0 | 8.3 | 2.5 | 2.5 | 1.3 |
aVariant ID’s as in Table 1, plus: DE (S239D/I332E) and AE (G236A/I332E). bPotency Fold = EC50 (Native IgG1)/EC50 (variant). cEfficacy Fold = Maximum Lysis (variant)/Maximum Lysis (Native IgG1).
Affinities of the variant anti-CD20 antibodies to activating and inhibitory human FcγRs were examined by SPR (Table 3). The FT double substitution marginally affected FcγR binding if at all, with the most significant perturbation being a slightly lower affinity for FcγRIIb. Addition of the DE and AE substitutions to this variant dramatically improved FcγR binding, resulting in variants with broad enhancement to FcγRs, particularly the isoforms of FcγRIIIa (FT + DE), or selective enhancement for FcγRIIa and FcγRIIIa relative to FcγRIIb (FT + AE). The EFT CDC-enhancing variant displayed reduced FcγRIIIa affinity and sharply increased binding to FcγRIIa R131 and FcγRIIb. This binding profile is similar to that observed in previous work with the S267E substitution.22 Combination with the AE substitutions produced a variant (EFT + AE) with substantially higher FcγRIIa affinity, a greater H131 FcγRIIa:FcγRIIb ratio yet high FcγRIIb affinity, and FcγRIIIa binding slightly better than native IgG1. The clearly superior CDC activity of this combination variant (Fig. 3B) made the EFT + AE variant our sole EFT combination choice for further characterization.
Table 3.
Binding affinities of Fc variant antibodies for human Fcγ receptors measured by SPR
| FcγRI | FcγRIIa H131 | FcγRIIa R131 | FcγRIIb | FcγRIIIa V158 | FcγRIIIa F158 | |||||||
| ID | Kd (nM) | Fold | Kd (µM) | Fold | Kd (µM) | Fold | Kd (µM) | Fold | Kd (µM) | Fold | Kd (µM) | Fold |
| IgG1 | 0.17 | 1 | 1.2 | 1 | 1.6 | 1 | 4.6 | 1 | 0.33 | 1 | 2.2 | 1 |
| FT | 0.25 | 0.70 | 0.80 | 1.5 | 2.4 | 0.67 | 12 | 0.38 | 0.33 | 1.0 | 2.2 | 1.0 |
| EFT | 0.11 | 1.6 | 0.35 | 3.5 | 0.045 | 37 | 0.25 | 18 | 1.0 | 0.32 | 11 | 0.21 |
| FT + DE | 0.021 | 8.2 | 0.21 | 5.8 | 0.18 | 9.2 | 0.47 | 9.9 | 0.0058 | 57 | 0.066 | 33 |
| FT + AE | 0.26 | 0.67 | 0.043 | 28 | 0.35 | 4.7 | 3.1 | 1.5 | 0.12 | 2.8 | 0.55 | 4.0 |
| EFT + AE | 0.10 | 1.8 | 0.035 | 35 | 0.0073 | 220 | 0.038 | 120 | 0.15 | 2.2 | 0.65 | 3.4 |
To examine the effect of these FcγR binding profiles on effector recruitment, the anti-CD20 variant antibodies were studied in cell-based ADCC (Fig. 3C and D) and ADCP assays (Fig. 3E and F). These experiments used the Ramos cell line as target cells, and either purified peripheral blood mononuclear cells (PBMCs) or monocytederived macrophages as effectors for ADCC and ADCP respectively. For both sets of experiments donor allotypes were determined to be heterozygous for both FcγRIIa (H131/R131) and FcγRIIIa (V158/F158). For ADCC, we and others have previously shown that FcγRIIIa-expressing natural killer cells are the primary effectors.19 For macrophage phagocytosis, we have demonstrated that FcγRIIa is the dominant receptor, with less prominent but still significant contributions from FcγRI and FcγRIIIa. The H268F/S324T variant had similar ADCC activity to native IgG1 and slightly improved ADCP (Fig. 3C and E). Combination with the AE and DE variants resulted in moderate (5.3-fold) and dramatic (22-fold) enhancements in ADCC activity, respectively, (Fig. 3C, Table 2) as a consequence of their increased binding to FcγRIIIa (Table 3). The FT + AE and FT + DE variants also showed 4- to 5-fold improvements in macrophage ADCP (Fig. 3E, Table 2), consistent with their greater binding to the activating receptors, particularly FcγRIIa (Table 3). The EFT variant, which has 70% reduced FcγRIIIa affinity (Table 3), mediated lower ADCC activity, both in terms of its potency and efficacy (Fig. 3D, Table 2), and ADCP comparable to native IgG1 (Fig. 3F, Table 2). Addition of the AE substitutions restored ADCC to IgG1 level (Fig. 3D, Table 2). Interestingly, the EFT + AE combination did not enhance phagocytosis (Fig. 3F, Table 2), despite its improved affinity for the activating receptors and particularly strong binding to FcγRIIa. This result may reflect a role of the inhibitory receptor FcγRIIb, which binds tightly to this variant, distinguishing this outcome from observations in previous work.19 Regardless, together the variants provide a range of effector function activities, including dramatically improved complement-mediated yet preserved FcγR-mediated activities (EFT + AE), and simultaneously enhanced CDC, ADCC and ADCP (FT + AE, FT + DE).
Engineering of CDC-enhanced anti-CD19 and -CD40 antibodies.
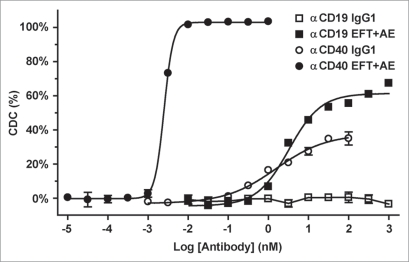
In a final experiment, we examined the transferability of the variants to other antibodies. We tested the substitution combination with the most potent CDC enhancement, EFT + AE, in the context of humanized anti-human CD19 and antihuman CD40 antibodies. Both native IgG1 and CDC-enhanced variants were examined in the CDC assay against Ramos cells as described above. The Fc variant antibodies exhibited improved CDC activity, both in terms of potency and efficacy (Fig. 4), consistent with the anti-CD20 results. Strikingly, the variant anti-CD19 antibody mediated complement activity even though the native IgG1 version was completely lacking, reaching approximately 60% lysis with an EC50 of 3.2 nM. The variant anti-CD40 showed remarkable gains in efficacy (2.5-fold) and potency (620-fold) relative to the native IgG1 version. These results demonstrated that the identified substitutions are not only broadly useful for anti-cancer antibodies, but can confer potent CDC activity even when it is absent or weak in a native IgG1.
Figure 4.
EFT + AE variant enhances CDC in anti-CD19 and anti-CD40 mAbs. CDC activity of variant and IgG1 versions of humanized anti-CD19 and anti-CD40 antibodies was tested against Ramos cells using human complement. Antibody-dependent % lysis was measured at multiple antibody concentrations by Alamar Blue-based detection (mean ± SE of duplicate wells).
Discussion
Over the past decade substantial progress has been made towards improving the cytotoxic potency of mAb drugs. The strongest motivation for the consideration of complement activity as an optimization goal is its contribution to the mechanism of action of one of the most successful antibody drugs—rituximab anti- CD20. Support includes the dependence of rituximab activity on complement in mouse models,23–25 the association between expression of complement regulatory proteins (CRPs) and resistance to rituximab therapy26,27 and the consumption of complement upon rituximab treatment in chronic lymphocytic leukemia (CLL) patients.28 Yet inconsistencies remain, including the absence of observed complement-mediated cytotoxicity in vitro using tumor cells from different response groups29 and the uncompromised activity of other anti-CD20 mAbs in complement deficient mice.30,31 Complicating the picture further is the recent observation that complement protein C3 can inhibit rituximab-mediated natural killer (NK) cell activation and ADCC,32 impacting antibody activity in vivo.33 Yet strong complementmediated activity is an argued benefit of the next generation anti- CD20 ofatumumab34,35 and the relevance of complement to the in vivo activity of mAbs targeting other tumor antigens36,37 suggests a broader role for complement in anti-cancer immunotherapy. Beyond oncology, complement intuitively plays a prominent mechanistic role for antibodies that target pathogens, particularly given the activation of complement pathways by, and the absence of CRPs on, microbial surfaces. The capacity of mAbs to destroy microbes using complement mechanisms is well-established by both in vitro38,39 and in vivo40,41 data. Unfortunately, little clinical data is available concerning the mechanisms of action of antipathogenic mAbs, due principally to the low number of such drugs that have progressed through clinical trials.42 Regardless, although antibodies modified for improved complement have not progressed into clinical development at the same rate as those enhanced for FcγR-mediated effector functions,1 optimization for complement has enormous potential for improving the next generation of mAbs.
Our objective was to engineer variants that improve Fc-mediated complement activity, and we were particularly interested in combining them with substitutions that improve FcγR engagement to optimize the entire repertoire of cytotoxic effector functions. Our CDC-enhancing modifications, consisting of various combinations of substitutions S267E, H268F and S324T, improve CDC up to 6.9-fold and C1q affinity up to 47-fold in the context of an anti-CD20 antibody. The direct relationship we observed between effector function and the effector ligand, an essential foundation for virtually all Fc engineering efforts, is consistent with previous work by others.7,11,14 The most potent single substitution we identified, S267E, modulates the charge of the CH2 domain, similar to other CDC-altering variants such as D270A,4 K322A,4 K326A,14 E333A14 and 113F7 (which includes K274Q and N276K). The Fc:C1q interaction has been shown to have a strong ionic component,43 potentially mediated through several exposed arginines on C1q subunit B.44–46 It is possible that S267E interacts with one of these arginines, a hypothesis that requires further studies to confirm.
Combination of CDC-enhancing substitutions with previously characterized substitutions generated a set of variants that improved affinity to both complement and FcγRs. The binding sites on the Fc region for C1q and FcγR are overlapping, illustrated by the different properties of the FT and EFT combinations. The FT variant possesses not only improved complement activity, but FcγR affinities favorable for ADCC and ADCP. The FcγR-binding properties of the AE and DE variants stacked additively on top of the complement improvements provided by the FT variant, generating variants (FT + AE and FT + DE) with simultaneously enhanced ADCC (up to 22-fold), ADCP (up to 4.7-fold), and CDC (up to 3.3-fold). In contrast, the S267E substitution acts as a trade-off between CDC activity and FcγR-mediated effector function. The poor ADCC activity of the EFT variant was not only rescued by the AE mutations, but the synergy between S267E and I332E provided an additional boost to complement-mediated effector function, thereby resulting in a variant (EFT + AE) with a native IgG1 level of ADCC activity and 23-fold enhancement in CDC.
An important result in the present study was the capacity of the variants to function not only in other antibodies, but in one case to impart CDC activity onto an antibody that otherwise did not mediate it. Whereas most mAbs mediate ADCC in vitro, fewer seem capable of mediating complement activity (unpublished results). This is likely a contributing factor to the lower emphasis on complement relative to FcγRs for mAb cancer therapy. One possible reason for the high bar for complement activity is that it requires high antibody opsonization density,47–49 which is consistent with the fact that pentameric IgM is the most active isotype for complement. The capacity of the EFT + AE variant to impart CDC onto anti-CD19, and moreover the dramatic (>600-fold) enhancement over the poor CDC activity of anti-CD40 are encouraging for the broad applicability of complement-enhancing approaches to antibody drug optimization.
There are multiple cytotoxic mechanisms accessible by the classical complement pathway. These include not only the noncellular CDC activity mediated by MAC as described here, but a number of cellular activities.15 Opsonic C3b, iC3b, and C4b proteins on target cells result in two cellular complement activities, both of which are mediated by interaction with complement receptors CR1 (CD35), CR3 (CD11b-CD18) and CR4 (CD11c-CD18) expressed on NK cells, neutrophils, and monocytic phagocytes. CR-dependent phagocytosis and cytotoxicity, also referred to as complement-dependent cellular cytotoxicity (CDCC), are analogs of the FcγR-dependent mechanisms of similar name, except that effector function is mediated directly by binding of CR to opsonin. The other mechanism involves enhancement of FcγR-mediated effector functions by the CR/opsonin interaction. CR-enhancement of cellular effector functions is activated by opsonic complement protein C5a, which is not only chemotactic for effector cells, but also selectively increases macrophage expression of activating FcγRs relative to FcγRIIb,16,50 providing additional crosstalk between FcγR and complement effector arms.
Two key issues related to these cellular mechanisms include the dependence of CR activation on binding to pathogenassociated molecular patterns (PAMPs),51 for example cell wall β-glucan,52–54 and the negative regulation of complement activation by CRPs,55 which include both soluble and cell surface proteins. While these regulatory mechanisms do not hinder antibodies that target pathogens, they pose barriers for anti-cancer antibodies. Addressing these issues will undoubtedly require a greater understanding of complement biology as it relates to immunotherapy. Yet our hope is that enhanced recruitment of complement, possibly synergistically with improved FcγR engagement, may help overcome these obstacles, analogous to the capacity of high FcγRIIIa affinity to induce activation of NK cells.56,57 Indeed, encouraging the immune system to deal with tumor cells as it would an invading microorganism is in essence the basis of the present strategy, and that of many others, for improving the therapeutic activity of anti-cancer antibodies.
Materials and Methods
Cells and reagents.
Burkitt lymphoma Ramos cell line was obtained from DSMZ (German Collection of Microorganisms and Cell Lines). Burkitt’s lymphoma Raji cell line was obtained from American Type Culture Collection. Human PBMCs were purified from leukapheresis of an anonymous healthy volunteer (HemaCare, VanNuys, CA) using Ficoll-Paque Plus density gradients (Amersham Biosciences, Newark, NJ). Monocyte-derived macrophages were generated as described.19
Human C1q was purchased from GenWay Biotech (San Diego, CA). Human Fc gamma receptor protein FcγRI was obtained from R&D Systems (Minneapolis, MN). Human FcγRIIa, FcγRIIb and FcγRIIIa receptor proteins were produced at Xencor. FcγRIIa, FcγRIIb and FcγRIIIa genes were obtained from the Mammalian Gene Collection (American Type Culture Collection). The extracellular domains of these were subcloned into the vector pcDNA3.1Zeo (Invitrogen) with a C-terminal 6xHis tag, transfected into HEK293T cells and purified using nickel affinity chromatography (Qiagen, Valencia, CA).
Construction, expression and purification of antibody variants.
The variable region VH and VL domains of ocrelizumab (also known as PRO70769 or rhuMAb 2H7) antihuman CD20 antibody20 were generated by gene synthesis (Blue Heron Biotechnology, Bothell, WA) and subcloned into the vector pTT5 (National Research Council, Canada)58 encoding human heavy IgG1 and light Cκ constant regions. Substitutions in the Fc domain were introduced using site-directed mutagenesis (QuikChange, Stratagene, Cedar Creek, TX). Positions are numbered according to the EU index.59 The 113F variant positive control for enhanced CDC activity was also constructed using site-directed mutagenesis (QuikChange, Stratagene, Cedar Creek, TX) as described.7 Heavy and light chain constructs were cotransfected into HEK293E cells (National Research Council, Canada)58 for expression, and antibodies were purified using protein A affinity chromatography (GE Healthcare).
Humanized, affinity-optimized 4G760 anti-human CD19 antibody and humanized S2C661,62 anti-human CD40 antibody were engineered as described.21,63 To serve as an Fc isotype control, the variable region domains of the anti-respiratory syncytial virus (RSV) antibody motavizumab64 were generated by gene synthesis (Blue Heron Biotechnology, Bothell, WA). Fc variant versions of the anti-CD19, CD40 and RSV antibodies were constructed by subcloning into the appropriate IgG1 and Cκ pTT5 vectors from ocrelizumab Fc variants.
Cell-based assays.
For CDC assays, target Ramos or Raji cells were washed 2x in RHB Buffer (RPMI Medium 1640 containing 20 mM HEPES, 2 mM glutamine, 0.1% BSA, pH 7.2) by centrifugation and resuspension and seeded at 40,000 cells per well. Native IgG1 or variant antibody was added at the indicated final concentrations. Human serum complement (Quidel, San Diego, CA) was diluted with RHB buffer and added to opsonized target cells. Final complement concentration was one-eighteenth original stock. Plates were incubated for 2 hr at 37°C, Alamar Blue was added, cells were cultured overnight, and fluorescence was measured in relative fluorescence units. Data were normalized to maximal (Triton X-100) and minimal (complement alone) lysis and fit to a four-parameter sigmoidal dose-response curve using GraphPad Prism (La Jolla, CA).
ADCC was determined by lactate dehydrogenase release as described,19 except that Ramos cells were used as targets (seeded at 10,000 per well) and effector cells were added at a 50:1 PBMC/target cell ratio. Macrophage ADCP was determined by flow cytometry as described,19 except that Ramos cells were used as targets and labeled with CFSE (Guava Technologies, Hayward, CA). Purified PBMCs used in these assays were DNA genotyped for Fcγ RIIa (position 131) and Fc?RIIIa (position 158) using methods by and as a commercial service at Gentris Clinical Genetics (Morrisville, NC).
Surface plasmon resonance determination of binding affinities.
SPR measurements were performed in HBS-EP running buffer (10 mM HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v surfactant P20, GE Healthcare) using a Biacore 3000 instrument (GE Healthcare). Fcγ R affinity was determined as described,19 and the results reported are the average obtained from separate Langmuir fittings of the data from the two independent flow cells of the biosensor chip. For determining C1q affinity, a protein A (Pierce Biotechnology) CM5 biosensor chip (GE Healthcare) was generated using a standard primary amine coupling protocol. The chip’s reference channel was coupled to bovine serum albumin (BSA) to minimize nonspecific binding of C1q. Antibodies at 50 nM were immobilized on the protein A surface for 0.5 or 1 min at 10 µ L/min. C1q in 2-fold serial dilutions (starting at 100 or 25 nM, 5 concentrations total) was injected over antibody-bound surface for 3 min at 30 µL/min followed by a 4.5 min dissociation phase. C1q molarity was calculated using the molecular weight of the C1q hexameric bundle, 410 kDa. Response units for C1q association and dissociation never dropped below the RU level of protein A-captured antibody for native IgG1 or any of the variants, suggesting that antibody was not displaced from the protein A chip upon binding to C1q and that protein A and C1q can be bound simultaneously. After each cycle, the surface was regenerated by injecting glycine buffer (10 mM, pH 1.5). In order to subtract nonspecific C1q binding to antibody-coated protein A surface, an Fc variant with greatly ablated CDC activity was included. Sensorgrams were processed by zeroing time and response before the injection of C1q and by subtracting appropriate nonspecific signals (response of BSA-blocked reference channel, response of an Fc variant with ablated CDC, and response of running buffer). Kinetic parameters were determined by global fitting of association and dissociation phase data with a two-state binding model (A + B ⇌ AB ⇌ AB*). Kd was calculated as Kd1/(1 + 1/Kd2).
Acknowledgements
We thank Cristina Bautista, Araz Eivazi, Jonathan Jacinto, Patrick Joyce, Umesh Muchhal, Duc-Hanh Nguyen, Erik Pong and Crystal Sheriff for technical contributions, Matthew Bernett and Jonathan Zalevsky for helpful discussions, and Bassil Dahiyat and John Desjarlais for encouragement.
Abbreviations
- ADCC
antibody-dependent cellular cytotoxicity
- ADCP
antibody-dependent cellular phagocytosis
- CDC
complement-dependent cytotoxicity
- CR
complement receptor
- CRP
complement regulatory protein
- FcγR
Fc gamma receptor
- mAb
monoclonal antibody
- MAC
membrane-attack complex
- NK
natural killer
- PBMC
peripheral blood mononuclear cell
- RSV
respiratory syncytial virus
- SPR
surface plasmon resonance
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/11158
Supplementary Material
References
- 1.Desjarlais JR, Lazar GA, Zhukovsky EA, Chu SY. Optimizing engagement of the immune system by anti-tumor antibodies: an engineer’s perspective. Drug Discov Today. 2007;12:898–910. doi: 10.1016/j.drudis.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Presta LG. Molecular engineering and design of therapeutic antibodies. Curr Opin Immunol. 2008;20:460–470. doi: 10.1016/j.coi.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Hu W, Qin X. The role of complement in the mechanism of action of rituximab for B-cell lymphoma: implications for therapy. Oncologist. 2008;13:954–966. doi: 10.1634/theoncologist.2008-0089. [DOI] [PubMed] [Google Scholar]
- 4.Idusogie EE, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, et al. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J Immunol. 2000;164:4178–4184. doi: 10.4049/jimmunol.164.8.4178. [DOI] [PubMed] [Google Scholar]
- 5.Thommesen JE, Michaelsen TE, Loset GA, Sandlie I, Brekke OH. Lysine 322 in the human IgG3 C(H)2 domain is crucial for antibody dependent complement activation. Mol Immunol. 2000;37:995–1004. doi: 10.1016/s0161-5890(01)00010-4. [DOI] [PubMed] [Google Scholar]
- 6.Dangl JL, Wensel TG, Morrison SL, Stryer L, Herzenberg LA, Oi VT. Segmental flexibility and complement fixation of genetically engineered chimeric human, rabbit and mouse antibodies. EMBO J. 1988;7:1989–1994. doi: 10.1002/j.1460-2075.1988.tb03037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natsume A, In M, Takamura H, Nakagawa T, Shimizu Y, Kitajima K, et al. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 2008;68:3863–3872. doi: 10.1158/0008-5472.CAN-07-6297. [DOI] [PubMed] [Google Scholar]
- 8.Natsume A, Shimizu-Yokoyama Y, Satoh M, Shitara K, Niwa R. Engineered anti-CD20 antibodies with enhanced complement-activating capacity mediate potent anti-lymphoma activity. Cancer Sci. 2009 doi: 10.1111/j.1349-7006.2009.01327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sensel MG, Kane LM, Morrison SL. Amino acid differences in the N-terminus of C(H)2 influence the relative abilities of IgG2 and IgG3 to activate complement. Mol Immunol. 1997;34:1019–1029. doi: 10.1016/s0161-5890(97)00112-0. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Oomen R, Klein MH. Residue at position 331 in the IgG1 and IgG4 CH2 domains contributes to their differential ability to bind and activate complement. J Biol Chem. 1994;269:3469–3474. [PubMed] [Google Scholar]
- 11.Dall’Acqua WF, Cook KE, Damschroder MM, Woods RM, Wu H. Modulation of the effector functions of a human IgG1 through engineering of its hinge region. J Immunol. 2006;177:1129–1138. doi: 10.4049/jimmunol.177.2.1129. [DOI] [PubMed] [Google Scholar]
- 12.Michaelsen TE, Sandlie I, Bratlie DB, Sandin RH, Ihle O. Structural difference in the complement activation site of human IgG1 and IgG3. Scand J Immunol. 2009;70:553–564. doi: 10.1111/j.1365-3083.2009.02338.x. [DOI] [PubMed] [Google Scholar]
- 13.Redpath S, Michaelsen T, Sandlie I, Clark MR. Activation of complement by human IgG1 and human IgG3 antibodies against the human leucocyte antigen CD52. Immunology. 1998;93:595–600. doi: 10.1046/j.1365-2567.1998.00472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idusogie EE, Wong PY, Presta LG, Gazzano-Santoro H, Totpal K, Ultsch M, et al. Engineered antibodies with increased activity to recruit complement. J Immunol. 2001;166:2571–2575. doi: 10.4049/jimmunol.166.4.2571. [DOI] [PubMed] [Google Scholar]
- 15.Gelderman KA, Tomlinson S, Ross GD, Gorter A. Complement function in mAb-mediated cancer immunotherapy. Trends Immunol. 2004;25:158–164. doi: 10.1016/j.it.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Godau J, Heller T, Hawlisch H, Trappe M, Howells E, Best J, et al. C5a initiates the inflammatory cascade in immune complex peritonitis. J Immunol. 2004;173:3437–3445. doi: 10.4049/jimmunol.173.5.3437. [DOI] [PubMed] [Google Scholar]
- 17.Shushakova N, Skokowa J, Schulman J, Baumann U, Zwirner J, Schmidt RE, et al. C5a anaphylatoxin is a major regulator of activating versus inhibitory FcgammaRs in immune complex-induced lung disease. J Clin Invest. 2002;110:1823–1830. doi: 10.1172/JCI200216577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci USA. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards JO, Karki S, Lazar GA, Chen H, Dang W, Desjarlais JR. Optimization of antibody binding to FcgammaRIIa enhances macrophage phagocytosis of tumor cells. Mol Cancer Ther. 2008;7:2517–2527. doi: 10.1158/1535-7163.MCT-08-0201. [DOI] [PubMed] [Google Scholar]
- 20.Vugmeyster Y, Beyer J, Howell K, Combs D, Fielder P, Yang J, et al. Depletion of B cells by a humanized anti-CD20 antibody PRO70769 in Macaca fascicularis. J Immunother. 2005;28:212–219. doi: 10.1097/01.cji.0000155050.03916.04. [DOI] [PubMed] [Google Scholar]
- 21.Lazar GA, Desjarlais JR, Jacinto J, Karki S, Hammond PW. A molecular immunology approach to antibody humanization and functional optimization. Mol Immunol. 2007;44:1986–1998. doi: 10.1016/j.molimm.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 22.Chu SY, Vostiar I, Karki S, Moore GL, Lazar GA, Pong E, et al. Inhibition of B cell receptor-mediated activation of primary human B cells by coengagement of CD19 and FcgammaRIIb with Fc-engineered antibodies. Mol Immunol. 2008;45:3926–3933. doi: 10.1016/j.molimm.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 23.Cragg MS, Glennie MJ. Antibody specificity controls in vivo effector mechanisms of anti-CD20 reagents. Blood. 2004;103:2738–2743. doi: 10.1182/blood-2003-06-2031. [DOI] [PubMed] [Google Scholar]
- 24.Di Gaetano N, Cittera E, Nota R, Vecchi A, Grieco V, Scanziani E, et al. Complement activation determines the therapeutic activity of rituximab in vivo. J Immunol. 2003;171:1581–1587. doi: 10.4049/jimmunol.171.3.1581. [DOI] [PubMed] [Google Scholar]
- 25.Golay J, Cittera E, Di Gaetano N, Manganini M, Mosca M, Nebuloni M, et al. The role of complement in the therapeutic activity of rituximab in a murine B lymphoma model homing in lymph nodes. Haematologica. 2006;91:176–183. [PubMed] [Google Scholar]
- 26.Bannerji R, Kitada S, Flinn IW, Pearson M, Young D, Reed JC, et al. Apoptotic-regulatory and complementprotecting protein expression in chronic lymphocytic leukemia: relationship to in vivo rituximab resistance. J Clin Oncol. 2003;21:1466–1471. doi: 10.1200/JCO.2003.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Treon SP, Mitsiades C, Mitsiades N, Young G, Doss D, Schlossman R, et al. Tumor cell expression of CD59 is associated with resistance to CD20 serotherapy in patients with B-cell malignancies. J Immunother. 2001;24:263–271. [PubMed] [Google Scholar]
- 28.Kennedy AD, Beum PV, Solga MD, DiLillo DJ, Lindorfer MA, Hess CE, et al. Rituximab infusion promotes rapid complement depletion and acute CD20 loss in chronic lymphocytic leukemia. J Immunol. 2004;172:3280–3288. doi: 10.4049/jimmunol.172.5.3280. [DOI] [PubMed] [Google Scholar]
- 29.Weng WK, Levy R. Expression of complement inhibitors CD46, CD55 and CD59 on tumor cells does not predict clinical outcome after rituximab treatment in follicular non-Hodgkin lymphoma. Blood. 2001;98:1352–1357. doi: 10.1182/blood.v98.5.1352. [DOI] [PubMed] [Google Scholar]
- 30.Hamaguchi Y, Uchida J, Cain DW, Venturi GM, Poe JC, Haas KM, et al. The peritoneal cavity provides a protective niche for B1 and conventional B lymphocytes during anti-CD20 immunotherapy in mice. J Immunol. 2005;174:4389–4399. doi: 10.4049/jimmunol.174.7.4389. [DOI] [PubMed] [Google Scholar]
- 31.Uchida J, Hamaguchi Y, Oliver JA, Ravetch JV, Poe JC, Haas KM, et al. The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptordependent mechanisms during anti-CD20 antibody immunotherapy. J Exp Med. 2004;199:1659–1669. doi: 10.1084/jem.20040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang SY, Racila E, Taylor RP, Weiner GJ. NK-cell activation and antibody-dependent cellular cytotoxicity induced by rituximab-coated target cells is inhibited by the C3b component of complement. Blood. 2008;111:1456–1463. doi: 10.1182/blood-2007-02-074716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SY, Veeramani S, Racila E, Cagley J, Fritzinger D, Vogel CW, et al. Depletion of the C3 component of complement enhances the ability of rituximab-coated target cells to activate human NK cells and improves the efficacy of monoclonal antibody therapy in an in vivo model. Blood. 2009;114:5322–5330. doi: 10.1182/blood-2009-01-200469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glennie MJ, French RR, Cragg MS, Taylor RP. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol Immunol. 2007;44:3823–3837. doi: 10.1016/j.molimm.2007.06.151. [DOI] [PubMed] [Google Scholar]
- 35.Pawluczkowycz AW, Beurskens FJ, Beum PV, Lindorfer MA, van de Winkel JG, Parren PW, et al. Binding of submaximal C1q promotes complement-dependent cytotoxicity (CDC) of B cells opsonized with anti- CD20 mAbs ofatumumab (OFA) or rituximab (RTX): considerably higher levels of CDC are induced by OFA than by RTX. J Immunol. 2009;183:749–758. doi: 10.4049/jimmunol.0900632. [DOI] [PubMed] [Google Scholar]
- 36.Imai M, Landen C, Ohta R, Cheung NK, Tomlinson S. Complement-mediated mechanisms in anti-GD2 monoclonal antibody therapy of murine metastatic cancer. Cancer Res. 2005;65:10562–10568. doi: 10.1158/0008-5472.CAN-05-1894. [DOI] [PubMed] [Google Scholar]
- 37.Zent CS, Secreto CR, LaPlant BR, Bone ND, Call TG, Shanafelt TD, et al. Direct and complement dependent cytotoxicity in CLL cells from patients with high-risk early-intermediate stage chronic lymphocytic leukemia (CLL) treated with alemtuzumab and rituximab. Leuk Res. 2008;32:1849–1856. doi: 10.1016/j.leukres.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston MJ, Gerceker AA, Reff ME, Pier GB. Production and characterization of a set of mousehuman chimeric immunoglobulin G (IgG) subclass and IgA monoclonal antibodies with identical variable regions specific for Pseudomonas aeruginosa serogroup O6 lipopolysaccharide. Infect Immun. 1998;66:4137–4142. doi: 10.1128/iai.66.9.4137-4142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly-Quintos C, Cavacini LA, Posner MR, Goldmann D, Pier GB. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect Immun. 2006;74:2742–2750. doi: 10.1128/IAI.74.5.2742-2750.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Y, Kozel TR, Zhang MX, MacGill RS, Carroll MC, Cutler JE. Complement is essential for protection by an IgM and an IgG3 monoclonal antibody against experimental, hematogenously disseminated candidiasis. J Immunol. 2001;167:1550–1557. doi: 10.4049/jimmunol.167.3.1550. [DOI] [PubMed] [Google Scholar]
- 41.Wells J, Haidaris CG, Wright TW, Gigliotti F. Complement and Fc function are required for optimal antibody prophylaxis against Pneumocystis carinii pneumonia. Infect Immun. 2006;74:390–393. doi: 10.1128/IAI.74.1.390-393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker M. Anti-infective antibodies: finding the path forward. Nat Biotechnol. 2006;24:1491–1493. doi: 10.1038/nbt1206-1491. [DOI] [PubMed] [Google Scholar]
- 43.Burton DR, Boyd J, Brampton AD, Easterbrook-Smith SB, Emanuel EJ, Novotny J, et al. The C1q receptor site on immunoglobulin G. Nature. 1980;288:338–344. doi: 10.1038/288338a0. [DOI] [PubMed] [Google Scholar]
- 44.Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, et al. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 45.Kojouharova MS, Gadjeva MG, Tsacheva IG, Zlatarova A, Roumenina LT, Tchorbadjieva MI, et al. Mutational analyses of the recombinant globular regions of human C1q A, B and C chains suggest an essential role for arginine and histidine residues in the C1q-IgG interaction. J Immunol. 2004;172:4351–4358. doi: 10.4049/jimmunol.172.7.4351. [DOI] [PubMed] [Google Scholar]
- 46.Marques G, Anton LC, Barrio E, Sanchez A, Ruiz S, Gavilanes F, et al. Arginine residues of the globular regions of human C1q involved in the interaction with immunoglobulin G. J Biol Chem. 1993;268:10393–10402. [PubMed] [Google Scholar]
- 47.Dechant M, Weisner W, Berger S, Peipp M, Beyer T, Schneider-Merck T, et al. Complement-dependent tumor cell lysis triggered by combinations of epidermal growth factor receptor antibodies. Cancer Res. 2008;68:4998–5003. doi: 10.1158/0008-5472.CAN-07-6226. [DOI] [PubMed] [Google Scholar]
- 48.Macor P, Tedesco F. Complement as effector system in cancer immunotherapy. Immunol Lett. 2007;111:6–13. doi: 10.1016/j.imlet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 49.Spiridon CI, Ghetie MA, Uhr J, Marches R, Li JL, Shen GL, et al. Targeting multiple Her-2 epitopes with monoclonal antibodies results in improved antigrowth activity of a human breast cancer cell line in vitro and in vivo. Clin Cancer Res. 2002;8:1720–1730. [PubMed] [Google Scholar]
- 50.Konrad S, Baumann U, Schmidt RE, Gessner JE. Intravenous immunoglobulin (IVIG)-mediated neutralisation of C5a: a direct mechanism of IVIG in the maintenance of a high Fc gammaRIIB to Fc gammaRIII expression ratio on macrophages. Br J Haematol. 2006;134:345–347. doi: 10.1111/j.1365-2141.2006.06185.x. [DOI] [PubMed] [Google Scholar]
- 51.Gasque P. Complement: a unique innate immune sensor for danger signals. Mol Immunol. 2004;41:1089–1098. doi: 10.1016/j.molimm.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 52.Vetvicka V, Thornton BP, Ross GD. Soluble betaglucan polysaccharide binding to the lectin site of neutrophil or natural killer cell complement receptor type 3 (CD11b/CD18) generates a primed state of the receptor capable of mediating cytotoxicity of iC3bopsonized target cells. J Clin Invest. 1996;98:50–61. doi: 10.1172/JCI118777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vetvicka V, Thornton BP, Wieman TJ, Ross GD. Targeting of natural killer cells to mammary carcinoma via naturally occurring tumor cell-bound iC3b and beta-glucan-primed CR3 (CD11b/CD18) J Immunol. 1997;159:599–605. [PubMed] [Google Scholar]
- 54.Yan J, Vetvicka V, Xia Y, Coxon A, Carroll MC, Mayadas TN, et al. Beta-glucan, a “specific“ biologic response modifier that uses antibodies to target tumors for cytotoxic recognition by leukocyte complement receptor type 3 (CD11b/CD18) J Immunol. 1999;163:3045–3052. [PubMed] [Google Scholar]
- 55.Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 56.Bowles JA, Wang SY, Link BK, Allan B, Beuerlein G, Campbell MA, et al. Anti-CD20 monoclonal antibody with enhanced affinity for CD16 activates NK cells at lower concentrations and more effectively than rituximab. Blood. 2006;108:2648–2654. doi: 10.1182/blood-2006-04-020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowles JA, Weiner GJ. CD16 polymorphisms and NK activation induced by monoclonal antibody-coated target cells. J Immunol Methods. 2005;304:88–99. doi: 10.1016/j.jim.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 58.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabat EA, Wu TT, Perry HM, Gottesmann KS, Foeller C. Sequences of Proteins of Immunological Interest. Bethesda, MD: U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 60.Meeker TC, Miller RA, Link MP, Bindl J, Warnke R, Levy R. A unique human B lymphocyte antigen defined by a monoclonal antibody. Hybridoma. 1984;3:305–320. doi: 10.1089/hyb.1984.3.305. [DOI] [PubMed] [Google Scholar]
- 61.Koho H, Paulie S, Ben-Aissa H, Jonsdottir I, Hansson Y, Lundblad ML, et al. Monoclonal antibodies to antigens associated with transitional cell carcinoma of the human urinary bladder I. Determination of the selectivity of six antibodies by cell ELISA and immunofluorescence. Cancer Immunol Immunother. 1984;17:165–172. doi: 10.1007/BF00205481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulie S, Koho H, Ben-Aissa H, Hansson Y, Lundblad ML, Perlmann P. Monoclonal antibodies to antigens associated with transitional cell carcinoma of the human urinary bladder II. Identification of the cellular target structures by immunoprecipitation and SDSPAGE analysis. Cancer Immunol Immunother. 1984;17:173–179. doi: 10.1007/BF00205482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horton HM, Bernett MJ, Pong E, Peipp M, Karki S, Chu SY, et al. Potent in vitro and in vivo activity of an Fc-engineered anti-CD19 monoclonal antibody against lymphoma and leukemia. Cancer Res. 2008;68:8049–8057. doi: 10.1158/0008-5472.CAN-08-2268. [DOI] [PubMed] [Google Scholar]
- 64.Mejias A, Chavez-Bueno S, Rios AM, Aten MF, Raynor B, Peromingo E, et al. Comparative effects of two neutralizing anti-respiratory syncytial virus (RSV) monoclonal antibodies in the RSV murine model: time versus potency. Antimicrob Agents Chemother. 2005;49:4700–4707. doi: 10.1128/AAC.49.11.4700-4707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.