Abstract
Changes in gene expression have been proposed to underlie many, or even most, adaptive differences between species. Despite the increasing acceptance of this view, only a handful of cases of adaptive gene expression evolution have been demonstrated. To address this discrepancy, we introduce a simple test for lineage-specific selection on gene expression. Applying the test to genome-wide gene expression data from the budding yeast Saccharomyces cerevisiae, we find that hundreds of gene expression levels have been subject to lineage-specific selection. Comparing these findings with independent population genetic evidence of selective sweeps suggests that this lineage-specific selection has resulted in recent sweeps at over a hundred genes, most of which led to increased transcript levels. Examination of the implicated genes revealed a specific biochemical pathway—ergosterol biosynthesis—where the expression of multiple genes has been subject to selection for reduced levels. In sum, these results suggest that adaptive evolution of gene expression is common in yeast, that regulatory adaptation can occur at the level of entire pathways, and that similar genome-wide scans may be possible in other species, including humans.
Changes in gene expression have long been theorized to play a major role in evolution (1, 2) and, more recently, have become a major focus of many studies on the evolution of development (3–8). In fact, it has recently been proposed that regulatory evolution “is pervasive and constitutes the primary fuel of the continuous morphological diversification of lineages and traits (3).” However, the strength of evidence supporting this view has been questioned (9) on the grounds that only a few unambiguous cases of regulatory adaptation have been demonstrated and that this small number has mostly been limited to just two model organisms, Drosophila fruit flies (4–6) and stickleback fish (7, 8) (we note that although many gene regulatory changes have been shown to affect phenotypes, very few of these have been shown to be adaptive). It is clear that what is needed to resolve this issue is a systematic, genome-scale approach to inferring regulatory adaptations, because only then can the question of their pervasiveness be rigorously addressed.
Although numerous studies of selection pressures acting on genome-wide gene expression levels have been reported (10), nearly all have found only negative selection and/or neutrality, consistent with the intuition that random changes to any functional system (including gene expression) will tend to be deleterious, and thus selected against. This is most likely because these studies were designed to detect the “average” mode of selection acting on new mutations that affect gene expression (SI Text). If adaptive mutations are rare, and usually occur in genes where most regulatory mutations are deleterious (and thus subject to negative selection)—as seems likely to be the case—then the average selection pressure on regulatory mutations will be dominated by negative selection, so methods measuring this average will have little power to reveal the occasional instances of positive selection.
An alternative is to specifically focus on what selective pressures have acted on differences that have accumulated between lineages or species. This approach was taken by Orr (11) in an elegant test of selection on quantitative traits: if alleles increasing the value of a trait are preferentially found in one lineage as compared to another, then neutrality can be rejected in favor of positive selection. Gene expression levels can be treated as quantitative traits (12), but, unfortunately, too few gene expression quantitative trait loci (eQTL) are known for any single gene to have any power to reject neutrality in Orr’s test. However, the sheer number of gene expression traits that can be measured provides a solution: by analyzing the directionality of hundreds or thousands of eQTL simultaneously, one can estimate a lower bound on the fraction that has been subject to lineage-specific selection.
Results and Discussion
A Systematic Test for Lineage-Specific Selection on Gene Expression.
For a gene whose expression is controlled by two independent eQTL (caused by distinct polymorphisms), under neutrality their directions of effect should be independent (11). In other words, in a genetic cross between parents A and B, knowledge that the allele from parent A at one of the gene’s eQTL leads to higher expression of the target gene does not provide any information about the effect of parent A’s allele at the second eQTL. In contrast, if the target gene has been selected for altered expression in the two lineages leading to A and B, then it is likely that A alleles at both eQTL will act in the same direction; thus, knowing that allele A at one eQTL increases expression indicates that the same is likely true at the other eQTL. A test of independence between pairs of eQTL controlling the same genes can therefore reveal if neutrality can be rejected en masse, and if so, approximately what fraction of genes show the effects of lineage-specific selection.
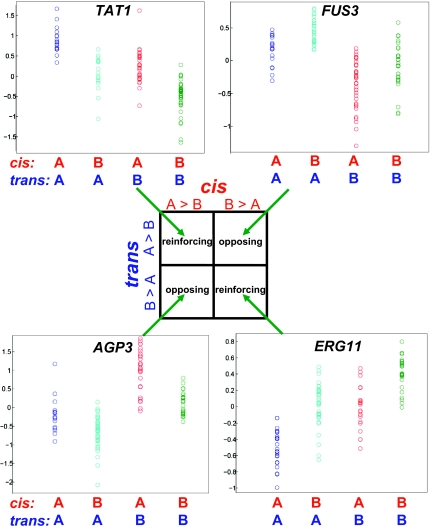
The vast majority of genes affected by multiple eQTL have at least one trans-acting, and one (presumably) cis-acting eQTL (although we refer to these as cis-acting, the assumption of a cis-mechanism is not necessary for our test; SI Text). There are two possible categories—allele A leads to higher or lower expression than allele B—for both cis and trans eQTL, leading to four classes in a 2 × 2 contingency table (Fig. 1). When A alleles have the same direction of effect on a gene in both cis and trans (and thus B alleles do as well), this is termed “reinforcing”; when the two A alleles have opposite effects, they are “opposing”. Directional selection in either lineage will tend to result in cis/trans changes in the same direction in the same lineage (Fig. S1) and an excess of reinforcing cases. The number of genes affected by two eQTL arising in two different lineages—which can give rise to either reinforcing or opposing cis/trans pairs, depending on the eQTL directionality (Fig. S1)—is expected to be approximately equal in all four classes (assuming the absence of massive convergent evolution) (SI Text), and thus will not affect the outcome of the test. Even though a single reinforcing case is not at all inconsistent with neutrality, a systematic excess of reinforcing cases can forcefully reject this null hypothesis. A standard χ2 test of independence on the 2 × 2 table constitutes the test.
Fig. 1.
A test of lineage-specific selection. For genes whose expression is affected by both cis and trans eQTL, there exist four classes of cis/trans effect directions and two general categories: reinforcing (where parent A’s alleles at cis and trans eQTL have the same direction of effect on the target gene) and opposing (where A alleles at cis and trans eQTL have opposite directions of effect on the target gene). An excess of the former implies lineage-specific selection—either positive or relaxed negative selection—whereas an excess of the latter implies stabilizing selection. An example of a gene belonging to each class is shown, using gene expression data from S. cerevisiae (14) analyzed in detail below; for each gene, four possible combinations of cis/trans genotypes exist, and the expression levels of individuals with each combination are shown (A alleles are from BY, B alleles are from RM; “A > B” indicates the A allele leads to higher expression). See also Fig. S1 for the possible evolutionary scenarios leading to each class of the 2 × 2 table.
Perhaps the most attractive aspect of the test is its lack of any assumptions about population demography or a subset of neutral sites. Nearly all previous tests of selection (on both gene expression and protein coding regions) (SI Text) require information from neutral sites to assess when neutrality can be rejected, which can lead to incorrect inferences (e.g., interpreting dN/dS > 1 as positive selection when synonymous sites are negatively selected) (13). Many tests also are sensitive to population bottlenecks or assumptions about effective population sizes, mutation rates, etc. Because the present test (like Orr’s; ref. 11) focuses only on the directionality of differences between lineages, it requires no such assumptions. In addition, it is insensitive to biases in the directionality (i.e., up- vs. down-regulation) of both new mutations and the subset of these that become fixed; biases in the directionality of parental alleles (e.g., if A alleles are more often up-regulate genes); and technical biases in the experiment, such as false cis eQTL due to microarray hybridization differences between alleles (SI Text).
Despite its robustness, care must be taken in applying the test and interpreting the results. In addition to independence of the cis and trans eQTL for each gene, no two genes being tested can have their cis and trans eQTL caused by the same polymorphisms (although genes may share one eQTL polymorphism) (SI Text). Trait ascertainment bias is an issue addressed by Orr (11), but not usually applicable to gene expression traits (SI Text). Epistasis between eQTL could pose a problem for our method if extremely strong and widespread, but no epistatic interactions were extreme enough to affect our results for even a single gene (SI Text). Furthermore, although an excess of reinforcing cases does indicate lineage-specific selection, it cannot distinguish whether that lineage-specificity is due to positive selection or relaxed negative selection; to demonstrate adaptive evolution, independent evidence is needed (SI Text). Finally, the test can only detect an enrichment of either reinforcing or opposing cases over the other (e.g., if 20% of genes are subject to positive selection and 15% to stabilizing, the test will only detect the 5% excess of reinforcing cases caused by positive selection). Therefore, it can only provide a conservative lower bound for estimates of genome-wide selection pressures on gene expression.
Applying the Selection Test to Yeast.
We applied the test to gene expression data (14) from 112 haploid segregants of a genetic cross between two strains of the budding yeast Saccharomyces cerevisiae. Sequence divergence suggests the strains (named RM, a wine strain, and BY, a laboratory strain nearly identical to S288C) diverged approximately 107 generations ago, and although they are fully capable of mating with other lineages of S. cerevisiae, they have done so only several hundred times since their divergence (15). Because these therefore represent largely distinct lineages, adaptive differences may have accumulated in each. Previous studies have used these expression data to map thousands of eQTL (14) and also to show that negative selection acts on many cis eQTL (16).
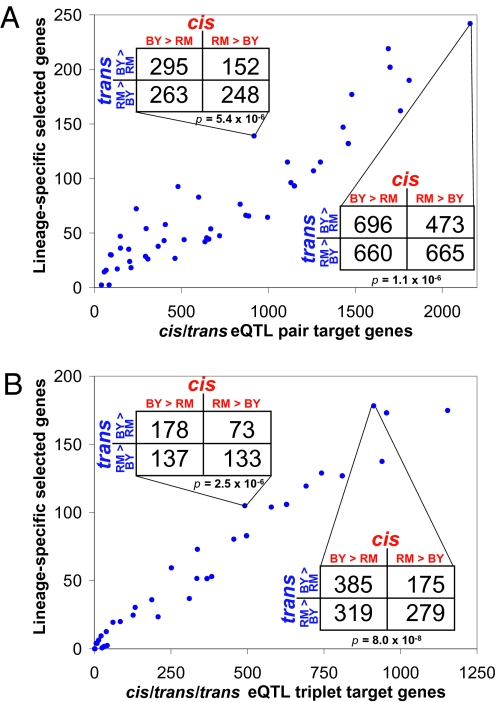
To map eQTL, we computed Pearson correlation coefficients (equivalent to two-sample t tests) between a set of 1,226 nonredundant genetic markers and expression levels of 6,215 genes. We tested a range of correlation cutoffs for the minimum strength of putative cis (defined as being within 50 kb of the gene itself) and trans (defined as being on a separate chromosome from the gene) eQTL. At each cutoff, we calculated the false discovery rate (FDR) by randomizing the genotypes 1,000 times and estimating the number of cis/trans eQTL pairs expected by chance. For each cutoff, a 2 × 2 contingency table was also constructed, as described above. For example, at cis |r| > 0.2 and trans |r| > 0.35 (Fig. 2A, Upper Left Inset), 958 genes were detected at an FDR = 4.1%, and the χ2 test P = 5.4 × 10−6. This significant P value indicates that the null hypothesis of neutrality is rejected for these gene expression levels. To ascertain the validity of this P value, we performed three controls: two different randomization-based tests, in which data were permuted either before or after the eQTL mapping step, and also a negative control of cis/trans eQTL pairs that cannot have been due to selection because the trans eQTL resulted from mutations engineered by experimenters. Our observed excess of reinforcing eQTL pairs fell well outside both randomized distributions, whereas the negative control set did not, demonstrating the absence of unexpected biases in the data (and although it may appear that the deficit of cases is specific to the upper right quadrant, this is not actually the case) (SI Text).
Fig. 2.
Results of selection test in yeast. (A) Applying the selection test on many cis/trans strength cutoffs yielded a nearly linear relationship between the number of true positive eQTL-regulated genes and the excess number of reinforcing cis/trans pairs (lineage-specific selected genes). Cutoffs with up to FDR = 30% were included, although most cutoffs (74%) had FDR < 5%. (Insets) 2 × 2 tables for two cutoffs, including χ2 P values. “RM > BY” indicates that the RM allele at a particular eQTL is associated with higher expression of the target gene. (B) Same as A, for cis/trans/trans triplets. (Insets) Same as A, except that “RM > BY” for trans indicates that RM alleles at both trans eQTL regulating a target gene lead to higher expression.
At each eQTL strength cutoff, we estimated both the number of “true positive” target genes—by subtracting the number expected by chance from that observed—as well as the number of genes subject to lineage-specific selection (i.e., the excess number of genes regulated by reinforcing cis/trans pairs). Plotting these against one another, a nearly linear relationship was observed (Fig. 2A), indicating that highly significant cis/trans eQTL pairs have approximately the same probability of being reinforcing as do weaker pairs. This probability is estimated by the slope of their relationship as 0.10, meaning at least approximately 10% of cis/trans pairs have been subject to lineage-specific selection between these two yeast strains. This number reaches as high as 242 genes in this data set (Fig. 2A, Lower Right Inset), although this is surely an underestimate because the slope shows no signs of saturation and the test will also be rendered conservative by any stabilizing selection.
An extension of the cis/trans test described above is to apply it to genes regulated by three eQTL: one cis and two independent trans. Because a reinforcing triplet is less likely to occur by chance than a reinforcing pair, this may reduce the level of background signal in the test. The appropriate comparison is between reinforcing triplets and triplets where the cis effect opposes both of the trans effects (genes regulated by two trans eQTL with opposite effects are not used in this test) (SI Text). Application of this test to a range of eQTL strength cutoffs resulted in an even more linear relationship than for eQTL pairs and greater significance of the χ2 test (Fig. 2B, Lower Right Inset). Although the requirement for three eQTL per gene decreased the number of genes detected, it increased the slope of the relationship to 0.17, indicating that at least approximately 17% of cis/trans/trans triplets tested have been subject to lineage-specific selection.
Identifying Regulatory Adaptations.
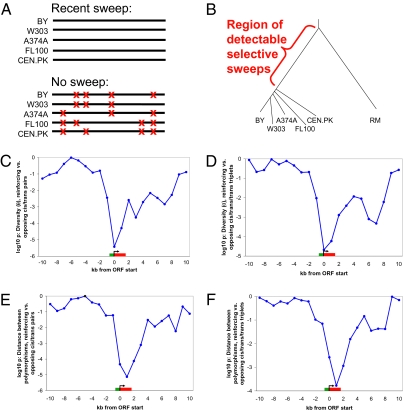
An important question is whether the extensive lineage-specific selection we detected reflects adaptive differences or simply relaxed selection on many genes. To address this, we analyzed a data set of genome-wide polymorphisms from 63 strains of S. cerevisiae (17). It is well established that adaptive evolution results in selective sweeps that leave a clear signature in genetic variation: complete sweeps erase all variation from a region, which is then only slowly reconstituted by new mutations (Fig. 3A). In contrast, relaxed purifying selection—the alternative explanation for the excess of reinforcing eQTLs (SI Text)—would, if anything, result in increased genetic variation. Polymorphism data were available for five closely related laboratory strains, including BY (Fig. 3B). Sweeps that occurred before the divergence of these strains may be detectable as regions of reduced genetic diversity among the five (unfortunately there were too few strains to apply other tests of selection that are based on allele frequency distributions). To compare genetic diversity (referred to here as θ, but also known as π or θπ) to the results from the test of lineage-specific selection, we calculated θ in 2-kb windows centered at each gene coding region’s 5′ end and repeated this every 1 kb for 10 kb in each direction (because S. cerevisiae is thought to lack long-range transcriptional regulation, the vast majority of cis eQTL are expected to be within several hundred bp of the ORF; therefore, this is testing for a signature of reduced genetic diversity at reinforcing cis eQTL). Comparing the distribution of θ values for genes regulated by reinforcing vs. opposing cis/trans pairs, we observed a striking deficit of variation in the reinforcing class, which was strongest in the window centered on each gene’s 5′ end (Fig. 3C). This central window was highly significant (Wilcoxon pw = 3.8 × 10−6), suggesting that many of the cis eQTL among reinforcing pairs have been subject to recent selective sweeps in the laboratory strain lineage. Repeating the θ distribution test for genes regulated by reinforcing vs. opposing eQTL triplets, a similar result was observed (Fig. 3D; pw = 2.1 × 10−5). Performing the same test using polymorphisms from strains far removed from BY and RM, no significant difference in θ was seen in the central window (pw = 0.83; Fig. S2) or any other, indicating that the deficit of variation seen is specific to the relevant strains (and thus unlikely to be caused by any hidden factors, e.g., mutation cold spots, that might lead to a deficit of genetic variation if systematically differing between reinforcing and opposing eQTL targets) (SI Text).
Fig. 3.
Population genetic analysis of lineage-specific selection. (A) The effects of a complete selective sweep on genetic variation. Each red “x” represents a polymorphism. (B) Phylogenetic tree of the five laboratory strains and RM (based on ref. 17). Power to detect sweeps is greatest just before the divergence of the laboratory strains; it drops off sharply after this (because sweeps will affect fewer strains) and more gradually (due to mutation accumulation) before it. Note branch lengths are not precisely to scale. (C) Results of Wilcoxon test in a sliding 2-kb window (step size = 1 kb) on the distribution of θ values near target genes of reinforcing vs. opposing cis/trans eQTL pairs. The average ORF length is shown in red, and approximate promoter length is shown in green. Cutoffs used are cis |r| > 0.2, trans |r| > 0.35 (corresponding to Fig. 2A, Upper Left Inset). (D) Same as C but for targets of cis/trans/trans triplets. Cutoffs used are cis |r| > 0.2, trans |r| > 0.3 (Fig. 2B, Upper Left Inset). (E) Same as C but testing the distribution of distances in between successive laboratory strain polymorphisms. A point was sampled every 1 kb, the distance separating the two closest flanking polymorphisms to that point was calculated, and then compared for reinforcing vs. opposing cis/trans eQTL pairs. (F) Same as E but for targets of cis/trans/trans triplets.
The fraction of reinforcing cis eQTL that have experienced recent selective sweeps in the laboratory strain lineage can be estimated from the distributions of θ values (Fig. S3). At the eQTL cutoffs used above, we found that θ was lower than expected by chance for approximately 78 genes (14.4% of reinforcing pairs), implying the action of selective sweeps in these regions. For eQTL triplets, approximately 56 genes (18.0% of reinforcing triplets) showed similar deficits of variation. At more permissive eQTL cutoffs, over 100 genes had lower θ than expected. Because many sweeps may be too recent (after divergence of the five strains) or too ancient (because of mutation accumulation) to be detected, these are likely substantial underestimates of the extent of sweeps affecting gene expression in the laboratory strain lineage.
Analyzing θ in the two classes of reinforcing eQTL (Fig. 1 and Fig. S1) separately is also informative. In this case, the appropriate set of opposing pairs for comparison is that with the same cis eQTL direction (SI Text). By keeping the cis eQTL direction constant in each comparison, the relationship between trans eQTL direction and θ at each (unlinked) cis eQTL can be studied. The genes regulated by reinforcing eQTL with higher expression in BY showed a clear deficit in θ (pairs pw = 8.3 × 10−5, triplets pw = 2.7 × 10−4), whereas those with lower expression in BY had only a marginally significant deficit (pairs pw = 0.013, triplets pw = 0.031). This suggests that most detectable sweeps of cis-acting variants in the laboratory strain lineage have increased the expression levels of their target genes (SI Text).
Another test of reduced diversity is to measure the distance between successive polymorphisms at different points in the genome; a greater distance is expected in regions with recent sweeps (Fig. 3A). When this distance is often greater than the window size used in the θ distribution test, this test may be a more sensitive method to detect sweeps. Applying this test in the same manner as the θ distribution test, a similar pattern was seen: a greater distance separated polymorphisms near reinforcing cis eQTL than opposing ones for both eQTL pairs (Fig. 3E; pw = 9.7 × 10−6) and triplets (Fig. 3F; pw = 1.2 × 10−4), and maximal significance was observed within the ORFs, adjacent to the central window at each ORF’s 5′ end.
Similar population genetic analyses can be applied to 23 strains (mostly annotated as wine strains) (17) closely related to RM. All four tests (θ and distance test on pairs and triplets) yielded similar results: there was a deficit of variation at the reinforcing target genes, although this was only marginally significant in each case (pw < 0.05 for each) (Fig. S4). Partial genome sequences exist for 10 wine strains (18) (plus a full sequence for RM), and applying the four tests to these data revealed similarly weak but significant deficits of variation at reinforcing cis eQTL (pw < 0.02 for each) (Fig. S5). The number of sweeps detected in the wine strain lineage was approximately 50 (9.2% of reinforcing pairs; for triplets, approximately 33 were detected, or 10.6% of reinforcing triplets). There was no significant difference when analyzing the two classes of reinforcing eQTL (higher vs. lower expression in RM) separately, perhaps because the signal was weak even with the full set. Taken together, these results suggest that either there is less power to detect wine strain sweeps (despite having more genotyped wine strains), or fewer cis eQTL have been subject to positive selection in the wine lineage than in the laboratory lineage. It is possible that a difference in the extent of sweeps is due to effects of artificial selection in the laboratory on the laboratory strains, although this selection would have had to have been extremely intense because the time between the initial “laboratory domestication” and divergence of these strains was short (19).
Multiple Adaptations in the Ergosterol Biosynthesis Pathway.
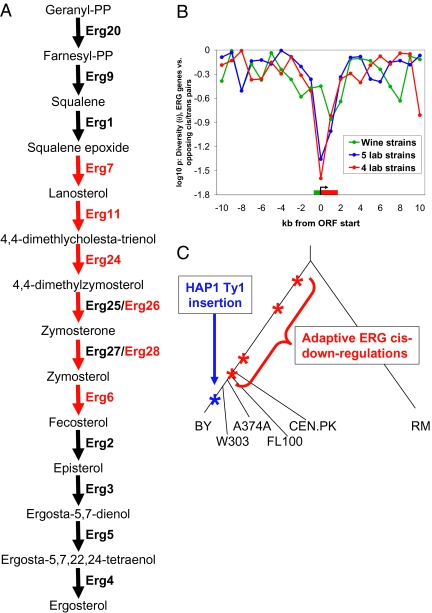
To gain insights into specific adaptations, it is not sufficient to simply know how many gene expression levels have been subject to positive selection; the affected processes must be discovered as well. To this end, we searched the list of genes regulated by reinforcing cis/trans eQTL pairs for enrichments in Gene Ontology annotations. Eight genes from the ergosterol (ERG) biosynthesis pathway passed a stringent cis |r| > 0.2, trans |r| > 0.5 cutoff (FDR < 0.001%); all eight had reinforcing eQTL with lower expression in BY, although only 47 genes in the entire genome satisfied this constraint (approximately 50-fold enrichment; hypergeometric P = 1.7 × 10−12). Remarkably, six of these ERG genes are responsible for a series of reactions clustered in the pathway (Fig. 4A; the other two, ERG8 and HMG1, catalyze steps preceding the reactions shown), suggesting that selection may have specifically targeted successive steps in this pathway.
Fig. 4.
Selection on the ERG pathway. (A) Final steps of the ergosterol biosynthesis pathway. Targets of reinforcing cis/trans pairs are shown in red. Note Erg25 and Erg26 actually catalyze a series of four reactions, shown here collapsed into one arrow, and Erg28 is not an enzyme but an important regulator of the reaction shown. (B) Results of θ distribution test, similar to Fig. 3C except using only the six ERG genes highlighted in A for the targets of reinforcing eQTL. (C) Phylogenetic tree of yeast strains, with the most parsimonious location of the HAP1 Ty1 insertion (blue) and possible times of ERG gene selective sweeps on down-regulating cis-regulatory variants (red). Note that other ERG gene sweeps may also have occurred more recently than shown; polymorphism data from additional laboratory strains will be needed to test this.
Applying the θ distribution test to these six genes, a marginally significant deficit of variation was seen in the five laboratory strains (Fig. 4B, blue line; pw = 0.044), but not the wine strains (Fig. 4B, green line; pw = 0.35), consistent with sweeps having occurred in the laboratory strain lineage. The trans eQTLs for all six genes map to the same marker on chromosome 12, and the likely causal polymorphism underlying this eQTL is already known, a Ty1 retrotransposon insertion in the activation domain of HAP1, a transcription factor known to regulate the ERG pathway. This insertion has been shown to substantially reduce the activity of HAP1 and to alter regulation of the HMG1 and CYC1 promoters (20, 21). Surprisingly, the insertion is quite recent: it is not present in either W303 or CEN.PK (20) (or any other of more than 70 partially sequenced yeast strains) (18), suggesting that it occurred in the BY-specific branch (Fig. 4C). Considering how recently this event occurred, we tested whether excluding CEN.PK (the most diverged laboratory strain) from the θ analysis might increase the power by improving detection of sweeps that occurred after the divergence of this lineage. Although most reinforcing genes lose significance when excluding CEN.PK, the ERG genes became approximately twofold more significant (Fig. 4B, red line; pw = 0.025). In particular, one gene (ERG28) had four CEN.PK-specific polymorphisms but no variants in any of the other four strains, consistent with a sweep occurring after CEN.PK’s divergence. In sum, these results suggest that the expression of multiple ERG genes may have been subject to recent selective sweeps (Fig. 4C). Because these genes all have lower expression in BY than RM, they likely represent cases of adaptive down-regulations—in contrast to the majority of detectable sweeps in the laboratory lineage, which are associated with up-regulation. The selective advantage of lower ERG gene expression is unknown, but could be tied to one or more of the diverse roles that ergosterol plays in a wide range of processes (signal transduction, membrane fluidity, mating, etc.) (22).
It is especially intriguing that the expression levels of so many genes have been subject to positive selection in a species where so few genes show any evidence of positive selection on protein sequences (23–25)—even when the McDonald-Kreitman test (26) was applied to over 1,000 genes in a collection of over 70 strains, no cases of positive selection were identified (18). Although still preliminary, this striking contrast suggests that budding yeast may represent a case where most adaptation has occurred at the level of gene expression—similar to what was proposed for humans and chimpanzees over 30 years ago (2). It will be interesting to apply the test of lineage-specific selection introduced here to detect selection in other species, and—with some modifications—in outbred groups, such as human populations.
Materials and Methods
A Pearson correlation (mathematically equivalent to a 2-sided t test because there are only two genotype classes in haploid yeast) was calculated for each gene-marker pair (coding genotypes as 0/1). For each gene, the strongest correlation in cis (defined as markers within 50 kb of the gene itself; although having a “local” eQTL is not proof of a cis-acting effect, the eQTL need not be cis-acting for the test of selection to be valid, it must only be caused by a polymorphism distinct from the local eQTLs of other genes that share the exact same trans eQTL) and the two strongest trans (defined as being on different chromosomes from the gene itself and from one another) were then recorded. Results from randomization tests are shown in Fig. S6.
Additional methods are explained in SI Text.
Supplementary Material
Acknowledgments
We thank T. Babak, D. Kingsley, M. Feldman, D. Petrov, and J. Plotkin for helpful discussions and advice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912245107/DCSupplemental.
References
- 1.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 2.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 3.Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sucena E, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc Natl Acad Sci USA. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prud’homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 6.Gompel N, et al. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 8.Miller CT, et al. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 10.Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- 11.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Parmley JL, Hurst LD. How common are intragene windows with KA > KS owing to purifying selection on synonymous mutations? J Mol Evol. 2007;64:646–655. doi: 10.1007/s00239-006-0207-7. [DOI] [PubMed] [Google Scholar]
- 14.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruderfer DM, Pratt SC, Seidel HS, Kruglyak L. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet. 2006;38:1077–1081. doi: 10.1038/ng1859. [DOI] [PubMed] [Google Scholar]
- 16.Ronald J, Akey JM. The evolution of gene expression QTL in Saccharomyces cerevisiae. PLoS One. 2007;2:e678. doi: 10.1371/journal.pone.0000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaisne M, Bécam AM, Verdière J, Herbert CJ. A “natural” mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1) Curr Genet. 1999;36:195–200. doi: 10.1007/s002940050490. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, et al. A hap1 mutation in a laboratory strain of Saccharomyces cerevisiae results in decreased expression of ergosterol-related genes and cellular ergosterol content compared to sake yeast. J Biosci Bioeng. 2004;98:159–166. doi: 10.1016/S1389-1723(04)00260-9. [DOI] [PubMed] [Google Scholar]
- 22.Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J Cell Biol. 2008;180:813–826. doi: 10.1083/jcb.200705076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, et al. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doniger SW, et al. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellis M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 26.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.