Abstract
The ubiquitin-proteasome system (UPS) and autophagy are two major intracellular protein degradation pathways. The UPS mediates the removal of soluble abnormal proteins as well as the targeted degradation of most normal proteins that are no longer needed. Autophagy is generally responsible for bulky removal of defective organelles and for sequestering portions of cytoplasm for lysosomal degradation during starvation. Impaired or inadequate protein degradation in the heart is associated with and may be a major pathogenic factor for a wide variety of cardiac dysfunctions, while enhanced protein degradation is also implicated in the development of cardiac pathology. It was generally assumed that the UPS and autophagy serve distinct functions. Therefore, the functional roles of the UPS and autophagy in the hearts have been largely investigated separately. However, recent advances in understanding the shared mechanisms contributing to UPS alteration and the induction of autophagy have helped reveal the link and interplay between the two proteolytic systems in the heart. These links are exemplified by scenarios in which inadequate UPS proteolytic function leads to activation of autophagy, helping alleviate proteotoxic stress. It is becoming increasingly clear that a coordinated and complementary relationship between the two systems is critical to protect cells against stress. Several proteins including p62, NBR1, HDAC6, and co-chaperones appear to play an important role in harmonizing and mobilizing the consortium formed by the UPS and autophagy.
Keywords: ubiquitin-proteasome system, autophagy, heart disease, p62, NBR1, HDAC6
1. Introduction
The ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway are the two major pathways responsible for the clearance of proteins and organelles in eukaryotic cells.1 UPS-mediated proteolysis generally consists of two steps: ubiquitination and proteasome-mediated degradation. Macroautophagy (commonly referred to as autophagy) is the process engulfing a portion of cytoplasm, often including organelles, in a membrane bound compartment for degradation by the lysosome. Therefore, autophagy mediates bulk degradation of cytoplasmic proteins and defective or surplus organelles.2-3 Earlier studies defined proteins into two categories including “short-lived” and “long-lived” proteins based on their degradation kinetics.4 The UPS predominantly degrades short-lived normal protein molecules after they have fulfilled their duty in the cell.5 On the other hand, autophagy is primarily responsible for degrading long-lived proteins.6 Notably, the distinction of substrate preference between the two proteolytic systems is relative. Recent studies indicate that the UPS can participate in the degradation of long-lived proteins while the autophagy can also be involved in the degradation of short-lived proteins.4, 7-8
An alternative way to categorize proteolysis is based on the functional outcome rather than the kinetics of degradation. From the functional point of view, the UPS degrades two types of proteins: (1) the fully functional proteins which are degraded as a regulatory mechanism, such as proteins involved in regulation of signal transduction, gene transcription, endocytosis, and cell division; and (2) abnormal or non-functional proteins whose degradation serves a critical step of posttranslational protein quality control (PQC) in the cell.9-10 Autophagy also degrades two types of proteins: functional proteins which are degraded in a bulk fashion as a nutrient recycling mechanism, and the aggregated misfolded proteins. Hence, the two protein degradation pathways cooperate with each other in the removal of misfolded proteins.11 A recent study with cultured cells has revealed that misfolded cytosolic proteins are partitioned into two distinct PQC compartments. Soluble misfolded proteins tend to migrate to a juxtanuclear compartment preferentially for degradation by the UPS. In contrast, terminally aggregated proteins are partitioned in a perivacuolar inclusion and are mainly degraded by autophagy because they can not be efficiently degraded by the proteasome.11 This review will highlight the alterations of the UPS and autophagy and their potential roles in cardiac disease, with an emphasis on the interplay between the two proteolytic pathways.
2. UPS dysfunction in cardiac disease
Marked increases in ubiquitinated proteins and accumulation of preamyloid oligomers in cardiomyocytes of human failing hearts suggest inadequate PQC in heart failure.12-15 Moreover, increased level of E3 ligase MDM2, ubiquitin carboxyl-terminal hydrolase (UCH), and enhanced 20S-proteasome chymotrypsin-like activities were also observed in human hearts with dilated cardiomyopathy.14, 16 Alteration of the UPS has also been observed in experimental animal models with various forms of cardiac dysfunction, including desmin-related cardiomyopathy (DRC),3 myocardial ischemia or ischemia/reperfusion (I/R),17-18 and load-induced heart disease.19-20
Desmin-related myopathy (DRM) is a family of myopathy caused by genetic mutations of desmin, αB-crystallin (CryAB), and other desmin partner proteins. Its pathological feature is the presence of aberrant desmin-positive protein aggregates in myocytes. DRC is the cardiac manifestation of DRM. Several transgenic (tg) mouse models of DRC were created by cardiomyocyte-restricted tg expression of human DRC-linked mutant genes, including a 7 amino acid deletion (R172~E178) mutation of the desmin gene (D7-des) and a missense mutation (R120G) of CryAB (CryABR120G).15, 21-22 Cardiac UPS proteolytic function has been evaluated in both CryABR120G and D7-des tg mice by our laboratory.23-25 The tremendous advantage of these studies is the employment of a tg mouse model with ubiquitous overexpression of a previously validated UPS surrogate substrate, a green fluorescence protein (GFP) modified by carboxyl fusion of degron CL1, referred to as GFPdgn.26 In both cases, an increased level of ubiquitinated proteins and significant accumulation of GFPdgn indicates proteasome functional insufficiency (PFI) in the DRC hearts despite marked increases in 20S proteasome peptidase activities. The reduction of key subunits of the 19S proteasome was observed and suggests that a key defect resides in the delivery of the targeted protein substrates by the19S into the 20S proteolytic chamber. Increased proteasome activities here are likely a compensatory mechanism for the cardiomyocytes to cope with increased misfolded proteins. When misfolded proteins exceed the degradation capacity of the UPS or escape from degradation they accumulate and trigger aberrant protein aggregation, which in turn impairs proteasome functions, forming a vicious cycle. Indeed, in vitro experiments have shown that aberrant protein aggregation is required by the DRC mutants to impair UPS in cardiomyocytes.23-25
In animal models of ischemic cardiomyopathy, declined peptidase activities accompanied by accumulation of oxidatively modified and ubiquitinated proteins were observed.27-28 Additionally, the decreased proteasome peptidase activity is associated with oxidative modification of several proteasome subunits and contributes to accumulation of ubiquitinated proteins in animal models of myocardial ischemia and I/R injury.27, 29-30
Accumulation of ubiquitinated proteins was also observed in pressure-overloaded mouse hearts.12, 31 Depressed proteasome activities occurred in pressure-overloaded mouse hearts prior to discernible cardiac dysfunction.12 However, some other studies showed elevated abundance of proteasome subunits and E3 ligases, as well as increased proteasome peptidase activities in transverse aortic constriction (TAC)-induced cardiac hypertrophy.31-33 Additionally, the transcript levels of ubiquitin B (UbB), ubiquitin ligases including atrogin-1 and muscle-specific RING finger 1 (MuRF-1), as well as the proteasomal subunit PSMB4 were increased in rat hypertrophic hearts.34 It should be pointed out that the observed up-regulation of UPS components and proteasome peptidase activities do not necessarily indicate that UPS function has surpassed the cell’s increased needs to deal with proteotoxic stress under these conditions. On the contrary, the increases in peptidase activities and in the abundance of UPS components, coupled with the elevated levels of ubiquitinated proteins, might reflect severe PFI and PQC inadequacy in the diseased hearts.31-32 It will be interesting and important to test this proposition by introducing a surrogate substrate into these disease models and assessing a change in its clearance.
3. Genetic deficiency of UPS components causes cardiac pathology
Recent studies involving genetic perturbation of specific components of the UPS provide valuable information about the role of the UPS in maintaining normal cardiac function and shine light on the poorly understood pathophysiological significance of UPS dysfunction in cardiac dysfunction.
Several observations suggest that some E3-ligases play a key role in myocardial ischemia. For instance, mice deficient in CHIP (carboxyl terminus of Hsc70-interacting protein), a co-chaperone with ubiquitin ligase properties, show accelerated age-related pathophysiological phenotypes and a heightened vulnerability to I/R injury.35-36 MDM2 has also been shown to protect cardiomyocytes in both cell cultures and intact mice. Isolated cardiomyocytes overexpressing MDM2 acquire resistance to hypoxia/reoxygenation-induced cell death and phenylephrine- and endothelin-1-induced hypertrophy.37 Conversely, inactivation of MDM2 by a peptide inhibitor promotes hypoxia/reoxygenation-induced apoptosis. Consistent with the in vitro findings, hypomorphic MDM2 expression increases mouse heart’s sensitivity to I/R injury.37 The ability of MDM2 to promote ubiquitination and proteasomal degradation of p53, an important effector of I/R, likely contributes to the protective effect.38
MuRFs are an important subfamily of RING-finger E3 ligases that are specifically expressed in striated muscle. MuRF1 is a key regulator of protein kinase C (PKC)-dependent hypertrophic responses.39 Overexpressing MuRF1 attenuated phenylephrine-induced hypertrophy in isolated cardiomyocytes.40 Compared with wild type (WT) mice, mice lacking MuRF1 were found to exacerbate cardiac hypertrophic responses to pressure overload.39 An important role of MuRF3 in maintaining cardiac integrity and function after acute myocardial infarction (AMI) is suggested by the finding that mice lacking MuRF3 display normal cardiac function but are prone to post-AMI cardiac rupture. MuRF3′s cardioprotection is likely attributed to the turnover of four and a half LIM domain protein (FHL2) and γ-filamin.41 Single knockout of MuRF1, MuRF2 or MuRF3 in mice does not cause any sign of cardiac disease at the baseline but sensitizes the hearts to pathologic stimuli. Double knockout of MuRF genes do yield phenotypes under basal conditions. For instance, MuRF1/MuRF3 double-knockout mice exhibit hypertrophic cardiomyopathy and decreased cardiac function.42 Similarly, MuRF1/MuRF2 double-knockout mice develop extreme cardiac hypertrophy.43 Taken together, these studies demonstrate that MuRF genes seem to function redundantly to protect the heart.
As an F-box protein in myocytes, atrogin-1 associates with Skp1, Cullin1, and Roc1 to assemble an SCFatrogin-1 complex with ubiquitin ligase activity.44 Overexpressing atrogin-1 attenuates, whereas down-regulation of atrogin-1 enhances, phenylephrine-induced hypertrophy in isolated cardiomyocytes.45 Consistent with the observations in cell cultures, tg overexpression of atrogin-1 in mouse hearts blunts TAC-induced cardiac hypertrophy by targeting calcineurin for ubiquitin-mediated degradation.45 Conversely, mice lacking atrogin-1 develop exaggerated cardiac hypertrophy in response to voluntary exercise through regulating FoxO1 ubiquitination.46
4. Pharmacological proteasome inhibition (PSMI) in cardiac diseases
Both detrimental and beneficial effects were reported with pharmacologically induced PSMI on myocardial ischemia. Recent data from both in vitro and in vivo experiments support the concept that short-term PSMI under certain conditions may play a protective role in myocardial I/R but sustained PSMI may be detrimental.18 The cardioprotection of PSMI may be mediated by several mechanisms including inhibition of the nuclear factor κ-B (NF-κB) activity, inactivation of G-protein-coupled receptor kinase 2 (GRK2), and inhibition of apoptosis.18
Given that NF-κB is a critical factor in the hypertrophic growth of cardiomyocytes and that the activation of NF-κB relies on the proteasome-dependent degradation of the inhibitory κB (IκB),47 it follows that PSMI likely suppresses cardiac hypertrophy through repressing NF-κB activity.48 This is confirmed by several studies. For instance, in neonatal rat cardiomyocytes, partial inhibition of the proteasome by low doses of proteasome inhibitor effectively suppressed cardiomyocyte hypertrophy.49 Moreover, in hypertensive Dahl-salt sensitive rats, low doses of PSMI significantly reduced hypertrophic heart growth.49 Similar findings have been observed in aorta-banded mouse models.32, 48 After the onset of pressure overload, the proteasome inhibitor epoxomicin decreased NF-κB activity and prevented or even reversed cardiac remodeling without discernibly affecting cardiac function.48 Further supports for a hypertrophy inhibition role of proteasome inhibitor treatment come from the findings that in a mouse model of isoproterenol-induced cardiac hypertrophy, the proteasome inhibitor PS-519 effectively prevented the development of hypertrophy and more importantly, promoted the regression of existing left ventricular hypertrophy.50 These studies with rodents provide compelling evidence that proteasome function is required for cardiac hypertrophic growth.20
However, the long term outcome of PSMI in human cardiac diseases remains to be tested. Cardiac complications including arrhythmias and heart failure have been reported in clinical use of proteasome inhibitor bortezomib in cancer patients.51-52 Interestingly, aging-associated decreases in proteasome activities do not appear to inhibit hypertension-induced cardiac hypertrophy in the human elderly.
5. Genetic perturbations of autophagy compromise cardiac function
Ablation of autophagic or lysosomal function can, under some circumstances, bedetrimental to the heart. For instance, dilated cardiomyopathy is developed by genetic impairment of autophagy in cathepsin L- and LAMP2-deficient mice.53-54 Conditional deletion of Atg5 in the adult heart leads to cardiac hypertrophy, left ventricular dilatation, contractile dysfunction, and heart failure in the absence of exogenous stress.55 Thus, basal level of autophagy is required to maintain homeostasis in cardiomyocytes.
6. Alteration and functional role of autophagy in cardiac dysfunction
Although it is quite clear that deficiency in autophagy can disrupt normal cardiac function, a critical question concerning the role of autophagy in the development of the cardiac diseases initiated by mutations or environmental stress, remains to be addressed. To answer this question, the autophagic activity has been evaluated in a wide range of cardiac diseases and more importantly, the effect of genetic and pharmacological manipulations of autophagy on cardiac diseases has been assessed.
6.1. Cardiac proteinopathy
Aberrant protein aggregation, in the form of pre-amyloid oligomer formation, has been observed in failing human hearts resulting from hypertrophic (HCM) or dilated cardiomyopathies (DCM).15 Therefore, at least a subset of HCM and DCM arguably belong to proteinopathy. Increased autophagic activity was reported in human hearts with DCM.56-57 Recently, the hallmark of autophagy has been detected in the heart of several mouse models of cardiac proteinopathy. For instance, significantly increased autophagosomes and lysosomal markers were detected in mouse hearts with tg expression of an 83 residue–long polyglutamine repeats (polyQ83),58 which is sufficient to cause amyloid-based cardiac dysfunction in mice.59 Similarly, increases in autophagosomes were also observed in CryABR120G-expressing cultured cardiomyocytes and CryABR120G tg mouse hearts.60 Ubiquitinated proteins, oxidized proteins, and lipofuscin accumulate in cardiomyocytes as the mouse ages. Delaying cardiac ageing in mice by suppression of phosphoinositide 3-kinase (PI3K) or its downstream effector (mTOR) was associated with increased autophagy in the heart.61 Unloading the heart of patients with DCM using the left ventricular assisting device down-regulates autophagic markers in myocardial biopsies.57 These findings suggest that autophagy activation in diseased hearts is an adaptive response to inadequate PQC.
The protective role of autophagy in CryABR120G based DRC is demonstrated by blunting autophagy by belcin1 haploinsufficiency. The latter induces greater accumulation of high molecular weight polyubiquitinated proteins in cardiomyocytes, accelerates heart failure progression, and causes early mortality in the mice expressing a human CryABR120G.60 Consistently, suppressing autophagy by 3-methyladenine (3-MA) increases aggresome accumulation in CryABR120G-expressing cultured cardiomyocytes.60 Thus, autophagy is activated and plays a compensatory role in removing misfolded proteins in cardiomyocytes of CryABR120G-based DRC.
6.2. Myocardial ischemia and I/R
Autophagy is induced in the heart during myocardial ischemia.62-63 Enhanced autophagy was also observed in mouse hearts and isolated cardiomyocytes subjected to simulated I/R (sI/R).63-64 The functional role of autophagy in these circumstances was investigated. In studying autophagy in an ischemic mouse model, Matsui et al. showed that attenuated autophagy enhanced myocardial injury in acute myocardial ischemia.63 Another study reported that the autophagy marker in pig hearts with repetitive myocardial ischemia was much more prevalent in areas of subendocardium where apoptosis was absent, but areas with increased apoptosis exhibited much less cathepsin B immunostaining.62 Thus, autophagy may contribute to the survival of cardiomyocytes in response to both acute and chronic myocardial ischemia.
In contrast, several reports suggest that enhanced autophagy may contribute to cell death in response to I/R and prolonged hypoxia. Matsui et al. found that autophagy was detrimental during reperfusion although it was protective during ischemia. In their study, myocardial I/R injury was attenuated in a mouse model with diminished autophagy.63 Prolonged hypoxia also induced autophagic cell death, which was reduced by treatment with 3-MA or by knockdown of beclin-1 or Atg5.65
Similarly, both detrimental and protective roles of autophagy were reported in cell cultured models subjected to sI/R. In vitro experiments of the study by Matsui et al. showed that inhibition of autophagy by beclin1 knockdown increased cell viability in response to H2O2, which is consistent with their in vivo experiments.63 The findings were verified by another study.64 However, autophagy has also been shown to protect against cell injury in cultured cardiac cells in response to sI/R. It was found that cell injury was decreased by enhanced autophagy through overexpression of beclin1 and increased by inhibition of autophagy through expression of a dominant-negative Atg5, knockdown of Beclin1, or treatment of 3-MA.66-67 These inconsistencies may arise from variations in the timing, extent, and duration of ischemia and perhaps in the methods used for autophagy manipulation.
Ischemic preconditioning (IPC) induces tolerance to subsequent ischemic episode in the heart. Several studies indicate that autophagy is involved in cardioprotection of IPC. Enhanced autophagy and up-regulated BAG-1 (Bcl-2-associated athanogene-1) were observed during hypoxia adaption both in vitro and in vivo. Furthermore, inhibition of autophagy by treating rats with either wortmannin or BAG-1 siRNA through intramyocardial injection abolished IPC-induced LC3I to LC3-II conversion and cardioprotection.68-69 These studies indicate that myocardial protection by IPC is mediated at least in part by up-regulation of autophagy in association with BAG-1 protein. Similarly, 2-chloro-N(6)-cyclopentyladenosine (CCPA), an adenosine A1 receptor agonist, markedly induces autophagy in short term and confers a preconditioning protection against sI/R injury in HL-1 cells. Moreover, the cardioprotective effect of CCPA on cells subjected to sI/R injury is abolished when autophagy is blocked with a dominant negative Atg5.63 The role of autophagy in preconditioning can also be inferred from agents known to activate autophagy also promote cardioprotection. For instance, rapamycin, an autophagy inducer, induces potent preconditioning-like effects against AMI in rat hearts.70
6.3. Cardiac hypertrophy
During the compensatory hypertrophy stage of pressure overload, autophagic activity is suppressed; however, in failing hearts, autophagic activity is induced by accumulated protein aggregates or damaged organelles.31, 55, 71
Disrupting autophagy by cardiomyocyte-restricted inactivation of Atg5 deteriorated TAC- or β-adrenergic stimulation- induced cardiac dysfunction.55 However, a maladaptive role of load-induced autophagy was implicated in a study using mice with genetic inhibition of beclin1, a gene required for the early events of autophagy. In this study, heterozygous ablation of the beclin1 gene blunted the load-induced autophagy activation and pathological remodeling; whereas tg mice with cardiomyocyte-restricted overexpression of beclin1 had amplified autophagy and enhanced pathological remodeling in response to stress.71 These two sets of experiments had different levels of autophagy at work in the pathogenesis of pressure overload. In Atg5-deficient hearts, the basal level of constitutive autophagy is lost. In contrast, the belcin1+/− mice have only a 50% reduction in autophagic flux.72 Therefore, it is likely that the basal level of constitutive autophagy in the heart is a homeostatic mechanism and is protective;55 whereas, stress-related increases in autophagy can be maladaptive in the setting of load-induced hypertrophy.71-72 An alternative interpretation is that manipulating belcin1 is less specific than inhibiting Atg5 in terms of impact on autophagy in the pressure overload setting.
Furthermore, in vitro studies suggest that the absence of basal levels of autophagic activity in cardiomyocytes contributes to cellular hypertrophy. For instance, Nakai A et al. found that inhibition of autophagy by Atg7 knockdown induced morphological and biochemical features of cardiomyocyte hypertrophy.55 Moreover, compared to cells from WT mice, cardiomyocytes isolated from Atg5-deficient mice showed higher sensitivity to isoproterenol.55
The roles of autophagy in cardiac hypertrophic growth can also be inferred from studies on agents known to pharmacologically manipulate autophagy. Rapamycin prevents cardiac hypertrophy induced by TAC or thyroid hormone treatment, and regresses established cardiac hypertrophy. 73-74 It is very likely that the induction of autophagy contributes to the cardioprotection of rapamycin.
7. Stimuli and pathways that alter the UPS and autophagy in cardiac diseases
The alterations of the UPS and autophagy in cardiac dysfunction reviewed so far suggest that UPS proteolytic function often becomes inadequate or impaired by severe cellular stress and under this circumstance autophagy may be increasingly activated to help ameliorate the stress. Understanding how the upstream stimuli and signal mechanisms lead to UPS alterations and induce autophagy would shine light on the interplay between the two proteolytic pathways.
7.1 Misfolded proteins and endoplasmic reticulum (ER) stress
Perturbation of ER environment induces ER stress and leads to the accumulation of unfolded/misfolded proteins. Cardiomyocytes have a PQC system to deal with the challenge. Unfolded protein response (UPR) triggered by ER stress is an integrated part of intracellular PQC.75 Terminally misfolded ER proteins are retrogradely transported out of the ER and immediately subjected to ubiquitination and proteasomal degradation via ER-associated protein degradation (ERAD) in the cytosol.76 However, sustained ER stress causes accumulation of UPS reporter substrates, which indicates that sustained ER stress has an inhibitory effect on the UPS.77 When ERAD is overloaded by ER inhibitors or blocked by proteasome inhibitors, autophagy is mobilized to degrade terminally misfolded ER proteins via the ER-activated autophagy (ERAA) pathway.78-81
In a recent study, Hill’s group reported that accumulation and aggregation of ubiquitinated proteins upregulated the UPR regulator Bip and triggered activation of autophagy in a mouse model of load-induced heart failure.31 Additionally, accumulation of misfolded proteins, such as polyQ72 aggregates, in the ER stimulated LC3 conversion from LC3-I to LC3-II through phosphorylation of PERK (RNA-dependent protein kinase-like ER kinase) and eukaryotic initiation factor 2α (eIF2α).79 A related study with human tumor cells reveals that UPR protects against hypoxic tumor cells through inducing LC3 and Atg5 gene expression via the PARK/eIF2α signaling branch.82
Hypoxia or sI/R also leads to ER stress and activates UPR in cultured cardiomyocytes.83-84 Moreover, UPR is also activated in cardiomyocytes adjacent to, but not in those distal to, the infarct zone in mouse hearts subjected to AMI.85 Additionally, it has been suggested that δPKC activity mediates I/R-induced ER stress in the myocardium.86
7.2 ATP level and AMP-activated protein kinase (AMPK)
Protein ubiquitination and unfolding during UPS-mediated proteolysis are ATP-dependent. ATP is also required to assemble the 26S proteasome complex.2 Therefore, it is likely that decreases in ATP levels contribute to diminished 26S proteasome activity and impaired UPS function in certain cardiac dysfunction.
When ATP levels in a cell decrease, various intracellular adaptive mechanisms are initiated, attempting to restore ATP levels. Among them, autophagy is one of the most important. In the study by Matsui et al.,63 glucose deprivation-induced autophagy in cardiomyocytes was mediated by activation of AMPK and inhibition of mTOR. Similarly, during myocardial ischemia, a rapid drop in ATP concentration also increases the AMP/ATP ratio and activates AMPK.87 Moreover, autophagosome formation in response to myocardial ischemia is decreased in tg mice with cardiac specific expression of a dominant negative AMPK, suggesting that endogenous AMPK is essential in mediating autophagy during myocardial ischemia.63 As a prominent energy sensor, AMPK is involved in activating energy generating pathways to maintain ATP levels and is required to activate autophagy.88
7.3 Hypoxia and mitochondrial stress
Bnip3 (Bcl2/E1B 19kDa interacting protein), a member of BH3-only proteins, is an important contributor to I/R injury and is induced in cultured ventricular myocytes and adult rat hearts subjected to hypoxia.89-90 Overexpression of Bnip3 caused significant induction of autophagy in HL-1 myocytes, while expression of a dominant-negative form of BNIP3 reduced hypoxia-induced autophagy.89 These studies suggest that Bnip3 likely contributes to the upregulation of autophagy during I/R and hypoxia.
Furthermore, the downstream players mediating Bnip3-induced autophagy have been identified. Bnip3 was shown to directly bind Rheb (Ras homolog enriched in brain) and inhibit its GTPase activity, thereby playing an important role in mTOR inactivation in response to hypoxia.91 This inactivation of mTOR might mediate Bnip3-induced autophagy. Alternatively, as an integral mitochondrial membrane protein, Bnip3 induces mitochondrial defects leading to the opening of the mitochondrial membrane permeability transition pore (mPTP).90 Given that mPTP opening induces autophagy in mammalian cells,92 it is possible that Bnip3 induces autophagy through damaging mitochondria. Reperfusion also triggers mPTP opening.93 Hence, mPTP opening may also contribute to autophagy activation in myocardial I/R.
7.4 Oxidative stress
Production of reactive oxygen species (ROS) is associated with a number of stimuli, inducing tumor necrosis factor (TNF) stimulation, ER stress, starvation, and inhibition of mitochondrial function.94 ROS are generated in the myocardium during I/R.95 Nitric oxide is released in heart failure and myocardial ischemia.96 It is known that the UPS is involved in removing mildly oxidized proteins. However, heavily oxidized proteins appear to form aggregates and then extensive covalent cross-linkage that make them highly resistant to UPS-mediated proteolysis.97 Moreover, some proteasome subunits may be suppressed or inactivated by oxidative stress. For instance, the 26S proteasome subunit S6 ATPase is very sensitive to oxidative inactivation.29 Oxidative modification of 20S proteasome subunits by the lipid peroxidation product 4-hydroxy-2-nonenal (HNE), which occurs in cardiac I/R, results in selective inactivation of 20S proteasome activity.30 Taken together, these findings explain the decreased peptidase activity of the proteasome in animal models of myocardial ischemia and I/R injury.27
On the other hand, ROS can activate autophagy in cardiomyocytes. For example, a study by Gottlieb’s group showed that lipopolysaccharide (LPS) induced autophagy through oxidative stress in both cardiomyocytes and mouse hearts. Nitric oxide synthase (NOS) inhibitor L-NMMA or potent antioxidant reagent N-acetyl-cysteine (NAC) reduced LPS-induced autophagy in cardiomyocytes.98 Furthermore, the ability of H2O2 and NO to directly activate autophagy was also tested in HL-1 cells in this study. It was observed that either H2O2 or SNP, a NO donor, was able to trigger autophagy in HL-1 cells.98 This suggests both reactive oxygen and nitrogen species induce autophagy directly.
Notably, a study by Scherz-Shouval and colleagues reveals a molecular underpinning of redox regulation of the autophagic process. They have shown ROS, specifically H2O2, are essential for starvation-induced autophagy because this oxidative signal leads to inactivation of Atg4. Atg4, a cysteine protease, is involved both in the initial lipidation processing of Atg8 to allow the conjugation of Atg8 to phosphatidylethanolamine (PE) and in the delipidation step by cleaving PE from Atg8-PE. This oxidative signal leads to inactivation of Atg4 at the site of autophagosome formation, thereby promoting lipidation of Atg8, an essential step in autophagosome formation.94, 99
7.5 Calcium overload
Another consequence of I/R injury is calcium overload. It has been established that increased cytosolic calcium is a potent inducer of autophagy. Various Ca2+ mobilizing agents inhibit the activity of mTOR and induce massive formation of autophagosomes.100
Therefore, as summarized in Figure 1, multiple factors are capable of impairing UPS function and triggering autophagy activation. The latter participate in regulating multiple cellular processes.
Figure 1. A summary of factors that impair the UPS and the potentially pathways that lead autophagy to participate in cellular processes.
AMPK, AMP dependent kinase; ER, endoplasmic reticulum.
8. Mechanism of the dual role of autophagy in cardiac disease
As mentioned earlier, autophagy plays a dual role in the pathogenesis of cardiac diseases. Autophagy is activated to protect cells against cellular stress, while excessive autophagy promotes cell death. Therefore, the extent to which autophagy is activated may affect the pathophysiological outcome of autophagy. It has also been suggested that whether autophagy plays a protective or detrimental role is etiology-dependent. Thus, it is important to understand how autophagy contributes to cell survival or cell death in response to different stimuli in the heart.
8.1. Aggregates clearance and ER stress alleviation
The autophagic clearance of aggregate-prone proteins is well-established in neurodegenerative diseases. For instance, activation of autophagy by rapamycin has been shown to increase clearance of aggregate-prone protein both in vitro and in vivo.101-103 Intrigued by the common features shared by neurodegenerative diseases and cardiac proteinopathy, cardiac biologists also tested the autophagic degradation of aggregate-prone proteins in the heart. Pattison et al. observed that polyQ83 aggresomes in the heart are frequently engulfed by autophagosomes, associating autophagy with protein aggregation.58-59 More importantly, inhibition of autophagy has been shown to increase the prevalence of protein aggregates in cardiomyocytes in CryABR120G-based DRC models both in vivo and in vitro.60
In both neurodegenerative diseases and DRC, it is generally believed that the mutant proteins exert particular toxicity in the oligomer form and that microscopically visible protein aggregates may be less toxic. Increased autophagic activity not only directly removes aggregates but also clears aggregate precursors, shifting the equilibrium away from aggregate formation and thereby attenuating its toxicity in the cell.11, 104
Moreover, under the condition that proteasome is inhibited, autophagy is likely activated in response to ER stress and plays a pro-survival role.105 The activated autophagy serves to compensate for the reduced proteasome function so that proteins that fail to be degraded by the UPS can be cleared via autophagy. By doing so, autophagy ameliorates ER stress and suppresses proteasome inhibitor-induced cell death at an upstream site of the death pathway.105
8.2. ATP maintenance
During ischemia and nutrient limitation, autophagy is perhaps the main mechanism for adaptation and salvage of ATP. Autolysosomal degradation of membrane lipids and proteins generates free fatty acids and amino acids that can be used as fuel for ATP generation.106
The study by Matsui et al. showed that inhibition of autophagy by 3-MA further reduced ATP levels and increased cell death in the setting of glucose deprivation.63 Takemura et al. confirmed this result and further showed that rapamycin mitigated the reduction in ATP level and improved the survival of glucose-starved cells.107 These findings are consistent with the notion that autophagy promotes cell survival through preserving cellular ATP content during glucose-deprivation. Further supporting this notion is the finding that treatment with bafilomycin A, a lysosome inhibitor, did not affect cardiac function in normally fed mice; but instead, bafilomycin A severely depressed cardiac function and led to significant left ventricular dilatation with severely reduced myocardial amino acid and ATP contents in the starved mice.108
8.3. Mitochondrial quality control
Since damaged mitochondria can release cytochrome c and other pro-apoptotic factors and generate ROS during I/R, their sequestration and degradation are cytoprotective. Autophagy is the only process known to degrade organelles in their entirety. Removal of ROS-producing mitochondria through autophagy would lower the oxidative stress experienced by a cell, facilitate mitochondrial biogenesis, and help with the mitochondrial quality control.
Several lines of evidence suggest that damaged mitochondria are selectively removed by autophagy. One example is that in rat hepatocytes, depolarized mitochondria are selectively sequestrated into autophagosomes.92 Similarly, the association between autophagosomes and damaged mitochondria is indicated by their colocalization in HL-1 cells where fragmentation of mitochondria is induced by overexpressing Bnip3.89 The damage in rat neonatal cardiomyocytes treated by LPS was limited by the rapid removal of damaged mitochondria triggered by the stimulation of autophagy.109 Likewise, during chronic hypoxia, Bnip3-mediated mitochondrial autophagy leads to reduction in mitochondrial DNA, mitochondrial mass, cell respiration and ROS production in response to hypoxia.110 Conversely, autophagy inhibition causes accumulation of damaged mitochondria. Ultrastructural analyses of Atg5-deficient hearts has revealed a disorganized sarcomere structure and the misalignment and aggregation of mitochondria.55 Moreover, suppression of autophagy by 3-MA in neonatal rat ventricular myocytes accumulates mitochondria with reduced membrane potential and mitochondria accumulation is partly reversed after 3-MA withdrawal.111 Taken together, upregulation of autophagy serves as a protective response likely by sequestering damaged mitochondria.
8.4. Cell death
8.4.1 Autophagic cell death
Previously, autophagic cell death was mainly a morphologic definition. Autophagic cell death is characterized by cell death with accumulation of vacuoles in physiological and pathological conditions.112 However, the question arises whether autophagic activity in dying cells is the cause of death or is actually an attempt to prevent it. Morphological definition can not prove a causative relationship between the autophagic process and cell death. The current definition of autophagic cell death includes an association with autophagosomes as well as dependence on autophagy proteins.113
Inhibition of apoptotic pathways can switch cellular response to stress from apoptosis to autophagic cell death. For instance, Bax/Bak-deficient mouse embryonic fibroblasts (MEFs), which fail to trigger apoptotic cell death, undergo non-apoptotic cell death when exposed to various cytotoxic agents, such as etoposide, staurosporine, and thapsigargin. This non-apoptotic form of cell death can be inhibited by suppressing autophagosome formation with autophagy inhibitors, such as 3-MA and wortmannin, or by silencing Atg5 and Atg6.114 Independently, other studies have also shown that with cytotoxic agents, autophagic cell death is induced with pan-caspase inhibitor, and this caspase-independent cell death can be inhibited by either chemical autophagy inhibitors or by the silencing of beclin1, Atg7 or Atg5.115-116
Excessive activation of autophagy itself can serve as a mechanism of cell death. Hyperactivation of autophagy, which destroys large proportions of the cytosol and organelles beyond a certain threshold, would lead to irreversible cellular atrophy with a consequential collapse of cell function.106 During reperfusion phase, low levels of ROS induce autophagy to degrade antioxidant catalase, thereby increasing the level of H2O2, which further induces autophagy. This positive feedback loop between autophagy and ROS accumulation resulting in a prolonged H2O2 signal is responsible for shifting the outcome from survival to death.117 Similarly, prolonged hypoxia induces autophagic cell death mediated through Bnip3.65 Therefore, autophagic cell death would explain the detrimental role of excessively activated autophagy under I/R or prolonged hypoxia, at least in some experimental conditions.
8.4.2. Autophagy-dependent apoptosis
Recent studies suggest a cross-talk between two self-destructive processes: autophagy and apoptosis. As mentioned in 8.4.1, inhibition of apoptotic pathways can elicit autophagic cell death in cells subjected to cellular stress. On the other hand, autophagy constitutes a stress adaptation to avoid apoptosis in some scenarios. One example is that nutrient depletion triggers apoptosis under autophagy suppression.118 Notably, Atg5 serves as a point of cross-talk between autophagic and apoptotic pathways. While it is a key player in autophagy, Atg5 may also play a role in apoptosis. Under lethal stress, Atg5 is cleaved by calpains to generate a 24kDa truncated Atg5. The truncated Atg5 loses its autophagy-inducing activity and instead acquires a proapoptotic one. It translocates from cytosol to the mitochondria and associates with the anti-apoptotic molecule Bcl-XL, provoking apoptotic cell death.119 Therefore, Atg5 serves as a “double-agent” in both autophagy and apoptosis, which is regulated by the proteolysis of ATG5. Furthermore, the finding that Atg5 mediates interferon-γ-induced cell death by interacting with FADD (Fas-associated protein with death domain) also suggests that Atg5 has pro-apoptotic effects. This Atg5-mediated cell death is both autophagy- and caspase-dependent.120 Hence, Atg5 can trigger apoptosis through two different mechanisms.
There is growing evidence to suggest that lysosomal proteases are actively involved in apoptosis. For example, cathepsin D exerts its apoptosis-stimulating effect upstream of caspase-3-like activation in NRVMs.121 Cathepsin G also induces SHP2 (SH2 domain-containing tyrosine phosphatase 2) activation, which leads to focal adhesion kinase (FAK) tyrosine dephosphorylation and promotes apoptosis in NRVMs.122 A study using Hela cells shows that disruption of lysosomes by a lysosomotropic agent results in translocation of lysosomal proteases to the cytosol and induction of cell death through a caspase-dependent mechanism, involving Bid cleavage by papain-like cysteine proteases.123
9. The UPS and autophagy form a consortium
9.1 Functional interactions
Pharmacologically induced PSMI induces autophagy in multiple mammalian cell types.31, 105 Genetic impairment of the proteasome also induces autophagy in Drosophila.124 Importantly, autophagy induction in the setting of proteasome impairment is likely protective. Supportive evidence comes from the reports suggesting that the degenerative phenotypes caused by proteasome impairment are enhanced in an autophagy-deficient background, whereas the degeneration is significantly alleviated with autophagy induction in Drosophila.124 In cultured cells, it has been shown that the pretreatment with rapamycin attenuates proteasome inhibitor lactacystin-induced apoptosis and ubiquitinated protein aggregates;125 whereas, suppression of autophagy by 3-MA or Atg knockdown enhances proteasome inhibitor-induced cell death and accumulation of ubiquitinated proteins.31, 105
On the other hand, multiple lines of evidence suggest that reduced autophagy also results in enhancement of proteasome-dependent protein degradation. One example is that autophagy inhibition by 3-MA results in a robust increase in proteasome activities in NRVMs.31 Moreover, elevated polyubiquitinated protein levels and increased proteasome activities have been observed in Atg5-ablated hearts both in the basal state and in response to TAC.71 Taken together, the UPS and autophagy seem to help each other in PQC.
9.2 The mechanistic link between the UPS and autophagy
Ubiquitination used to be the hallmark of protein degradation by 26S proteasome. Modification of a protein with K48-linked polyubiquitin chains targets the substrate to the proteasomal degradation. However, recent findings reveal the involvement of K63-linked polyubiquitin chains in autophagy pathway.126 Adaptor molecules such as p62/SQSTM1, NBR1 simultaneously bind both ubiquitin and autophagy machinery. Identification of these adaptors provides a mechanistic link between the UPS and autophagy (Figure 2).
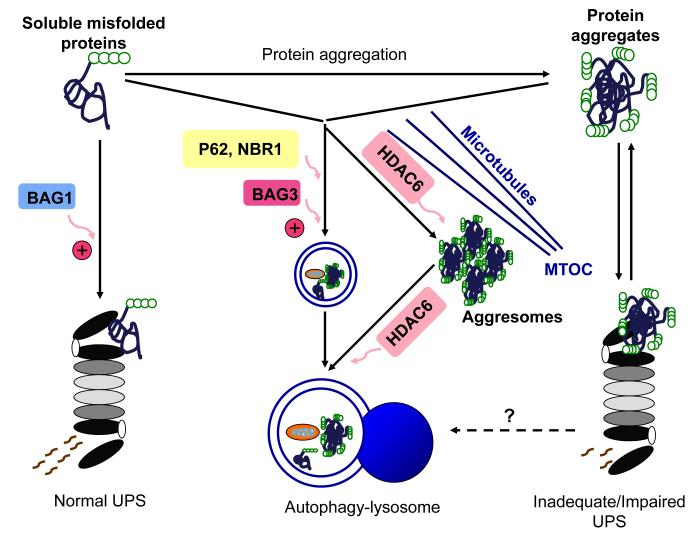
Figure 2. A schematic illustration of the interplay between the ubiquitin-proteasome system (UPS) and autophagy in removal of misfolded proteins.
The UPS degrades soluble misfolded proteins. Aberrant protein aggregation impairs proteasome proteolytic function and insufficient proteasome function in turn further accumulates misfolded proteins, thereby forming a vicious cycle. The inadequate or impaired UPS might activate the autophagy-lysosome pathway as a compensatory mechanism. The autophagy-lysosome pathway removes both soluble oligomers of misfolded proteins and protein aggregates. p62 and NBR1 recognize ubiquitinated proteins and mediate their autophagic degradation. HDAC6 binds to ubiquitinated proteins and facilitates their transportation along microtubules toward the microtubule organizing center (MTOC), where ubiquitinated proteins are organized into aggresomes. Moreover, the autophagic degradation of aggresomes is promoted by HDAC6. As co-chaperones, BAG1 and BAG3 regulate proteasomal and autophagic pathways, respectively.
9.2.1. p62/SQSTM1
p62/SQSTM1(hereafter referred to as p62) appears to be an adaptor molecule linking ubiquitinated proteins to the autophagic machinery.127 The C-terminal portion of p62 binds polyubiquitinated substrates through its ubiquitin-associated (UBA) domain and directly binds to LC3 via the LC3 interacting region (LIR) motif.128 p62 can also polymerize via its N-terminal Phox/ Bem1p (PB1) domain and interact with proteasome via an N-terminal ubiquitin-like (UBL) domain. Interaction of p62 UBL domain with proteasome may be involved in shuttling substrates for proteasomal degradation.129
The increase in both transcript and protein levels of p62 in response to PSMI indicates that p62 may sense the proteolytic stress with PSMI and be involved in mediating the alternative degradation pathway to alleviate the proteolytic stress.130 It has been suggested that p62 provides a key link between autophagy and the UPS by facilitating autophagic degradation of ubiquitinated proteins.131
p62 also co-localizes with autophagosomes and can be degraded by autophagy.128 The degradation of p62 by autophagy is evidenced by the elevated level of p62 following autophagy inhibition and the decreased level in rapamycin treatment.1, 127, 132 Besides co-localizing with autophagosomes, p62 also localizes in a variety of ubiquitin-positive inclusion bodies in many neurodegenerative diseases, including Lewy bodies in Parkinson’s disease, neurofilbrillary tangles in tauopathies, huntingtin aggregates in Huntington’s disease, and aggregates seen in familial amyotrophic lateral sclerosis.133-136
Notably, p62 is required for the formation and autophagic degradation of polyubiquitin-positive inclusion bodies in response to stress.128 The aggregate-promoting property of p62 is suggested by the observation that depletion of p62 diminishes the formation of ubiquitin-positive inclusions upon puromycin treatment.128 Moreover, p62-null mice fail to form ubiquitin-positive inclusions in hepatocytes and neurons in response to misfolded protein stress in an autophagy-deficient background.137
Several lines of evidence also suggest the involvement of p62 in autophagic degradation of ubiquitinated proteins in response to misfolded protein stress. For instance, in neurons of p62-null mouse brains, there is a detectable increase in ubiquitin staining paralleled by accumulation of insoluble ubiquitinated proteins.138 Similarly, hyperphosphorylated tau, neurofibrillary tangles, and neurodegeneration are observed in p62-null mice.139 Reduction of p62 protein levels or interference with p62 function significantly increases cell death induced by the expression of mutant huntingtin.127 However, one study using mice with genetic inactivation of p62 and Atg7 found that loss of p62 markedly attenuated liver injury caused by autophagy deficiency; whereas p62 deficiency had little effect on neuronal degeneration. This study indicates that the pathologic process associated with p62 deficiency in autophagy-deficient tissues is cell-type specific.137 Taken together, p62 plays a critical role in the formation of ubiquitin-positive inclusions, degradation of ubiquitinated proteins under stress and is a perfect candidate for a messenger between the UPS and autophagy.
9.2.2. Neighbor of BRCA1 gene1 (NBR1)
The relatively mild phenotypes as well as the cell-type specific phenotypes in p62 knockdown or knockout experiments raise the possibility of the existence of other p62-like proteins that might compensate for the lack of p62.133 NBR1 is the candidate for another autophagic receptor for ubiquitinated cargo.
NBR1 shares similar domain architecture with p62 and contains both a LIR motif interacting with ATG8 family proteins and a C-terminal UBA domain interacting with ubiquitin. NBR1 also has a PB1 domain and this region of NBR1 interacts directly with p62 in a canonical PB1-PB1 fashion. NBR1 can also homodimerise via the N-terminal two coiled-coil domains.140 The similar domain architecture between p62 and NBR1 raises the possibility for NBR1 to be another autophagic adaptor for ubiquitinated proteins.
On one hand, NBR1 co-localizes with p62 and Atg8-family proteins in the cell and can be degraded by autophagy in an LIR-dependent manner. This autophagic degradation is independent of p62.141 On the other hand, NBR1 co-localizes with p62 in the ubiquitin-positive aggregates in response to autophagy inhibition as well as in Mallory bodies in the liver of a patient with alcoholic steatohepatitis. More importantly, knockdown experiments suggest that both p62 and NBR1 are required for the formation of ubiquitin-positive aggregates upon autophagy inhibition and puromycin treatment. These findings lead to the conclusion that p62 and NBR1 cooperate in sequestration and degradation of ubiquitinated proteins.141
It is still unclear whether NBR1 can function as a substitute for p62 to perform some of the functions of p62. The recent finding that aggregation of NBR1 is dramatically increased under the condition that both p62 and Atg7 are deficient raises the possibility that the increased NBR1 can partially compensate for the loss of p62 to form aggregates.141 The involvement of autophagy receptor p62 and NBR1 in the formation of aggregates suggests a possible link between aggregate formation and autophagy.
9.2.3. Histone Deacetylase 6 (HDAC6)
Aggresome formation may help detoxify misfolded proteins through packaging more toxic and active oligomers of misfolded proteins into large inclusion bodies so that fewer vital cellular constituents would come into contact with them. The transportation of aggresome precursors via microtubules and their motor proteins to the vicinity of microtubule organizing center (MTOC) is critical to aggresome formation and subsequent removal by autophagy.142
HDAC6 is a microtubule- and dynein- associated deacetylase. It binds ubiquitin via the C-terminal BUZ domain and interacts directly with dynein motors, thereby serving as a linker coupling protein aggregates to the retrograde microtubule motor. HDAC6 has been found to localize to the experimentally induced ubiquitin-positive aggresomes.143-144 Moreover, HDAC6 deficiency results in defects in aggresome formation in response to experimentally induced misfolded protein stress.144 HDAC6 may therefore regulate the retrograde transport of misfolded proteins to MTOC to form aggresomes. More strikingly, HDAC6 is essential for retrograde transport of autophagosomes and lysosomes to MTOC where the aggresomes are mainly located, thereby facilitating the autophagic degradation of aggresome.143 The two functions of HDAC6 in promoting aggresome formation and autophagic degradation could be coupled and represent one integrated mechanism. From this point of view, efficient autophagic degradation of aggregated proteins may be promoted by HDAC6.11 This notion is supported by the finding that HDAC6 is required for autophagic clearance of aggregated mutant huntingtin in cultured cells and the mutant androgen receptor in a fly model of Kennedy’s disease.124, 143 Similarly, autophagy is induced by, and compensates for, UPS impairment in a fruitfly model of spinobulbar muscular atrophy in an HDAC6-depedent manner.124
p62, NBR1, and HDAC6 may work in synergy to pack the misfolded proteins together and facilitate their interactions with the phagophore, thus providing the specificity required for the degradation.11 It has also been suggested that K63-linked polyubiquitin chains interacts with p62, NBR1, and HDAC6, thereby providing a signal for selective autophagic degradation.145-146
9.2.4. Co-chaperones
CHIP can mediate α-synuclein degradation by both degradation pathways: the tetratricopeptide repeat domain is critical for proteasomal degradation, whereas the U-box domain is sufficient to direct α-synuclein toward the lysosomal degradation pathway. This study suggests that CHIP acts as a molecular switch between proteasomal and lysosomal degradation pathways.147
The Hsc/Hsp70 co-chaperones of the BAG protein family are modulators of PQC. BAG1 acts as a physical link between the Hsp70 and the proteasome. Moreover, BAG1 can accept substrates from Hsc/Hsp70 and direct them to proteasomal degradation through the direct interaction with CHIP.148 In contrast, BAG3-HSPB8 complex facilitates the degradation of Htt43Q through the activation of autophagy.149 In cell models of ageing, it has also been shown that BAG1 and BAG3 regulate proteasomal and autophagic pathways, respectively, for the degradation of polyubiquitinated proteins. More importantly, due to the increased ratio of BAG3/BAG1 in aged cells compared with young cells, the aged cells depend more intensively on autophagy to degrade polyubiquitinated proteins.150 Moreover, BAG3 interacts with p62 to promote p62-dependent autophagic degradation.150
Thus, as highlighted in this section and illustrated in Figure 2, a number of factors and signaling events orchestrate the UPS and autophagy to perform PQC in the heart. Each of the two proteolytic pathways has its own responsibilities at the baseline but they attempt to assist each other during cardiac dysfunction.
10. Conclusions and future prospective
PQC and protein degradation are extremely important for maintaining normal cardiac function. PQC inadequacy occurs in the progression of a wide range of cardiac diseases, as reflected by elevated levels of ubiquitinated proteins and sustained UPR in failing human hearts and in many animal models of cardiac dysfunction. The accumulation of misfolded proteins resulting from inadequate PQC in turn impairs PQC in the heart, forming a vicious cycle. Very few studies have shed light on the feasibility and effect of enhancing UPS function. This is probably because enhancing UPS function may accelerate the degradation of critical short-lived intracellular regulators, thereby being detrimental to the cell.151
In contrast, autophagy seems to be more amenable to manipulation. In most scenarios of cardiac dysfunction, autophagy protects cells by: (1) removing misfolded proteins and alleviating ER stress; (2) maintaining the ATP level in the cell; and (3) removing damaged organelles such as damaged mitochondria. Growing evidence links autophagy with the pathogenesis of multiple forms of heart disease, suggesting that autophagy may be therapeutically effective in treating the heart disease.71, 152 However, the role of autophagy is dose- and context-dependent, which poses special challenges. Excessive autophagy would cause cell death through excessive self-digestion of essential organelles and proteins. Therefore, in the clinical setting, physiologically beneficial autophagy should be preserved, whereas excessive autophagy should be avoided.
A growing body of evidence indicates that pharmacological upregulation of autophagy is indeed beneficial in animal models of a large subset of cardiac dysfunction, such as myocardial ischemia, cardiac hypertrophy, cardiac ageing. Given that drugs mitigating autophagy are already in clinical use,104 substantial advances in attaining more effective treatment for human heart disease by manipulating autophagy are on the horizon.
Additionally, caloric restriction and exercise are known to activate autophagy.153-154 Moreover, the up-regulated autophagy mediated by caloric restriction and exercise shows cardioprotection. For instance, removal of ROS-producing mitochondria through autophagy is associated with less oxidative stress and results in high-quality mitochondria which exhibit efficient ATP production and better resistance to stress.153, 155 Therefore, lifestyle change may show great promise in alleviating cardiac disease.
Acknowledgement
Dr. X. Wang is an established investigator of the American Heart Association (AHA). Research in his laboratory is supported in part by grants R01HL072166, R01HL085629, and R01HL068936 from the NIH and grant 0740025N from the AHA (to X. W.) and by the MD/PhD Program of the University of South Dakota. Dr. Q. Zheng is a recipient of AHA Predoctoral Fellowship (Reference # 0815571G). We thank Ms. Emily McDowell for her assistance in preparing this manuscript.
Footnotes
Disclosure The authors declared no conflict of interest.
Reference
- 1.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. J Mol Cell Cardiol. 2008;45:11–27. doi: 10.1016/j.yjmcc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: A quality control perspective. Cardiovasc Res. 2010;85:253–262. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuertes G, Villarroya A, Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35:651–664. doi: 10.1016/s1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 5.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimori T. Autophagy: A regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 7.Fuertes G, De Llano JJ Martin, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes AV, Zong C, Ping P. Protein degradation by the 26s proteasome system in the normal and stressed myocardium. Antioxid Redox Signal. 2006;8:1677–1691. doi: 10.1089/ars.2006.8.1677. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 11.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 12.Tsukamoto O, Minamino T, Okada K, Shintani Y, Takashima S, Kato H, Liao Y, Okazaki H, Asai M, Hirata A, Fujita M, Asano Y, Yamazaki S, Asanuma H, Hori M, Kitakaze M. Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Commun. 2006;340:1125–1133. doi: 10.1016/j.bbrc.2005.12.120. [DOI] [PubMed] [Google Scholar]
- 13.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 14.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 15.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: A cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birks EJ, Latif N, Enesa K, Folkvang T, Luong le A, Sarathchandra P, Khan M, Ovaa H, Terracciano CM, Barton PJ, Yacoub MH, Evans PC. Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc Res. 2008;79:472–480. doi: 10.1093/cvr/cvn083. [DOI] [PubMed] [Google Scholar]
- 17.Powell SR, Divald A. The ubiquitin-proteasome system in myocardial ischaemia and preconditioning. Cardiovasc Res. 2010;85:303–311. doi: 10.1093/cvr/cvp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Kem DC. Proteasome inhibition during myocardial infarction. Cardiovasc Res. 2010;85:312–320. doi: 10.1093/cvr/cvp309. [DOI] [PubMed] [Google Scholar]
- 19.Tsukamoto O, Minamino T, Kitakaze M. Functional alterations of cardiac proteasomes under physiological and pathological conditions. Cardiovasc Res. 2010;85:339–346. doi: 10.1093/cvr/cvp282. [DOI] [PubMed] [Google Scholar]
- 20.Hedhli N, Depre C. Proteasome inhibitors and cardiac cell growth. Cardiovasc Res. 2010;85:321–329. doi: 10.1093/cvr/cvp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, Hewett T, Robbins J. Expression of r120g-alphab-crystallin causes aberrant desmin and alphab-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. Faseb J. 2006;20:362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, Liu J, Zheng H, Wang X. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. Faseb J. 2005;19:2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 27.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 28.Powell SR, Wang P, Katzeff H, Shringarpure R, Teoh C, Khaliulin I, Das DK, Davies KJ, Schwalb H. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: Essential role of the proteasome. Antioxid Redox Signal. 2005;7:538–546. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- 29.Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: Identification of an oxidation-sensitive subunit in 26 s proteasome. Biochemistry. 2005;44:13893–13901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 30.Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20s proteasome subtypes. Arch Biochem Biophys. 2006;453:135–142. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr., Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, Hong C, Hittinger L, Ghaleh B, Sadoshima J, Vatner DE, Vatner SF, Madura K. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 33.Balasubramanian S, Mani S, Shiraishi H, Johnston RK, Yamane K, Willey CD, Cooper Gt, Tuxworth WJ, Kuppuswamy D. Enhanced ubiquitination of cytoskeletal proteins in pressure overloaded myocardium is accompanied by changes in specific e3 ligases. J Mol Cell Cardiol. 2006;41:669–679. doi: 10.1016/j.yjmcc.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 34.Razeghi P, Baskin KK, Sharma S, Young ME, Stepkowski S, Essop MF, Taegtmeyer H. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem Biophys Res Commun. 2006;342:361–364. doi: 10.1016/j.bbrc.2006.01.163. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Xu Z, He XR, Michael LH, Patterson C. Chip, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. 2005;288:H2836–2842. doi: 10.1152/ajpheart.01122.2004. [DOI] [PubMed] [Google Scholar]
- 36.Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. Chip deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28:4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toth A, Nickson P, Qin LL, Erhardt P. Differential regulation of cardiomyocyte survival and hypertrophy by mdm2, an e3 ubiquitin ligase. J Biol Chem. 2006;281:3679–3689. doi: 10.1074/jbc.M509630200. [DOI] [PubMed] [Google Scholar]
- 38.Clegg HV, Itahana K, Zhang Y. Unlocking the mdm2-p53 loop: Ubiquitin is the key. Cell Cycle. 2008;7:287–292. doi: 10.4161/cc.7.3.5358. [DOI] [PubMed] [Google Scholar]
- 39.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, Patterson C. Muscle ring finger protein-1 inhibits pkc{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004;167:1147–1159. doi: 10.1083/jcb.200402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fielitz J, van Rooij E, Spencer JA, Shelton JM, Latif S, van der Nagel R, Bezprozvannaya S, de Windt L, Richardson JA, Bassel-Duby R, Olson EN. Loss of muscle-specific ring-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci U S A. 2007;104:4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, Richardson JA, Bassel-Duby R, Olson EN. Myosin accumulation and striated muscle myopathy result from the loss of muscle ring finger 1 and 3. J Clin Invest. 2007;117:2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by murf1 and murf2. Embo J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J Biol Chem. 2007;282:17032–17040. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- 45.Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy f-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an scf ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of forkhead proteins. J Clin Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell NH, Tang G, Yu C, Mercurio F, DiDonato JA, Lin A. Activation of nf-kappa b is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc Natl Acad Sci U S A. 2001;98:6668–6673. doi: 10.1073/pnas.111155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hedhli N, Lizano P, Hong C, Fritzky LF, Dhar SK, Liu H, Tian Y, Gao S, Madura K, Vatner SF, Depre C. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am J Physiol Heart Circ Physiol. 2008;295:H1385–1393. doi: 10.1152/ajpheart.00532.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meiners S, Dreger H, Fechner M, Bieler S, Rother W, Gunther C, Baumann G, Stangl V, Stangl K. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension. 2008;51:302–308. doi: 10.1161/HYPERTENSIONAHA.107.097816. [DOI] [PubMed] [Google Scholar]
- 50.Stansfield WE, Tang RH, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294:H645–650. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 51.Hacihanefioglu A, Tarkun P, Gonullu E. Acute severe cardiac failure in a myeloma patient due to proteasome inhibitor bortezomib. Int J Hematol. 2008;88:219–222. doi: 10.1007/s12185-008-0139-7. [DOI] [PubMed] [Google Scholar]
- 52.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: A case report. BMC Cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in lamp-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 54.Stypmann J, Glaser K, Roth W, Tobin DJ, Petermann I, Matthias R, Monnig G, Haverkamp W, Breithardt G, Schmahl W, Peters C, Reinheckel T. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin l. Proc Natl Acad Sci U S A. 2002;99:6234–6239. doi: 10.1073/pnas.092637699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 56.Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65:965–968. doi: 10.1253/jcj.65.965. [DOI] [PubMed] [Google Scholar]
- 57.Kassiotis C, Ballal K, Wellnitz K, Vela D, Gong M, Salazar R, Frazier OH, Taegtmeyer H. Markers of autophagy are downregulated in failing human heart after mechanical unloading. Circulation. 2009;120:S191–197. doi: 10.1161/CIRCULATIONAHA.108.842252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pattison JS, Robbins J. Protein misfolding and cardiac disease: Establishing cause and effect. Autophagy. 2008;4:821–823. doi: 10.4161/auto.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, Robbins J. Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation. 2008;117:2743–2751. doi: 10.1161/CIRCULATIONAHA.107.750232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inuzuka Y, Okuda J, Kawashima T, Kato T, Niizuma S, Tamaki Y, Iwanaga Y, Yoshida Y, Kosugi R, Watanabe-Maeda K, Machida Y, Tsuji S, Aburatani H, Izumi T, Kita T, Shioi T. Suppression of phosphoinositide 3-kinase prevents cardiac aging in mice. Circulation. 2009;120:1695–1703. doi: 10.1161/CIRCULATIONAHA.109.871137. [DOI] [PubMed] [Google Scholar]
- 62.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 64.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 65.Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, Gibson SB. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving bnip3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 67.Dosenko VE, Nagibin VS, Tumanovska LV, Moibenko AA. Protective effect of autophagy in anoxia-reoxygenation of isolated cardiomyocyte? Autophagy. 2006;2:305–306. doi: 10.4161/auto.2946. [DOI] [PubMed] [Google Scholar]
- 68.Gurusamy N, Lekli I, Gorbunov NV, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by adaptation to ischaemia augments autophagy in association with bag-1 protein. J Cell Mol Med. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gurusamy N, Lekli I, Gherghiceanu M, Popescu LM, Das DK. Bag-1 induces autophagy for cardiac cell survival. Autophagy. 2009;5:120–121. doi: 10.4161/auto.5.1.7303. [DOI] [PubMed] [Google Scholar]
- 70.Khan S, Salloum F, Das A, Xi L, Vetrovec GW, Kukreja RC. Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. J Mol Cell Cardiol. 2006;41:256–264. doi: 10.1016/j.yjmcc.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 71.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ha T, Li Y, Gao X, McMullen JR, Shioi T, Izumo S, Kelley JL, Zhao A, Haddad GE, Williams DL, Browder IW, Kao RL, Li C. Attenuation of cardiac hypertrophy by inhibiting both mtor and nfkappab activation in vivo. Free Radic Biol Med. 2005;39:1570–1580. doi: 10.1016/j.freeradbiomed.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 74.McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, Izumo S. Inhibition of mtor signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation. 2004;109:3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 75.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 76.Hebert DN, Molinari M. In and out of the er: Protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 77.Menendez-Benito V, Verhoef LG, Masucci MG, Dantuma NP. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum Mol Genet. 2005;14:2787–2799. doi: 10.1093/hmg/ddi312. [DOI] [PubMed] [Google Scholar]
- 78.Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (erad) systems for the novel variant of the mutant dysferlin: Ubiquitin/proteasome erad(i) and autophagy/lysosome erad(ii) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- 79.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. Er stress (perk/eif2alpha phosphorylation) mediates the polyglutamine-induced lc3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 80.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 81.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes map1lc3b and atg5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H. Amp-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol. 2005;25:9554–9575. doi: 10.1128/MCB.25.21.9554-9575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of atf6. Circ Res. 2006;98:1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 85.Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res. 2006;99:275–282. doi: 10.1161/01.RES.0000233317.70421.03. [DOI] [PubMed] [Google Scholar]
- 86.Qi X, Vallentin A, Churchill E, Mochly-Rosen D. Deltapkc participates in the endoplasmic reticulum stress-induced response in cultured cardiac myocytes and ischemic heart. J Mol Cell Cardiol. 2007;43:420–428. doi: 10.1016/j.yjmcc.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takagi H, Matsui Y, Hirotani S, Sakoda H, Asano T, Sadoshima J. Ampk mediates autophagy during myocardial ischemia in vivo. Autophagy. 2007;3:405–407. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 88.Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. Amp-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 89.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 90.Regula KM, Ens K, Kirshenbaum LA. Inducible expression of bnip3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- 91.Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, You M, Guan KL. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with rheb. J Biol Chem. 2007;282:35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 92.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. Faseb J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 93.Halestrap AP, Clarke SJ, Javadov SA. Mitochondrial permeability transition pore opening during myocardial reperfusion--a target for cardioprotection. Cardiovasc Res. 2004;61:372–385. doi: 10.1016/S0008-6363(03)00533-9. [DOI] [PubMed] [Google Scholar]
- 94.Liu Z, Lenardo MJ. Reactive oxygen species regulate autophagy through redox-sensitive proteases. Dev Cell. 2007;12:484–485. doi: 10.1016/j.devcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 95.Becker LB, vanden Hoek TL, Shao ZH, Li CQ, Schumacker PT. Generation of superoxide in cardiomyocytes during ischemia before reperfusion. Am J Physiol. 1999;277:H2240–2246. doi: 10.1152/ajpheart.1999.277.6.H2240. [DOI] [PubMed] [Google Scholar]
- 96.Rabkin SW. Nitric oxide-induced cell death in the heart: The role of autophagy. Autophagy. 2007;3:347–349. doi: 10.4161/auto.4054. [DOI] [PubMed] [Google Scholar]
- 97.Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 98.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. Lps-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jaattela M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 101.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 102.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mtor induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 103.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 104.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 105.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: Cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 107.Takemura G, Maruyama R, Goto K, Kanamori H, Tsujimoto A, Minatoguchi S, Fujiwara H. Fate of isolated adult cardiomyocytes undergoing starvation-induced autophagic degeneration. Autophagy. 2009;5:90–92. doi: 10.4161/auto.5.1.7206. [DOI] [PubMed] [Google Scholar]
- 108.Takemura G, Kanamori H, Goto K, Maruyama R, Tsujimoto A, Fujiwara H, Seishima M, Minatoguchi S. Autophagy maintains cardiac function in the starved adult. Autophagy. 2009;5:1034–1036. doi: 10.4161/auto.5.7.9297. [DOI] [PubMed] [Google Scholar]
- 109.Hickson-Bick DL, Jones C, Buja LM. Stimulation of mitochondrial biogenesis and autophagy by lipopolysaccharide in the neonatal rat cardiomyocyte protects against programmed cell death. J Mol Cell Cardiol. 2008;44:411–418. doi: 10.1016/j.yjmcc.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 110.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an hif-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Terman A, Dalen H, Eaton JW, Neuzil J, Brunk UT. Mitochondrial recycling and aging of cardiac myocytes: The role of autophagocytosis. Exp Gerontol. 2003;38:863–876. doi: 10.1016/s0531-5565(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 112.Bursch W. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 2001;8:569–581. doi: 10.1038/sj.cdd.4400852. [DOI] [PubMed] [Google Scholar]
- 113.Tsujimoto Y, Shimizu S. Another way to die: Autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 114.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 115.Xu Y, Kim SO, Li Y, Han J. Autophagy contributes to caspase-independent macrophage cell death. J Biol Chem. 2006;281:19179–19187. doi: 10.1074/jbc.M513377200. [DOI] [PubMed] [Google Scholar]
- 116.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an atg7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 117.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 120.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK. Essential roles of atg5 and fadd in autophagic cell death: Dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 121.Ollinger K. Inhibition of cathepsin d prevents free-radical-induced apoptosis in rat cardiomyocytes. Arch Biochem Biophys. 2000;373:346–351. doi: 10.1006/abbi.1999.1567. [DOI] [PubMed] [Google Scholar]
- 122.Rafiq K, Kolpakov MA, Abdelfettah M, Streblow DN, Hassid A, Dell’Italia LJ, Sabri A. Role of protein-tyrosine phosphatase shp2 in focal adhesion kinase down-regulation during neutrophil cathepsin g-induced cardiomyocytes anoikis. J Biol Chem. 2006;281:19781–19792. doi: 10.1074/jbc.M513040200. [DOI] [PubMed] [Google Scholar]
- 123.Cirman T, Oresic K, Mazovec GD, Turk V, Reed JC, Myers RM, Salvesen GS, Turk B. Selective disruption of lysosomes in hela cells triggers apoptosis mediated by cleavage of bid by multiple papain-like lysosomal cathepsins. J Biol Chem. 2004;279:3578–3587. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- 124.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. Hdac6 rescues neurodegeneration and provides an essential link between autophagy and the ups. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 125.Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W. Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis. 2008;32:16–25. doi: 10.1016/j.nbd.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 126.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 127.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. P62/sqstm1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pankiv S, Clausen T Hoyvarde, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. P62/sqstm1 binds directly to atg8/lc3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;19:19. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 129.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuusisto E, Suuronen T, Salminen A. Ubiquitin-binding protein p62 expression is induced during apoptosis and proteasomal inhibition in neuronal cells. Biochem Biophys Res Commun. 2001;280:223–228. doi: 10.1006/bbrc.2000.4107. [DOI] [PubMed] [Google Scholar]
- 131.Bjorkoy G, Lamark T, Johansen T. P62/sqstm1: A missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 132.Yue Z. Regulation of neuronal autophagy in axon: Implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3:139–141. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- 133.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 134.Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in alzheimer’s disease: Possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 135.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 136.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. P62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura SI, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 138.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 139.Babu J Ramesh, Seibenhener M Lamar, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 140.Lamark T, Kirkin V, Dikic I, Johansen T. Nbr1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 141.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for nbr1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 142.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 143.Iwata A, Riley BE, Johnston JA, Kopito RR. Hdac6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 144.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase hdac6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 145.Tan JM, Wong ES, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy. 2007;4:2. [Google Scholar]
- 146.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated k63-linked polyubiquitination targets misfolded dj-1 to aggresomes via binding to hdac6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of hsp70-interacting protein (chip) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 148.Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an e3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 149.Carra S, Seguin SJ, Lambert H, Landry J. Hspb8 chaperone activity toward poly(q)-containing proteins depends on its association with bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 150.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by bag3. Embo J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of huntington’s disease neuronal model cells. PLoS ONE. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ravikumar B, Berger Z, Vacher C, O’Kane CJ, Rubinsztein DC. Rapamycin pretreatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 153.Franch HA. Nutrition and muscle catabolism in maintenance hemodialysis: Does feeding make muscle cells selective self-eaters? J Ren Nutr. 2009;19:86–90. doi: 10.1053/j.jrn.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mackenzie MG, Hamilton DL, Murray JT, Baar K. Mvps34 is activated by an acute bout of resistance exercise. Biochem Soc Trans. 2007;35:1314–1316. doi: 10.1042/BST0351314. [DOI] [PubMed] [Google Scholar]
- 155.Powers SK, Quindry JC, Kavazis AN. Exercise-induced cardioprotection against myocardial ischemia-reperfusion injury. Free Radic Biol Med. 2008;44:193–201. doi: 10.1016/j.freeradbiomed.2007.02.006. [DOI] [PubMed] [Google Scholar]