Abstract
Alternate frame folding (AFF) is a novel mechanism by which allostery can be introduced into a protein where none may have existed previously. We employ this technology to convert the cytotoxic ribonuclease barnase into an artificial zymogen that is activated by HIV-1 protease. The AFF modification entails partial duplication of the polypeptide chain and mutation of a key catalytic residue in one of the duplicated segments. The resulting molecule can fold in one of two “frames” to yield the wild-type structure or a circularly permuted form in which the positions of the N- and C-termini are exchanged with a surface loop. It cannot take on both structures simultaneously because each competes for a shared amino acid sequence. An HIV-1 protease recognition sequence is inserted into one of the surface loops in the nonpermuted frame, and cleavage induces a shift from the nonpermuted fold to the permuted fold. Using the AFF mechanism, we were able to suppress kcat/KM by 250-fold in the proenzyme relative to wild-type barnase. HIV-1 protease cleavage subsequently increases kcat/KM by 130-fold. AFF is significant because it is general and can in principle be used to control activity of many enzymes, including those whose functions are not regulated by any existing mechanism.
Keywords: mutually exclusive folding, design, HIV protease, molecular switch, barnase
Zymogens are inactive enzyme precursors that are activated by proteolytic cleavage. Natural zymogens play essential roles in apoptosis, blood coagulation, digestion, viral maturation, and other cellular processes. Artificial zymogens are of broad interest, with potential applications that include diagnostic tools and toxins that attack pathogens or diseased cells. Here we introduce a unique technique (alternate frame protein folding, or AFF) to convert the cytotoxic ribonuclease barnase (Bn) into an artificial zymogen that is activated by HIV-1 protease (PR). The significance of AFF is that it is general and can thus be applied to many enzymes, including those whose activities are not regulated by any existing mechanism.
Several groups have previously engineered artificial zymogens. Raines and colleagues circularly permuted ribonuclease A (RNase A) and bridged the original N- and C-termini with a peptide linker that contained cleavage sequences for PR, the NS3 protease from hepatitis C, and plasmepsin II from Plasmodium falciparium (1–3). The linkers inhibited enzymatic activity by partially occluding the active site. Cleavage with the respective protease increased kcat/KM by approximately 100-fold. In a recent study we took a related approach by permuting Bn with a short linker peptide intended to force the termini together, thereby distorting the active site or unfolding the enzyme (4). Cleaving the linker with a chemical reagent relieved conformational strain and increased RNase activity. Although these strategies successfully created artificial zymogens, they rely on specific properties of the target enzyme and are therefore not general. Cyclic permutation typically does not result in active site occlusion, nor is proteolytic cleavage guaranteed to remove it. Similarly, N-to-C-terminal distances in many proteins are too short to induce significant conformational strain when linked.
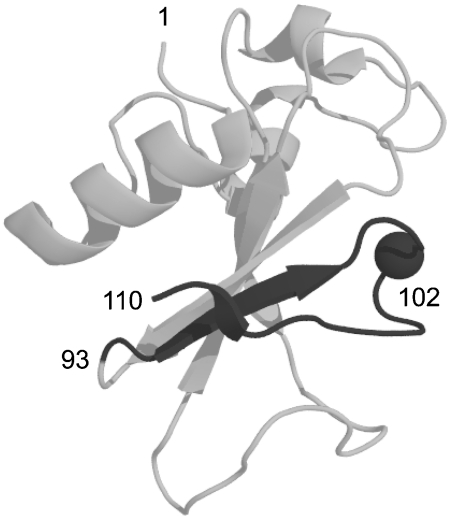
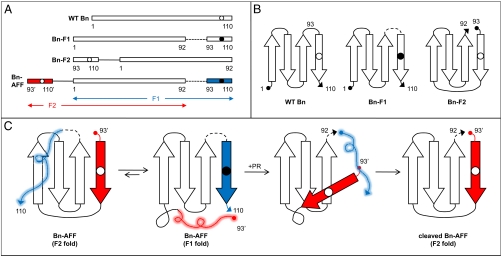
Here we employ the alternate frame folding mechanism to regulate the enzymatic activity of Bn. AFF entails two modifications. First, the segment of polypeptide from a chosen surface loop to the C-terminus of the protein is duplicated and appended to the N-terminus. That stretch of amino acids must contain a key catalytic residue. Once that criterion is satisfied, one can select the surface loop to vary the length of the duplicated segment. His102 is the general acid in the reaction catalyzed by Bn. Thus, to make the duplication as short as possible, we chose the loop immediately N-terminal to His102 (Fig. 1). Amino acids 93–110 were duplicated and fused to the N-terminus using a peptide linker long enough to comfortably span the distance between original N- and C-termini (27 Å). This fusion creates two native conformations (frames) to which the protein can fold. Frame 1 (F1) corresponds to the normal sequence (amino acids 1–110). Frame 2 (F2) is a circularly permuted version of F1. Amino acid sequences of Bn variants are shown in Fig. 2A, and the topological change between F1 and F2 is depicted schematically in Fig. 2B. Using the wild-type (WT) numbering scheme, the F2 sequence begins at Asp93, continues through the linker peptide, and ends at Ser92. New termini are created at the original surface loop, and a new surface loop bridges the original termini. We previously crystallized a similar variant in which Bn was permuted at position 67 and its termini linked by a single amino acid (4). The X-ray structure confirmed that the structure of the permutant is virtually identical to that of WT except for local changes in the immediate vicinity of the new loop and termini.
Fig. 1.
X-ray crystal structure of WT Bn. The dark gray region corresponds to the C-terminal segment of Bn-AFF that is duplicated and fused to the N-terminus. Amino acid positions 1 (N-terminus), 93 (site of circular permutation), 102 (catalytic acid; alpha carbon shown), and 110 (C-terminus) are indicated.
Fig. 2.
Amino acid sequences of Bn variants and mechanism of AFF switching. (A) Amino acid sequences of enzymes created in this study. Numbering is according to the WT Bn sequence; prime superscripts indicate amino acids in the F2 segment of Bn-AFF. Duplicate segments are shown in red and blue. Open and closed circles denote the presence of His102 or Ala102, respectively. Dashed lines represent the PR cleavage site (IFLETS). Solid lines symbolize the flexible linker used to bridge the original N- and C-termini in Bn-F2 and Bn-AFF. (B) Schematic illustrating the topological change that underlies the F1⇆F2 fold shift. F1 and F2 analogs are shown. Dashed line and open/closed circles are the same as in panel A. (C) Schematic of AFF switching mechanism. Wavy lines indicate unfolded regions; colors, dashed line, and open/closed circles are the same as in panel A.
The second modification eliminates catalytic activity of one of the folds but not the other. To do so we mutated His102 to Ala (5) in the F1 sequence but left His102 in place in the F2 sequence. His102 and Ala102 are indicated by open and closed circles, respectively, in Fig. 2. Because F1 and F2 share residues 1–92, folding is mutually exclusive. F1 and F2 are populated according to their thermodynamic stabilities (6) as described by 1, where U is the unfolded state:
| [1] |
Equilibrium is generally expected to favor F1 over F2 because circular permutation is known to destabilize Bn (4) as well as other proteins.
Fig. 2C illustrates how proteolytic cleavage induces a fold shift between the F1 and F2 conformations. An artificial PR cleavage sequence (IFLETS) (7) is inserted into the F1 sequence, at the same surface loop that served as the permutation site (between residues 92–93). The majority of full-length molecules adopts the F1 fold, with the red duplicate segment extending from the N-terminus as an unstructured tail. The enzyme is catalytically inert in this state. A small population of molecules is folded in the F2 conformation, in which the blue duplicate segment continues from the C-terminus as a disordered tail. Proteolytic action by PR cleaves F1 into two fragments. Dissociation of the 93–110 peptide (Blue) triggers the fold shift to the catalytically active F2 conformation. Although the F1 to F2 conversion involves folding of the red peptide and unfolding of the blue peptide, the molecule does not undergo net folding or unfolding. Rather, the active site is remodeled such that His102 becomes presented for catalysis.
A key event in the fold shift is dissociation of the cleaved peptide. One can envision two limiting scenarios in which the shift is governed by thermodynamic principles (stability of the cleaved F1 complex vs. stability of the intact F2 domain) or kinetic considerations (peptide off-rate). Cleaving Bn into two fragments is known to destabilize the molecule substantially and this result is to be expected for other proteins (4, 8). It is therefore likely that the conformational change will typically be under kinetic control at most protein concentrations. As remarked in Discussion, however, a thorough understanding of the AFF mechanism should allow the energetics of the fold shift to be manipulated (e.g. by mutation).
Results
Design and Nomenclature of Bn Variants.
Fig. 2A diagrams the enzymes constructed for this study. The alternate frame folded construct of Bn is designated Bn-AFF. Amino acids are numbered according to the WT Bn sequence. Residues in F2 of Bn-AFF are indicated by prime superscripts but are otherwise numbered identically to the corresponding amino acids in F1. Some variants contain a destabilizing mutation in F1 or F2 to modulate the F1⇆F2 equilibrium. The presence of such a mutation is indicated in parentheses. For example, Bn-AFF(A96) contains the I96A mutation in F1 and Bn-AFF(P94′) contains the W94′P mutation in F2. To mimic the individual components of the switching mechanism in isolation, we constructed analogs of F1 and F2 that lack the duplicated peptides and hence cannot fold shift. Bn-F1 represents F1 and is comprised of WT Bn with the IFLETS sequence (Fig. 2, Dashed Lines) inserted between Ser92–Asp93, and the His to Ala subsitution at position 102. Bn-F2 embodies F2 and consists of Bn circularly permuted at position 93 with a 24-amino acid linker between the original N- and C-termini.
F1 and F2 Analogs.
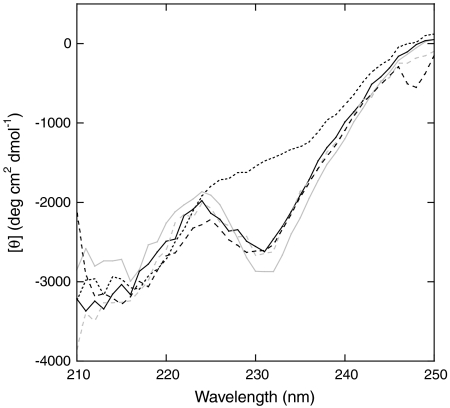
To elucidate the structural and thermodynamic bases of the AFF switching mechanism we first characterized the structures of F1 and F2 analogs by circular dichroism (CD). The WT CD spectrum is noteworthy for its unusually low spectral intensity and an atypical minimum at 231 nm. Fersht and coworkers attributed this minimum to tertiary interactions surrounding Trp94; it is thus a diagnostic feature of the native fold (9). CD spectra of Bn-F1 and Bn-F2 (Fig. 3) retain these characteristics and are very similar to that of WT Bn (9), suggesting that all three proteins adopt similar structures. As expected, Bn-F2 retains WT-like catalytic efficiency whereas Bn-F1 is inactive due to the presence of the H102A mutation (Table 1). Mutation of Trp94 to Pro eliminates the 231 nm minimum, as evident from the spectrum of Bn-F2(P94). However, Bn-F2(P94) is stable and possesses full enzymatic activity, indicating that it is well-folded (Table 1).
Fig. 3.
CD spectra of Bn-AFF variants and their F1/F2 analogs. Lines are as follows: Bn-F1 (Solid Gray), Bn-F2 (Dashed Gray), Bn-F2(P94) (Dotted Black), Bn-AFF(P94 + A96) (Dashed Black), Bn-AFF(P94′) (Solid Black).
Table 1.
Thermodynamic and catalytic parameters of Bn variants. Errors are standard deviations of at least three measurements.
| F1 or F2 analog | ΔGH2O (kcal/mol) | m (kcal/mol/M) | Cm (M) | kcat/KM (× 10-7 M-1 s-1) |
| WT Bn | 8.4 ± 0.8 | 1.8 ± 0.2 | 4.8 ± 0.02 | 10.0 ± 0.19 |
| Bn-F1 | 6.8 ± 0.2 | 2.1 ± 0.06 | 3.2 ± 0.04 | Not detected |
| Bn-F1(P94 + A96) | 2.2 ± 0.2 | 2.1 ± 0.03 | 1.0 ± 0.06 | Not detected |
| Bn-F2 | 5.7 ± 0.3 | 2.2 ± 0.1 | 2.6 ± 0.02 | 5.7 ± 0.1 |
| Bn-F2(P94) | 4.3 ± 0.3 | 2.5 ± 0.1 | 1.7 ± 0.03 | 7.4 ± 0.6 |
| AFF variant |
ΔGH2O (kcal/mol) |
m (kcal/mol/M) |
Cm (M) |
kcat/KM (× 10-7 M-1 s-1) |
| Bn-AFF | 5.3 ± 0.3 | 1.7 ± 0.07 | 3.2 ± 0.03 | Not measured |
| Bn-AFF(P94′) | 7.0 ± 0.3 | 2.1 ± 0.09 | 3.3 ± 0.08 | 0.03 ± 0.06 |
| Bn-AFF(P94′) + PR | Not measured | Not measured | Not measured | 3.9 ± 0.3 |
| Bn-AFF(P94 + A96) | 3.4 ± 0.6 | 1.5 ± 0.2 | 2.2 ± 0.1 | 6.0 ± 0.5 |
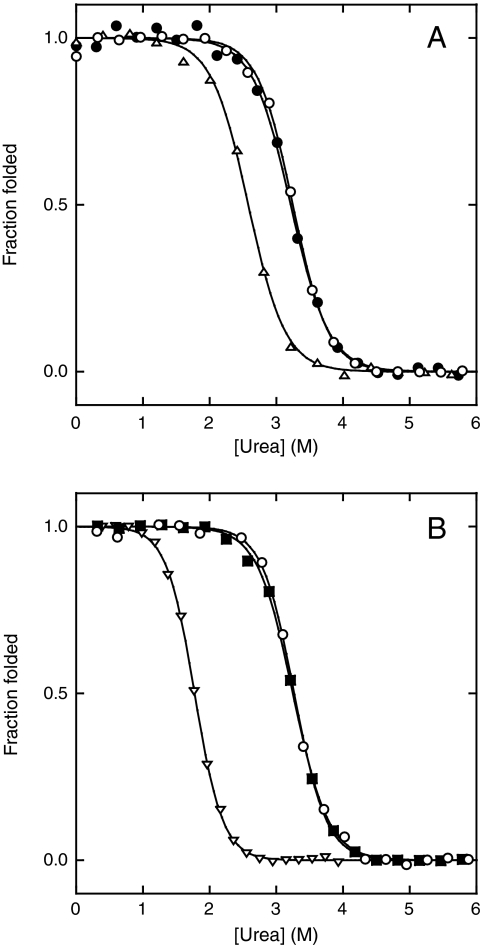
We next determined stabilities of F1 and F2 analogs by equilibrium unfolding experiments. Urea-induced unfolding is monitored by fluorescence of Trp35, Trp71, and Trp94. The maximum emission wavelength shifts from 335 nm to 355 nm upon unfolding and fluorescence decreases robustly below 330 nm (10). We simultaneously fit data from 315–329 nm to the linear extrapolation equation ΔG = ΔGH2O - m[urea], where ΔGH2O is the folding free energy in the absence of denaturant and m is the cooperativity parameter. All Bn variants in this study unfold reversibly and data fit adequately to the two-state equation (Fig. 4). Thermodynamic parameters are summarized in Table 1. Fig. 4A reveals that Bn-F1 and Bn-F2 are both stable with the former being slightly more stable than the latter. The difference in stability can be used to approximate the populations of F1 and F2 in Bn-AFF according to 1. The ΔΔGH2O value of 1.1 kcal/mol predicts that 86% of Bn-AFF molecules will exist in F1 and 14% will populate F2. Bn-F1 and Bn-F2, however, are not perfect analogs of the F1 and F2 conformations of Bn-AFF. The free energy difference between Bn-F1 and Bn-F2 consequently provides only an estimate of the F1 and F2 populations.
Fig. 4.
Urea-induced denaturation of Bn-AFF before stability optimization (A), and after stability optimization (B). Symbols in panel A are: Bn-F1 (Open Circles), Bn-AFF (Closed Circles), Bn-F2 (Triangles). Symbols in panel B are: Bn-F1 (Open Circles), Bn-AFF(P94′) (Closed Squares), Bn-F2(P94) (Inverted Triangles). Lines are best fits of the data to the linear extrapolation equation.
Bn-AFF.
The structure of Bn-AFF is expected to consist of the folded Bn-F1 domain plus an unstructured N-terminal tail (residues 93′–110′), with a minor population of folded Bn-F2 domain plus a disordered C-terminal tail (residues 93–110) (Fig. 2C). We recorded CD spectra to test that hypothesis. The presence of a minimum at 231 nm (Fig. 3) confirms that Bn-AFF(P94′) contains native Bn structure and that it is folded primarily in F1. To determine whether the 17-residue tails are structured or disordered it is necessary to scan below 210 nm, where unstructured peptides display strongly negative ellipticities. Excessive buffer absorbance prevented us from doing so. Nevertheless, Trp fluorescence and limited proteolysis data (vide infra) support the view that the N- and C-terminal tails of Bn-AFF are unstructured.
The AFF mechanism can be tested by comparing the stability of Bn-AFF with those of its isolated frames. The expectation is that the unfolding transition of Bn-AFF will match that of the most stable F1 or F2 analog. The duplicated segment of Bn-AFF contains one extra tryptophan (Trp94′). Unfolding, however, remains well-fit by the linear extrapolation equation (Fig. 4A), indicating that this segment is disordered and that the extra Trp residue contributes only to baseline fluorescence. Inspecting the primary data reveals that the unfolding transition of Bn-AFF is similar to that of Bn-F1 but not to that of Bn-F2 (Fig. 4A). This observation suggests that Bn-AFF is folded predominantly in F1. A more quantitative test is to compare midpoints of denaturation (Cm). Cm is preferable to ΔGH2O in this instance because the presence of the expected approximately 14% F2 population is likely to broaden the observed transition and artificially reduce ΔGH2O. In agreement with the model, the Cm value of Bn-AFF is approximately equal to that of Bn-F1 (Table 1).
“Tuning” Mutations.
We next optimized the switching mechanism by introducing destabilizing mutations into one of the duplicated segments of Bn-AFF but not the other. The purpose of such tuning mutations is to perturb the F1⇆F2 equilibrium. In this way the enzymatic activities of the full-length and cleaved forms can be modulated. For example, to suppress activity of the former to 1% of the latter, Bn-F1 must be more stable than Bn-F2 by RT· ln(100) = 2.7 kcal/mol. We chose W94P as the primary tuning mutation because, in addition to destabilizing Bn, it abolishes the 231 nm CD minimum (Fig. 3) and thus provides a means to establish in which frame Bn-AFF is folded. The W94P mutation reduces stability of Bn-F2 by 1.4 kcal/mol, making it 2.5 kcal/mol less stable than Bn-F1 (Table 1). In contrast, the W94′P mutation does not destabilize Bn-AFF because position 94′ is in the region that is already principally unfolded. Rather, Cm, m-value, and ΔGH2O are slightly higher for Bn-AFF(P94′) than they are for Bn-AFF. These changes are consistent with the view that the W94′P mutation causes nearly all molecules to adopt the F1 conformation. The CD spectrum of Bn-AFF(P94′) (Fig. 3) and the superimposability of the Bn-AFF(P94′) and Bn-F1 urea denaturation curves (Fig. 4B; Table 1) support that conclusion. Most importantly, enzymatic activity of Bn-AFF(P94′) is almost completely suppressed (Table 1). Bn-AFF(P94′) therefore represents a stability-optimized form of the Bn zymogen.
Tuning mutations also provide a robust test of the AFF mechanism. The reaction in 1 predicts that the switch can be driven in the reverse direction by destabilizing F1. The I96A mutation destabilizes WT Bn by 3.17 kcal/mol (11) and Bn-F1(P94 + A96) is 3.5 kcal/mol less stable than Bn-F2 (Table 1). The W94P and I96A mutations are therefore expected to induce Bn-AFF(P94 + A96) to undergo a fold shift from F1 to F2. Three lines of evidence support that conclusion. First, the CD spectrum of Bn-AFF(P94 + A96) shows a pronounced minimum at 231 nm (Fig. 3). Second, the Cm value of Bn-AFF(P94 + A96) (2.2 M) is closer to that of Bn-F2 (2.6 M) then to that of Bn-F1(P94 + A96) (1.0 M) (Table 1). Third, catalytic efficiencies of Bn-AFF(P94 + A96) and Bn-F2 are identical within error, whereas that of Bn-AFF(P94′) is almost immeasurably low (Table 1).
Protease Digestion.
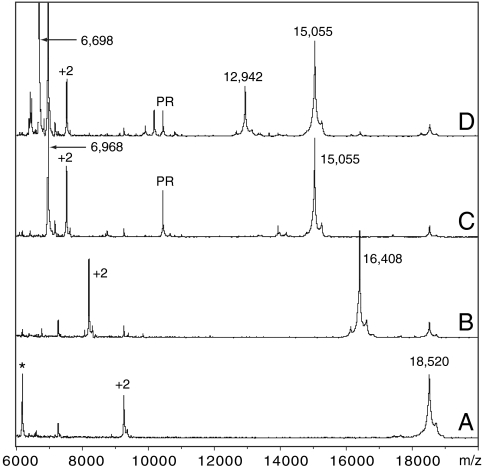
Cleaving Bn-AFF(P94′) (18,520 Da) with PR is predicted to produce fragments of 15,054 Da and 3,484 Da. SDS-PAGE (Fig. S1) and MALDI mass spectrometry (Fig. 5) confirm the expected products. An additional peak at 6,968 Da is observed in MALDI spectra (Fig. 5C). This species appears to be the product of a secondary PR cleavage event. It is relatively minor, however, as it is not detected by SDS-PAGE.
Fig. 5.
Limited trypsin digestion of Bn-AFF(P94′) characterized by mass spectrometry. Peaks marked with an asterisk or +2 indicate an impurity or the +2 charge species of a parent peak, respectively. Samples are as follows: Bn-AFF(P94′) (A), Bn-AFF(P94′) + trypsin (B), PR-digested Bn-AFF(P94′) (C), PR-digested Bn-AFF(P94′) + trypsin (D).
To test whether PR cleavage reverses the F1⇆F2 equilibrium to favor F2, we asked whether the F2 analog of Bn-AFF(P94′) is more stable than the PR-cleaved F1 analog. Thermal-induced denaturation curves, monitored by circular dichroism and fluorescence, are shown in Fig. S2. Bn-F1 and Bn-F2(P94) exhibit melting temperatures (Tm) of 46.5 °C and 39.5 °C, respectively. Cleaving Bn-F1 lowers its Tm to 34.0 °C (measured at 1 μM enzyme concentration) and to 35.0 °C at 10 μM concentration. This shift suggests that F2 is more stable than cleaved F1 at all protein concentrations employed, and that PR processing will likely induce the F1 to F2 fold shift.
As an independent structural test of the switching mechanism we characterized the PR-cleaved and uncleaved states of Bn-AFF(P94′) by partial trypsin digestion. If the N-terminal tail of Bn-AFF(P94′) is unstructured, then trypsin is expected to cleave exposed sites in the tail (Lys98′ and Arg108′; Fig. 2C, Wavy Red Lines) prior to proteolyzing the folded F1 domain. One major cleavage product is observed after 20 m of digestion (Fig. 5B). The mass-to-charge ratio (16,408 m/z) confirms that the major species is the intact F1 domain with an N-terminal tail truncated at Arg108′ (theoretical MW = 16,408 m/z). If PR processing induces the fold shift as envisioned, then Lys98′ and Arg108′ will be protected from trypsin. Trypsin will instead preferentially attack Lys98 and Arg108 in the dissociated peptide (Fig. 2C, Wavy Blue Lines). The 15,055 m/z PR-digested fragment is relatively resistant to trypsin cleavage, as expected for the folded F2 domain, but additional peaks at 12,942 m/z and 6,698 m/z are observed (Fig. 5D). The 12,942 m/z peak likely arises from a minor population of PR-cleaved Bn-AFF(P94′) in which the blue peptide (Fig. 2C) has not dissociated. This species would persist in conformation F1 and as such would be cleaved by trypsin at Arg108′, yielding a fragment of 12,942 Da (theoretical MW). The identity of the 6,698 m/z peak is not known; it may represent trypsin digestion of one of the minor species. The relevant point is that cleavage at Lys98′ or Arg108′ is prevented, indicating that these residues have become shielded by the folded F2 domain. These data provide evidence that the F1 to F2 conformational change involves the folding and unfolding events depicted in Fig. 2C.
Bn-AFF(P94′): An Artificial Zymogen.
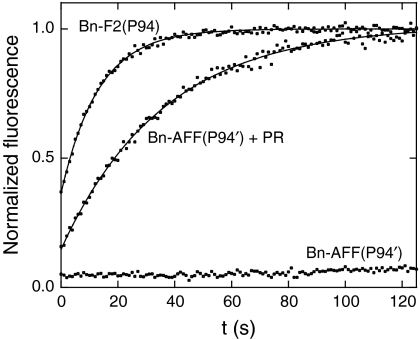
Fig. 6 shows that the AFF mechanism suppresses the rate of RNA hydrolysis by Bn-AFF(P94′) nearly to background levels. kcat/KM for the substrate 5′ 6-FAM-ArGAA -3′ TAMRA (12) is inhibited by 250-fold relative to the F2 analog Bn-F2(P94) (Table 1). This figure is in reasonable agreement with the F2 population calculated from thermodynamic data. PR cleavage increases kcat/KM by 130-fold, almost fully restoring RNase activity. This result suggests that the PR-induced fold shift from F1 to F2 is nearly complete. One reason for why activity is not completely restored may be that the cleaved peptide does not dissociate completely. It is also possible that PR cleaves the active enzyme at a second, uncharacterized site (Fig. 5). Nevertheless, the 130-fold increase in catalytic efficiency is comparable to or greater than that of some natural zymogens such as caspases (13) and serine proteases (14).
Fig. 6.
Hydrolysis of the fluorogenic RNA substrate 5′ 6-FAM-ArGAA-3′TAMRA by Bn-AFF(P94′). Lines are best fits of the data to equations in ref. 12.
Discussion
In this study we demonstrate that catalytic activity of an enzyme can be effectively regulated by the AFF mechanism. The unique advantage of AFF is that it can potentially be applied to any enzyme, with the exception of those that contain disulfide cross links that could prevent the F1 to F2 transition. The validity of this assertion depends on two factors: Whether the target protein is amenable to the requisite modifications (permutation and partial sequence duplication), and whether the switching mechanism can be controlled by known thermodynamic and kinetic considerations. With respect to the first point, proteins including GFP, SH3 domains, PDZ domains, antibody light chains, lysozyme, calbindin, and many others have been shown to retain structure and function after circular permutation (6, 15–21). A useful feature of AFF is that the length of the duplicated segment can be adjusted to minimize potential misfolding, degradation, or solubility problems. The main requirement is that the duplicated region must contain at least one key catalytic residue. Beyond that, length can be chosen to pinpoint the permutation site to a particular surface loop or reduce the length of the tails. As part of this study, we created an additional AFF variant in which amino acids 67–110 were duplicated and joined to the N-terminus. This variant was similar to Bn-AFF in its conformational switching behavior, but the additional duplicated sequence uncovered a cryptic PR cleavage site centered at Trp94. These experiments are detailed in SI Text. The cryptic site exposes a potential complication with our approach, but the fact that we were able to eliminate secondary cleavage by shifting the permutation site to position 93 demonstrates the flexibility inherent in the AFF design.
It should be further noted that the replicated peptide can be appended to either the N-terminus or the C-terminus of the parent protein to minimize overall length of the chimera. A key catalytic residue of Bn (His102) is near the C-terminus; thus, we fused the C-terminal peptide to the N-terminus. For enzymes with catalytic residues near the N-terminus it may be advantageous to duplicate the N-terminal region and append it to the C-terminus.
To engineer the switching mechanism for optimal response it is necessary to establish the extent to which it is governed by known thermodynamic and kinetic principles. For toxic enzymes such as Bn, it is especially important to be able to modulate the F1⇆F2 equilibrium to reduce unwanted lethality. Available evidence indicates that this equilibrium is under thermodynamic control. When mutations known to destabilize Bn are placed in F1 or F2 of Bn-AFF, fluorescence and CD experiments find that the populations are perturbed in the expected manner. PR cleavage of Bn-F1 destabilizes it significantly and cleaving Bn-AFF(P94′) induces the F1 to F2 fold shift. This structural rearrangement is reflected by a 130-fold increase in catalytic efficiency. Together, these data suggest that the conformational equilibrium can be controlled and fine-tuned in a rational manner. Whereas tuning mutations can often achieve the desired balance, a limitation with this approach is that known mutations are typically destabilizing rather that stabilizing. Accordingly, the extent to which the switching mechanism can be manipulated can be constrained by global stability in the case of relatively unstable proteins.
It is reasonable to surmise that the rate of the fold shift is limited by dissociation of the cleaved peptide. This rate is relatively fast in the case of Bn, as RNase activity is detected immediately after PR cleavage. For highly stable enzymes, however, the dissociation rate may be inordinately slow. In such cases one can introduce a destabilizing mutation into the region shared by F1 and F2. This mutation should destabilize F1 and F2 to similar extents and thus increase the rate of interconversion without affecting the equilibrium distribution.
We previously designed an optical biosensor by using AFF to couple ligand binding to a fluorescence-detected conformational change (6). The present work demonstrates that a similar conformational change can be triggered by proteolytic cleavage. This study paves the way for future designs that convert protein toxins into zymogens, to be activated by pathogens expressing unique proteases.
Materials and Methods
Protein Construction and Purification.
Bn-AFF and Bn-F2 contain a flexible peptide linker (GSGAGSGAGSGAGSGAGSGAGSGT) between the duplicated sequence and the parent sequence to maintain stability of the permuted fold. The synthetic PR cleavage sequence IFLETS was inserted between Ser92-Asp93 of Bn-AFF and Bn-F1. To ensure full access to PR, we flanked the IFLETS sequence with GGSGSGSG at its N-terminus and LGTGAGSS at its C-terminus. All variants were expressed in Escherichia coli and purified as full-length, nondegraded proteins. Protein expression and protein purification are described in SI Text.
Spectroscopic Measurements.
Samples were prepared by first dissolving lyophilized protein in 3 M guanidine hydrochloride to dissociate any aggregates. Proteins were then refolded by rapid dilution (typically 50–200-fold) into 10 mM sodium acetate (pH 5.0), 0.1 M NaCl. Protein concentration used in fluorescence and CD experiments was 1 μM and 5 μM, respectively. Data were collected on a Horiba/Jovin Yvon Fluoromax-3 fluorimeter and an Aviv Model 202 CD spectrometer. Experiments were performed at 20 °C. Samples for urea denaturation experiments were prepared as described (22). Fluorescence spectra were fit using the global fitting package of IGOR PRO (Wavemetrics). Wavelengths from 315 nm–329 nm were fit simultaneously using linked ΔGH2O and m values.
PR Digestion and Enzymatic Assays.
Samples (5–10 μM Bn variants) were digested for 2 h in the presence of 1/50 molar ratio of PR in 25 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8), 0.1 M NaCl, 1 mM EDTA. Cleavage took place at 25 °C because digestion at 37 °C resulted in significant degradation of Bn-AFF. Digestion was terminated by addition of 1 μM Ritonavir (National Insitutes of Health AIDS Research and Reference Reagent Program). RNase assays were performed at 37 °C in the same buffer supplemented with 0.1 mg/mL BSA. Enzyme assays contained 1–2 nM enzyme and 25 nM substrate (5′ 6-FAM-ArGAA-3′TAMRA, Integrated DNA Technologies) (12). Hydrolysis was monitored by the increase in fluorescence intensity at 515 nm with excitation at 490 nm. kcat/KM values were calculated by fitting the data to equations in ref. 12. Uncleaved Bn-AFF(P94′) hydrolyzes the substrate too slowly to achieve completion; consequently, kcat/KM was determined by increasing the enzyme concentration and fitting the initial velocities to equations in ref. 12.
Trypsin Digests and Mass Spectrometry.
Bn-AFF(P94′) (5 μM) was digested with Trypsin Gold (Promega) (0.1 μM) for 20 m at 25 °C. Proteolysis was quenched by lowering pH to 2.5 with acetic acid. Samples were analyzed on a Bruker Autoflex III MALDI-TOF mass spectrometer.
Supplementary Material
Acknowledgments.
We thank C. Schiffer for the gift of purified HIV-1 protease and J.-H. Ha for discussions. This work was supported by National Institutes of Health Grant GM069755 (S.N.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907668107/DCSupplemental.
References
- 1.Plainkum P, Fuchs SM, Wiyakrutta S, Raines RT. Creation of a zymogen. Nat Struct Biol. 2003;10:115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turcotte RF, Raines RT. Design and characterization of an HIV-specific ribonuclease zymogen. AIDS Res Hum Retro. 2008;24:1357–1363. doi: 10.1089/aid.2008.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson RJ, Lin SR, Raines RT. A ribonuclease zymogen activated by the NS3 protease of the hepatitis C virus. FEBS J. 2006;273:5457–5465. doi: 10.1111/j.1742-4658.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- 4.Butler JS, Mitrea DM, Mitrousis G, Cingolani G, Loh SN. Structural and thermodynamic analysis of a conformationally-strained circular permutant of barnase. Biochemistry. 2009;48:3497–3507. doi: 10.1021/bi900039e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mossakowska DE, Nyberg K, Fersht AR. Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquifaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry. 1989;28:3843–3850. doi: 10.1021/bi00435a033. [DOI] [PubMed] [Google Scholar]
- 6.Stratton MM, Mitrea DM, Loh SN. A Ca2+-sensing molecular switch based on alternate frame protein folding. ACS Chem Biol. 2008;3:723–732. doi: 10.1021/cb800177f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck ZQ, Hervio L, Dawson PE, Elder JH, Madison EL. Identification of efficiently cleaved substrates for HIV-1 protease using a phage display library and use in inhibitor development. Virology. 2000;274:391–401. doi: 10.1006/viro.2000.0420. [DOI] [PubMed] [Google Scholar]
- 8.Neira JL, Vázquez E, Fersht AR. Stability and folding of the protein complexes of barnase. Eur J Biochem. 2000;267:2859–2870. doi: 10.1046/j.1432-1327.2000.01290.x. [DOI] [PubMed] [Google Scholar]
- 9.Vuilleumier S, Sancho J, Loewenthal R, Fersht AR. Circular dichroism studies of barnase and its mutants: Characterization of the contribution of aromatic side chains. Biochemistry. 1993;32:10303–10313. doi: 10.1021/bi00090a005. [DOI] [PubMed] [Google Scholar]
- 10.Ha J-H, Butler JS, Mitrea DM, Loh SN. Modular enzyme design: Regulation by mutually exclusive protein folding. J Mol Biol. 2006;357:1058–1062. doi: 10.1016/j.jmb.2006.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrano L, Kellis JTJ, Cann P, Matouschek A, Fersht AR. The folding of an enzyme II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 12.Park C, et al. Fast, facile, hypersensitive assays for ribonucleolytic activity. Methods Enzymol. 2001;341:81–94. doi: 10.1016/s0076-6879(01)41146-3. [DOI] [PubMed] [Google Scholar]
- 13.Elliott JM, Rouge L, Wiesmann C, Scheer JM. Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation. J Biol Chem. 2009;284(10):6546–6553. doi: 10.1074/jbc.M806121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonsdale-Eccles JD, Neurath H, Walsh KA. Probes of the mechanism of zymogen catalysis. Biochemistry. 1978;17:2805–2809. doi: 10.1021/bi00607a016. [DOI] [PubMed] [Google Scholar]
- 15.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent protein. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cellitti J, et al. Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment. Protein Sci. 2007;16(5):842–851. doi: 10.1110/ps.062628607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Jiminez R, Garcia-Manyes S, Ainavarapu SRK, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J Biol Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- 18.Viguera AR, Serrano L, Wilmanns M. Different folding transition states may result in the same native structure. Nat Struct Biol. 1996;3:874–880. doi: 10.1038/nsb1096-874. [DOI] [PubMed] [Google Scholar]
- 19.Brinkmann U, et al. Stabilization of a recombinant Fv fragment by base-loop interconnection and VH-VL permutation. J Mol Biol. 1997;268:107–117. doi: 10.1006/jmbi.1996.0850. [DOI] [PubMed] [Google Scholar]
- 20.Haglund E, Lindberg MO, Oliveberg M. Changes of protein-folding pathways by circular permutation: Overlapping nuclei promote global cooperativity. J Biol Chem. 2008;283:27904–27915. doi: 10.1074/jbc.M801776200. [DOI] [PubMed] [Google Scholar]
- 21.Buchwalder A, Szadkowski H, Kirschner K. A fully active variant of dihydrofolate reductase with a circularly permuted sequence. Biochemistry. 1992;31:1621–1630. doi: 10.1021/bi00121a006. [DOI] [PubMed] [Google Scholar]
- 22.Radley TL, Markowska AI, Bettinger BT, Ha J-H, Loh SN. Allosteric switching by mutually exclusive folding of protein domains. J Mol Biol. 2003;332:529–536. doi: 10.1016/s0022-2836(03)00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.