Abstract
The mammalian innate immune system uses Toll-like receptors (TLRs) and Nod-LRRs (NLRs) to detect microbial components during infection. Often these molecules work in concert; for example, the TLRs can stimulate the production of the proforms of the cytokines IL-1β and IL-18, whereas certain NLRs trigger their subsequent proteolytic processing via caspase 1. Gram-negative bacteria use type III secretion systems (T3SS) to deliver virulence factors to the cytosol of host cells, where they modulate cell physiology to favor the pathogen. We show here that NLRC4/Ipaf detects the basal body rod component of the T3SS apparatus (rod protein) from S. typhimurium (PrgJ), Burkholderia pseudomallei (BsaK), Escherichia coli (EprJ and EscI), Shigella flexneri (MxiI), and Pseudomonas aeruginosa (PscI). These rod proteins share a sequence motif that is essential for detection by NLRC4; a similar motif is found in flagellin that is also detected by NLRC4. S. typhimurium has two T3SS: Salmonella pathogenicity island-1 (SPI1), which encodes the rod protein PrgJ, and SPI2, which encodes the rod protein SsaI. Although PrgJ is detected by NLRC4, SsaI is not, and this evasion is required for virulence in mice. The detection of a conserved component of the T3SS apparatus enables innate immune responses to virulent bacteria through a single pathway, a strategy that is divergent from that used by plants in which multiple NB-LRR proteins are used to detect T3SS effectors or their effects on cells. Furthermore, the specific detection of the virulence machinery permits the discrimination between pathogenic and nonpathogenic bacteria.
Keywords: caspase 1, inflammation, type III secretion, IL-1beta
Inflammation is required for effective host defense, but uncontrolled, results in disease. It is therefore critical that the innate immune system titrate its response to match the pathogenic potential of the infecting microorganism. An appropriate response is largely modulated by the selective secretion of cytokines and chemokines. These immune regulators are often produced and secreted in response to activated pattern recognition receptors (PRRs) such as the TLRs and NLRs (1 –3). However, in the case of IL-1β and IL-18 the secretion is regulated by a two-step process. TLR signaling first induces the transcription and translation of proforms of these cytokines. Certain NLRs then regulate the proteolytic maturation of proIL-1β and proIL-18 via activation of the caspase 1 inflammasome; only the processed cytokines are secreted and biologically active (2). This two-step regulation is one mechanism whereby macrophages discriminate between bacteria that encode virulence factors and bacteria that do not.
Many Gram-negative pathogens encode type III secretion systems (T3SS) with conserved structural features that promote virulence by injecting bacterial effector proteins directly into the cytosol of host cells (4). These effector proteins reprogram host cell physiology to benefit the specific pathogen and are generally much less conserved between pathogens than the T3SS apparatus. It would therefore be advantageous to the host to directly detect the T3SS, thereby circumventing the need to respond to the injected virulence factors on a case by case basis. We show here that macrophages detect a component of the T3SS apparatus that forms the periplasm-spanning rod. In response to the presence of rod proteins from many T3SS expressing bacteria within the cytosol, macrophages activate caspase 1 through NLRC4 and secrete IL-1β. Such a detection system has not previously been demonstrated.
Results and Discussion
SPI1 T3SS Rod Protein PrgJ Activates Caspase 1 Through NLRC4.
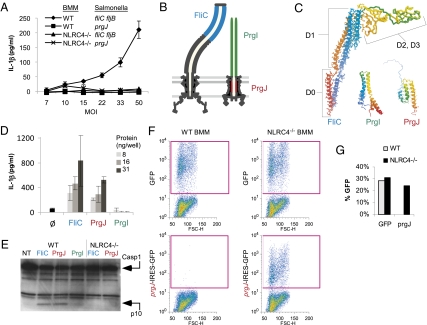
We and others have previously shown that cytosolic flagellin activates caspase 1 via NLRC4/Ipaf (3, 5 –13) (Fig. S1). In the case of S. typhimurium, flagellin is promiscuously secreted by the SPI1 T3SS because of conservation between the flagellar apparatus and the virulence-associated secretion system (14) (Fig. S2). We also observed activation of IL-1β secretion in bone marrow–derived macrophages (BMM) that was flagellin independent (15). We found that fliC fljB mutants, which carry mutations in both flagellin genes, as well as flgB mutants, which do not express flagellin monomers because of translational repression by FlgM, were detected by macrophages that secreted IL-1β in response (5) (Fig. 1A and Fig. S3A). Detection of these strains by macrophages requires bacterial expression of prgJ, which encodes an essential component of the SPI1 T3SS (5) (Fig. 1A and Fig. S3A), and signaling through NLRC4 (Fig. 1A). The observed IL-1β secretion cannot be accounted for by differential bacterial uptake by macrophages (Fig. S3B). Flagellin (FliC) polymerizes into a hollow tube that forms the flagellar filament (Fig. 1B); PrgJ and PrgI similarly polymerize into a hollow tube that form the rod and needle of the SPI1 T3SS (Fig. 1B) (16, 17). To further compare these proteins at a structural level, we examined published three-dimensional structures for FliC and PrgI (18, 19), and used two different approaches to predict the structure of PrgJ (we used our structure prediction pipeline (20, 21) as well as the 3D-jury fold recognition server (22) (Methods). The tertiary structure of FliC, PrgI, and our predicted structure of PrgJ, all contain a similar simple fold dominated by two long helices (Fig. 1C). Furthermore, like FliC, both PrgI and PrgJ are secreted by the SPI1 T3SS (23, 24). We therefore examined PrgI and PrgJ for their capacity to induce IL-1β secretion. Delivery of purified PrgJ protein, but not PrgI protein, to the macrophage cytosol via protein transfection triggered rapid caspase 1 processing and secretion of IL-1β, similar to that induced by flagellin (Fig. 1 D and E). To verify that PrgJ induces caspase 1 activation, and to avoid potential complications from contaminants in purified protein preparations, we used a retroviral lethality screen (25) as an alternative method of delivering heterologous proteins to the macrophage cytosol. This assay takes advantage of the fact that NLRC4-dependent caspase 1 activation in BMM results in pyroptosis (26), a rapid nonapoptotic form of programmed cell death. Retroviruses encoding control GFP, prgJ-IRES-GFP, or fliC-IRES-GFP were transduced into WT and NLRC4 null macrophages. Comparable GFP expression was observed in WT and NLRC4 null macrophages transduced with control GFP (Fig. 1 F and G). In contrast, prgJ-IRES-GFP (Fig. 1 F and G) and fliC-IRES-GFP–expressing cells (Fig. S3C) could be recovered only from NLRC4 null transductions. The inability to recover WT cells expressing prgJ or flagellin is consistent with NLRC4-dependent caspase 1 activation and subsequent cell death by pyroptosis. This experiment demonstrates both that the prgJ transgene is detected and induces cell death, and that this cell death is strictly dependent upon NLRC4 signaling.
Fig. 1.
PrgJ activates NLRC4. (A) WT or NLRC4−/− BMM were primed with LPS for 4 h, then infected with flagellin mutant (fliC fljB) or SPI1 mutant (prgJ) S. typhimurium strain under SPI1-inducing conditions at the indicated multiplicity of infection (MOI) for 2 h and IL-1β secretion determined by ELISA. (B) Cross-sectional schematic of structure of flagellar and SPI1 T3SS, with location of flagellar filament (FliC, blue), needle (PrgI, green), and internal rod (PrgJ, red) indicated. (C) Published structures of FliC (1UCU) and PrgI (2JOW) are presented in comparison with our predicted structure of PrgJ. The N-terminal and C-terminal regions of PrgI are not well resolved; the top three models from the published data are shown superimposed. The N-terminal region of PrgJ is not well resolved and is indicated only for reference. (D) Quantities of 8, 16, or 31 ng/well purified FliC, PrgI, or PrgJ or no protein control (Ø) were delivered to the cytosol of LPS primed BMM using a protein transfection reagent (Profect P1). IL-1β secretion was determined by ELISA 1 h later. Standard deviations are indicated. (E) BMM from WT or NLRC4 null mice were transfected with purified FliC, PrgJ, or PrgI protein for 1 h, and caspase 1 processing was determined by Western blot. (F) To perform a retroviral lethality screen, BMM were infected with transgenic retroviruses expressing GFP alone, or prgJ followed by an IRES-GFP element. GFP positive cells were identified by flow cytometry 2 days after infection. (G) Quantitation of F.
PrgJ and Flagellin Share Amino Acid Motif Critical for Caspase 1 Activation.
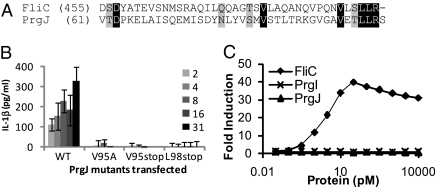
Because PrgJ and FliC are both protein activators of NLRC4, we examined whether they share structural features, and found identity in the carboxyl-terminal 7 amino acids (Fig. 2A) that was not shared by PrgI. To determine whether this sequence is required for NLRC4 activation in response to PrgJ, we used deletional mutagenesis and point mutations to probe the contribution of this motif to NLRC4 activation. Truncations within the last seven amino acids of PrgJ abrogated IL-1β secretion by macrophages in response to transfection of purified protein (Fig. 2B), and truncated PrgJ did not activate NLRC4 in the retroviral lethality screen (Fig. 3B). These results demonstrate that this region of the protein is necessary for activation of NLRC4. Point mutations within this domain identified V95 as the critical residue for PrgJ recognition (Fig. 2B). Unbiased as well as directed mutational analysis of FliC also identified the carboxy terminal region of identity and the homologous valine as critical for flagellin recognition by NLRC4 (25) (Fig. S4). Importantly, in contrast to flagellin, which is recognized by both TLR5 and NLRC4, PrgJ activates only NLRC4 (Fig. 2C). This result implies that NLRC4 and TLR5 are activated by different determinants of flagellin. We propose that these conserved residues are critical to the function of both FliC and PrgJ and thus are not easily mutated to enable NLRC4 evasion. It is interesting that PrgJ and FliC share structural features that are detected by NLRC4, but that although they both form parts of the hollow tube comprising the T3SS export apparatus, they are located in different places. Other proteins that also form parts of this tube, including the flagellar hook (FlgE) and the SPI1 needle (PrgI) do not contain these sequences and are not detected.
Fig. 2.
PrgJ and FliC share sequence identity within an NLRC4 activating motif. (A) Alignment of FliC and PrgJ starting at the indicated amino acid through the carboxy terminal residue. Identity (black) and similarity (gray) are indicated. (B) 6xHis-tagged PrgJ or protein carrying the indicated mutations were purified and delivered to the cytosol of LPS primed BMM. IL-1β secretion was determined 1 h later. (C) CHO cells expressing TLR5 were stimulated with the indicated protein and expression of firefly luciferase under the control of NF-κB was measured.
Fig 3.
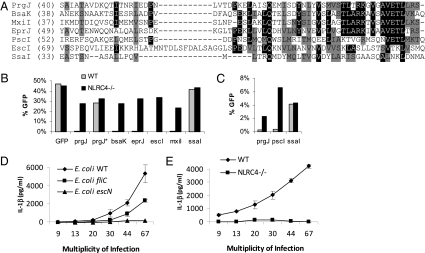
PrgJ homologs activate NLRC4. (A) Alignment of the carboxy terminal regions of PrgJ (S. typhimurium SPI1), BsaK (B. pseudomallei), MxiI (S. flexneri), EprJ (EHEC), PscI (P. aeruginosa), EscI (EHEC and EPEC), and SsaI (S. typhimurium SPI2) from the indicated amino acid residue through the carboxy terminal residue. Residues identical in four of the seven sequences (black) or similar residues (gray) are indicated. (B and C) Retrovirus lethality screen as described in Fig. 1; percentage of GFP-positive cells was determined by flow cytometry. (D and E) WT or NLRC4 null BMM were infected with WT, flagellin (fliC), or T3SS (escN) mutant strains of EPEC at the indicated MOI for 1 h. Gentamicin was added to the media, and the infection was continued for 3 h. IL-1β secretion was determined by ELISA. Standard deviations are shown.
T3SS Rod Proteins Are Broadly Detected Through NLRC4.
PrgJ homologs are found in most T3SS and fall into four large clades (Fig. S5). We examined several homologs of PrgJ, including BsaK (Burkholderia pseudomallei), EprJ (enterohemorrhagic Escherichia coli, EHEC), EscI (present in both EHEC, which encodes two T3SS, and enteropathogenic E. coli, EPEC), MxiI (Shigella flexneri), and PscI (Pseudomonas aeruginosa), which share varying sequence similarity to PrgJ (Fig. 3A). By using the retroviral lethality screen, we determined that BsaK, EprJ, EscI, MxiI, and PscI all activated NLRC4 (Fig. 3 B and C). S. flexneri and P. aeruginosa have previously been shown to induce NLRC4 dependent caspase 1 activation in macrophages independent of flagellin (10, 27), although the mechanism was not defined. Our results suggest that this detection is via their respective rod proteins, MxiI in S. flexneri, and PscI in P. aeruginosa.
Caspase 1 Activation in Response to EPEC.
Our finding that EscI is detected by macrophages implied that NLRC4 can promote IL-1β secretion in response to the EPEC T3SS encoded by the locus of enterocyte effacement (LEE). We therefore infected macrophages with wild-type EPEC or strains carrying mutations in flagellin (fliC) or the T3SS (escN, which ablates all T3SS translocation activity). In agreement with our S. typhimurium results, IL-1β secretion was triggered by both a flagellin-dependent and a flagellin-independent pathway (Fig. 3D); both pathways required expression of NLRC4 in macrophages and T3SS in bacteria (Fig. 3D and E). Thus, NLRC4 responds to T3SS activity via the two conserved protein agonists: flagellin and the rod protein of the T3SS apparatus. To date, direct interactions between NLRC4 and flagellin or rod proteins have not been demonstrated suggesting the possibility of an unidentified coreceptor. This possibility is also likely to apply to NLRP3 signaling in that multiple agonists activate NLRP3 but direct interactions have not been demonstrated (2).
S. typhimurium SPI2 T3SS Rod Protein SsaI Is Not Detected by NLRC4.
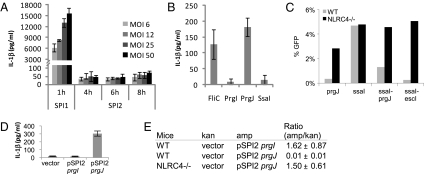
In addition to SPI1, S. typhimurium also encodes a second T3SS, SPI2, which promotes a different aspect of virulence (28). Interestingly, whereas S. typhimurium expressing SPI1 activated IL-1β secretion in macrophages, SPI2 expressing bacteria did not (Fig. 4A), and this was not due to increased entry of SPI1 induced bacteria (Fig. S3D). We hypothesized that this lack of activation could originate from mutation of SsaI, the PrgJ homolog in SPI2 T3SS (Fig. S6), and indeed, SsaI did not activate NLRC4 in protein transfection (Fig. 4B) or retroviral lethality experiments (Fig. 3B). There are several amino acid substitutions between SsaI and other detected rod proteins; notably, the essential valine residue within PrgJ is substituted for a leucine in SsaI (Fig. 3A). We examined the ability of NLRC4 to detect proteins in which the carboxyl-terminal eight amino acid residues of SsaI (-LNKLDNMA) were swapped with the homologous residues from PrgJ (-VETLLRS) or EscI (-VTKLVSMQ), and found that both of the hybrid proteins were detected by NLRC4 in the retroviral lethality screen (Fig. 4C). This suggests that the lack of NLRC4 detection can be mapped to amino acid substitutions within the carboxy-terimal eight residues of SsaI. Interestingly, Helicobacter pylori flagellin contains mutations that confer TLR5 evasion, however, compensatory mutations elsewhere within the flagellin protein are required to restore the ability of the protein to polymerize (29).
Fig. 4.
Lack of detection of SPI2 T3SS by NLRC4 is beneficial for S. typhimurium. (A) BMM were infected with S. typhimurium grown under SPI1 or SPI2 inducing conditions for the indicated time and MOI before IL-1β secretion was determined. (B) A 63-ng quantity of the indicated purified proteins was delivered to LPS primed BMM by protein transfection, and IL-1β secretion was determined. (C) Retroviral lethality screen was performed as in Fig. 3 for prgJ, ssaI, ssaI-prgJ, and ssaI-escI hybrid proteins. (D) BMM were infected with S. typhimurium grown under SPI2-inducing conditions for 8 h (1 h infection followed by 7 h treatement with gentamicin) at MOI 50, and IL-1β secretion was determined by ELISA. (E) Mice were infected with equal numbers of wild-type S. typhimurium carrying empty vector (pWSK129, kanamycin resistant) or a plasmid-expressing SPI2 controlled prgI or prgJ (ampicillin resistant). Competitive index was determined 48 h after i.p.injection of wild-type (BL6) or NLRC4 null mice by determining colony forming units in the spleen. The ratio of ampicillin/kanamycin-resistant colonies is shown with standard deviation from three mice (ratio of 0.01 represents a 100-fold reduction in recovered ampicillin-resistant bacteria compared with wild-type).
PrgJ Expression Permits NLRC4 Detection and S. typhimurium Clearance.
NLRC4 null mice do not have significantly increased susceptibility to S. typhimurium systemic infection (30) (Fig. S7A). We hypothesized that the lack of SPI2 detection by NLRC4 is beneficial to the bacteria. To investigate this hypothesis, we ectopically expressed prgJ, or prgI as a control, in S. typhimurium using a SPI2 promoter. prgJ expression resulted in detection of SPI2 induced S. typhimurium by macrophages that secreted IL-1β in response (Fig. 4D). In vivo, this detection resulted in the clearance of prgJ expressing S. typhimurium from the mouse spleen in a competitive index assay, whereas prgI expression had no effect on virulence (Fig. 4E). The clearance of these bacteria was dependent upon NLRC4, as equivalent numbers of WT and prgJ expressing bacteria were recovered from NLRC4-deficient mice (Fig. 4E), indicating both that NLRC4 detects PrgJ and that PrgJ expression does not compromise SPI2 T3SS promoted bacterial virulence (Fig. 4E and Fig. S7B). This effect was not due to any inherent problem in NLRC4-deficient mice since these mice have no alteration in their susceptibility to S. typhimurium (Fig. S7A).
Based on the lack of IL-1β secretion in response to SPI2 in vitro, the transcriptional repression of flagellin and prgJ that occurs after phagocytosis, the lack of detection of SsaI by NLRC4, the lack of contribution of NLRC4 to S. typhimurium resistance, and the ability of NLRC4 to clear prgJ expressing S. typhimurium, we conclude that NLRC4 fails to detect SPI2 T3SS and that this is beneficial for the pathogen. The apparent dichotomy between detection of the SPI1 rod and failure to detect the SPI2 rod reveals the constraints placed on S. typhimurium by the innate defenses of the cell type targeted by each T3SS. S. typhimurium typically use the SPI1 system to invade epithelial cells, which lack NLRC4. On the other hand, SPI2 is used within macrophages that express NLRC4.
Our results demonstrate that NLRC4 dependent activation of caspase 1 provides a wide net of protection against a large array of translocated virulence factors by responding to a component of the machinery through which they are injected into host cells (Fig. S1). Importantly, this method of innate immune detection permits the macrophage to discriminate pathogenic from avirulent bacteria.
Materials and Methods
Mice and Tissue Culture.
Bone marrow–derived macrophages (BMM) were prepared from the femur C57BL/6 (Charles River Laboratories) or NLRC4−/−(31) mice by culturing with L-cell supernatant. Mice were housed in a specific pathogen-free environment, with approval and supervision from the Institutional Animal Care and Use Committee at the Institute for Systems Biology.
Bacterial Infections.
BMM mice were primed with 50 ng/mL LPS for 3–4 h to induce proIL-1β expression and IL-1β determined by ELISA (R&D). Infections were performed for 1 h after centrifugation as previously described (5), followed by 15 μg/mL gentamicin treatment. EPEC were diluted 1:50 and grown at 37 °C with shaking for 3–4 h. S. typhimurium strains are derived from ATCC 14028s, which naturally lacks sopE. For SPI1 induction, 13-mL standing cultures were diluted 1:100 for 4 h in 13 mL LB without shaking at 37 °C. Alternately, cultures were diluted 1:40 for 3 h with shaking (Fig. 4A and Fig. S4). For SPI2 infections, overnight S. typhimurium that do not express SPI1 T3SS were used. Mice were infected intraperitoneally with 1 × 105 S. typhimurium equally divided between control and experimental strains.
Protein Transfections.
Amino terminal 6xhis tag fusion proteins were purified using Talon beads (Clontech). Point mutations in PrgJ or FliC were introduced by strand overlap exchange PCR, and the resulting proteins were equally expressed and soluble when compared with the wild-type 6xhis-tagged protein. Protein transfections were performed as previously described (13). Caspase 1 processing was assayed as described elsewhere (5)
Retroviral Lethality Screen.
Bacterial genes were cloned into pMXsIG (32), which contains an IRES-GFP element to track retroviral infection. Retroviruses were generated in Ecotropic Phoenix cells (American Type Culture Collection) as described elsewhere (5), and spinfections were performed into day 2–4 BMM. GFP expression was determined 2 days later.
Structural Predictions.
Given that many of the detectable sequence-based relationships between prgJ and other proteins of interest were weak (and likely too distant to be properly detected with purely sequence based), we used protein structure prediction to search for 3D-structure similarities between PrgJ and other T3SS apparatus proteins (21). We used a both the Human Proteome Folding (HPF) pipeline (20, 21) and the 3D-jury fold-recognition server (22) to predict the structure of PrgJ. We found that multiple fold recognition methods (used by both pipelines) found strong hits to the experimentally determined structure of other known type III secretion apparatus proteins PrgI (PDB code 2jow) and BsaL (PDB code 2g0u, the PrgI homolog from B. pseudomallei) (33). Furthermore, both pipelines found similar simple three-helix folds, suggesting that the fold identification is correct. Models resulting from FFAS (34) hits to 2g0u chain A and 2jow were refined using Modeler (35), resulting in confident structure predictions for residues 23–98of PrgJ. The 23 N-terminal residues were not well aligned to the template structures, and the N-terminal residues of 2jow and 2g0u are not well resolved in the experimental structure, suggesting that the N-terminal region of PrgJ and other related proteins are not ordered in solution. These residues could be ordered on polymerization or on interaction with other T3SS apparatus components.
Supplementary Material
Acknowledgments
The authors thank Bruce Vallance for supplying EPEC strains, Vishva Dixit and Richard Flavell for supplying mice, and members of the Aderem laboratory for discussion and critique of this manuscript. This research was supported by National Institutes of Health Grants AI065878, AI052286, AI032972, AI025032, and U54AI057141.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913087107/DCSupplemental.
*This Direct Submission article had a prearranged editor.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Lamkanfi M, Dixit VM. Inflammasomes: Guardians of cytosolic sanctity. Immunol Rev. 2009;227:95–105. doi: 10.1111/j.1600-065X.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 3.Miao EA, Andersen-Nissen E, Warren SE, Aderem A. TLR5 and Ipaf: Dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol. 2007;29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 4.Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 5.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 6.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 7.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 8.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2(e18):0175–0183. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchi L, et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 12.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci USA. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun YH, Rolán HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 15.Kimbrough TG, Miller SI. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 2002;4:75–82. doi: 10.1016/s1286-4579(01)01512-x. [DOI] [PubMed] [Google Scholar]
- 16.Kubori T, Sukhan A, Aizawa SI, Galán JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marlovits TC, et al. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, et al. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J Mol Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonneau R, Baliga NS, Deutsch EW, Shannon P, Hood L. Comprehensive de novo structure prediction in a systems-biology context for the archaea Halobacterium sp. NRC-1. Genome Biol. 2004;5:R52–R52.15. doi: 10.1186/gb-2004-5-8-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonneau R, Tsai J, Ruczinski I, Baker D. Functional inferences from blind ab initio protein structure predictions. J Struct Biol. 2001;134:186–190. doi: 10.1006/jsbi.2000.4370. [DOI] [PubMed] [Google Scholar]
- 22.Kaján L, Rychlewski L. Evaluation of 3D-Jury on CASP7 models. BMC Bioinformatics. 2007;8:304. doi: 10.1186/1471-2105-8-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sukhan A, Kubori T, Galán JE. Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J Bacteriol. 2003;185:3480–3483. doi: 10.1128/JB.185.11.3480-3483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3(e111):1082–1091. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galán JE. Salmonella interactions with host cells: Type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 29.Andersen-Nissen E, et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura T, et al. Retrovirus-mediated gene transfer and expression cloning: Powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 33.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 34.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: A server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33(Web Server issue):W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.