Abstract
Gram-negative bacteria deliver a cadre of virulence factors directly into the cytoplasm of eukaryotic host cells to promote pathogenesis and/or commensalism. Recently, families of virulence proteins have been recognized that function as E3 Ubiquitin-ligases. How these bacterial ligases integrate into the ubiquitin (Ub) signaling pathways of the host and how they differ functionally from endogenous eukaryotic E3s is not known. Here we show that the bacterial E3 SspH2 from S. typhimurium selectively binds the human UbcH5 ∼ Ub conjugate recognizing regions of both UbcH5 and Ub subunits. The surface of the E2 UbcH5 involved in this interaction differs substantially from that defined for other E2/E3 complexes involving eukaryotic E3-ligases. In vitro, SspH2 directs the synthesis of K48-linked poly-Ub chains, suggesting that cellular protein targets of SspH2-catalyzed Ub transfer are destined for proteasomal destruction. Unexpectedly, we found that intermediates in SspH2-directed reactions are activated poly-Ub chains directly tethered to the UbcH5 active site (UbcH5 ∼ Ubn). Rapid generation of UbcH5 ∼ Ubn may allow for bacterially directed modification of eukaryotic target proteins with a completed poly-Ub chain, efficiently tagging host targets for destruction.
Keywords: E2 ubiquitin conjugating enzyme, ubiquitin ligase, HECT, Salmonella SspH2 effector, ubiquitination
Modification of proteins by ubiquitin (Ub) is a powerful and flexible signaling/regulatory mechanism in eukaryotes. Protein ubiquitylation may serve to target a protein for proteasomal destruction, recruit other protein-binding partners, alter protein localization, or change protein activity (1–3). Ubiquitylation may involve conjugation of a single Ub onto a protein target (monoubiquitylation). This type of signaling is important for the regulation of cellular processes such as protein trafficking, endocytosis, and gene expression (4). Alternatively, a target protein may be modified by a polymer of Ub (poly ubiquitylation). Additional flexibility in Ub signaling comes from the diversity of Ub chains that can be synthesized by using one of several different Lys residues available on the Ub surface. For instance, Ub chains built using K48-linkages generally sentence a protein to destruction by the proteasome. Alternatively, K63-linked chains produce nonproteolytic signals that function in DNA damage repair, the inflammatory response, protein trafficking, and ribosomal protein synthesis (1).
Ubiquitylation of a target protein is the culmination of three sequential enzyme activities involving 1) an E1 Ub-activating enzyme, 2) an E2 Ub-conjugating enzyme, and 3) an E3 Ub-ligase (1). Two main classes of eukaryotic E3 Ub-ligases have been described, RING/U-box and HECT E3s, which differ structurally and mechanistically. RING and U-box E3s are topologically similar and are thought to serve as scaffolds that facilitate transfer of Ub directly from an E2 ∼ Ub conjugate to a target protein (5, 6). In contrast, HECT (Homologous to E6-AP Carboxy Terminus) domain E3 ligases form a thioester E3 ∼ Ub intermediate prior to modification of a protein target.
Given the central importance of Ub-signaling in eukaryotes it is not surprising that pathogenic bacteria have found ways to target Ub-transfer pathways to facilitate entry into host cells and establish environments suitable for replication (7–11). Gram-negative pathogens like Salmonella can introduce bacterial effector proteins directly into the cytosol of eukaryotic cells using a Type III Secretion System, an interspecies protein transport apparatus (12). In Salmonella, over 30 different effector proteins are introduced at different stages of pathogenesis and, recently, a number of effectors have been shown to have E3 Ub-ligase activity (7–11). To function as a Ub-ligase in a eukaryotic environment, bacterial E3s must directly interact with the Ub-transfer machinery provided by their host. However, the bacterial E3s found in Salmonella have very little sequence or structural homology to eukaryotic Ub-ligases (8, 13–15). Presumably, these proteins evolved via convergent evolution to manipulate and exploit this eukaryotic signaling pathway.
Previous studies of S. typhimurium SspH2 demonstrated robust E3 ligase activity with the human E2 UbcH5 (13). Therefore, we investigated the interactions and biochemical activities of purified SspH2 and human UbcH5c. In contrast with previous studies of E2/E3 interactions in eukaryotes, SspH2 has no detectable affinity for free UbcH5. Instead SspH2 selectively binds the UbcH5 ∼ Ub conjugate, recognizing regions of UbcH5 not previously thought important for governing interactions between eukaryotic E2s and E3s. In vitro, SspH2 directs the synthesis of K48-linked poly-Ub chains that are directly tethered to the active site of UbcH5. Rapid formation of activated poly-Ub chains tethered to the E2 may provide a pathway for bacterially directed modification of eukaryotic target proteins with a completed poly-Ub chain in a single encounter.
Results
Biochemical Features of SspH2.
The effector proteins SspH2, SspH1, and SlrP from Salmonella and the Shigella IpaH proteins are homologous with respect to primary sequence and domain architecture. Three-dimensional structures of IpaH1.4, IpaH3, and SspH2 have recently been reported and all have similar tertiary structures (13–15). The Ub-ligase activity of this family depends on a conserved Cys residue located in the C-terminal region of each protein that is essential for catalysis. DTT-sensitive E3 ∼ Ub adducts have been reported for IpaH3, IpaH9.8 (14, 15), and we have also detected SspH2 ∼ Ub adducts that are DTT-sensitive (Fig. S1). Mutation of the presumptive active site Cys (i.e., C580 of SspH2) abolishes activity (Fig. S2) (13). Therefore, these bacterial effectors have been classified as HECT-type E3 ligases whose defining characteristic is the obligate formation of an E3 ∼ Ub thioester intermediate during Ub-transfer reactions (10, 13–15).
SspH/IpaH effectors share a common domain arrangement with an N-terminal localization domain, a central Leucine-Rich Repeat (LRR) domain, and a C-terminal E3-ligase domain (Fig. 1A). We examined three different constructs of SspH2: full-length, SspH2171–788, and SspH2477–788. All exhibit Ub-ligase activity (Fig. S2), and the C-terminal domain (SspH2477–788) is sufficient for the synthesis of poly-Ub chains (Fig. 1B). SspH2, like other SspH/IpaH E3s, exhibits robust activity with UbcH5c relative to other human E2s such as UbcH7 (Fig. 1B). Use of single-site Lys to Arg Ub mutants and LC/MS analysis of SspH2 reaction products showed that SspH2, like IpaH3, preferentially catalyzes the synthesis of K48-linked poly-Ub chains (Fig. S3). However, other ubiquitylated species are observed if K48 of Ub is absent (Fig. S3A). Self-ubiquitylation of SspH2 is occasionally observed but represents only a minor product of in vitro reactions.
Fig. 1.
SspH2 constructs and activity of the SspH2 C-terminal domain. (A) Domain architecture of SspH2. SspH2 residues 10–149 encompass the N-terminal domain, 180–479 the LRR domain, and 494–788 the C-terminal E3-ligase domain. Constructs encompassing residues 171–788 (SspH2171–788) or 477–788 (SspH2477–788) as derived from the full-length protein. (B) The C-terminal domain of SspH2 (SspH2477–788) is sufficient for activity and is highly active with UbcH5 relative to UbcH7. 2 μM SspH2477–788 was mixed with 1 μM E1, 10 μM E2, 20 μM Ub, and 10 mM Mg2+ at 30 °C. The Ub contained an N-terminal T7-epitope tag (UbT7). Samples were collected at 0, 10, 20, and 40 m after addition of 5 mM ATP. After SDS–PAGE reaction products were visualized by western analysis using αT7 antibody.
SspH2 Selectively Binds the UbcH5 ∼ Ub Conjugate.
Heteronuclear 2D NMR spectroscopy was used to detect and characterize interactions between UbcH5 and SspH2. Resonances observed in 1H-15N TROSY NMR spectra are sensitive indicators of their environment and interaction with another protein will perturb the resonances of residues at the protein interface (16, 17). NMR mapping experiments to determine the surface of UbcH5 involved in binding to SspH2477–788 yielded an unexpected result. Previous NMR and crystallographic studies of E2 interactions with eukaryotic HECT, RING, or U-box E3s observed binding of a free E2 to an E3 (18–21). However, despite the robust Ub-ligase activity of SspH2477–788 with UbcH5 in in vitro assays, we found no detectable interaction with either free UbcH5 or Ub, even at protein concentrations that exceed 300 μM. This is evident by the lack of chemical shift perturbations and the near perfect overlay of 15N-UbcH5 and 15N-Ub resonances in the absence (Black peaks) and presence (Green peaks) of an equimolar amount of SspH2477–788 (Fig. 2A).
Fig. 2.
SspH2 selectively binds the UbcH5 ∼ Ub conjugate. Overlay of 1H, 15N- TROSY spectra of (A) free 15N-UbcH5 and 15N-Ub (300 μM each) in the absence (Black) and presence (Green) of 300 μM SspH2477–788, (B) 300 μM 15N-UbcH5-O-Ub in the absence (Black) and presence (Red) of 300 μM SspH2477–788, (C) 300 μM 15N-UbcH5-O-Ub in the absence (Black) and presence (Cyan) of 150 μM C580S-SspH2171–788, and (D) 300 μM 15N-A96D-UbcH5-O-Ub in the absence (Black) and presence (Magenta) of 300 μM SspH2477–788. In A and D the overlaid spectra are nearly identical indicating little or no interaction whereas significant perturbations are observed in B and C. Resonances of 15N-labeled UbcH5-O-Ub significantly affected by addition of SspH2477–788 (B) are mapped in red onto a ribbon structure (E) and a surface representation (F) of a model of UbcH5 ∼ Ub based upon the Ubc13 ∼ Ub conjugate (PDB ID 2 gmi). Topological features of UbcH5 described in the text are labeled in E. Residues that correspond to the canonical eukaryotic E3 binding surface located in Helix-1, Loop 4, and Loop 7 and are shown in light pink. The region on UbcH5 where eukaryotic E3 and SspH2 binding surfaces overlap is shown in orange.
The reaction mixtures described in Figure 1B also contain E2 ∼ Ub conjugates, and SspH2 may selectively bind this species. To examine binding of UbcH5 ∼ Ub conjugates to SspH2477–788 (Fig. 2B), we used a mutant of UbcH5 in which the active site Cys is replaced with Ser. Activation with Ub yields an ester-linked UbcH5-O-Ub conjugate that differs by only a single atom from the wild-type thioester complex and is an excellent structural mimic of the wild-type conjugate. UbcH5-O-Ub is stable, does not transfer its Ub to SspH2, and allows NMR investigation of an SspH2/UbcH5-O-Ub enzyme/substrate complex without complications arising from UbcH5-O-Ub turnover. Upon addition of SspH2477–788, a subset of resonances in the spectrum of uniformly 15N-labeled UbcH5-O-Ub either disappears or is substantially reduced in intensity. Similar spectral changes were also observed upon addition of a larger SspH2171–788 construct (Fig. 2C) indicating that the LRR domain of SspH2 does not prevent this interaction. These results demonstrate that SspH2 preferentially binds the substrate (i.e., UbcH5 ∼ Ub) as opposed to the product (i.e., free UbcH5) of the Ub-transfer reaction.
SspH2 Binds Regions of UbcH5 ∼ Ub Not Previously Recognized As Critical for Interactions with E3-Ligases.
The stability of the UbcH5-O-Ub conjugate allowed us to determine backbone resonance assignments using standard multidimensional NMR experiments (22). Though chemical shift perturbations are observed for UbcH5 amide resonances upon activation with Ub (23), there are only minor changes in the chemical shifts of 13Cα carbon resonances, and these are localized near the UbcH5c active site (Fig. S4). Whereas amide chemical shifts can be very sensitive to changes in environment, carbon backbone chemical shifts are primarily dependent on the amino acid and its structural context (i.e., whether a residue resides in an α-helix, β-strand, or random coil). The absence of large changes in 13Cα chemical shifts suggests that there are no significant conformational changes in UbcH5c upon activation with Ub. This result is similar to that found for Ubc13 that readily forms a heterodimeric complex with the E2 variant protein Mms2. No major structural changes were observed in the Ubc13 subunit upon formation of an Mms2/Ubc13 ∼ Ub conjugate complex (24).
Resonance assignments of UbcH5-O-Ub make it possible to map the surface(s) of UbcH5-O-Ub that bind SspH2. Residues in both UbcH5 and Ub whose resonances undergo a substantial reduction in intensity (Fig. S5) were mapped onto a model of the UbcH5c ∼ Ub conjugate (Figs. 2E and F). Significant changes are observed for residues surrounding the UbcH5 active site, Loop 7, and Helix 2. In contrast, interaction of UbcH7 with the HECT domain of E6-AP primarily involves residues in Helix 1, Loop 4, and Loop 7 (Fig. 2F) (20) and similar E2 surfaces have been defined for other eukaryotic E2/E3 complexes (19, 21, 25). The canonical E3-binding surface, depicted on a model of the UbcH5 ∼ Ub conjugate (Figs. 2E and F, Light Pink regions), only slightly overlaps (Orange region) with the SspH2 binding surface. Thus, SspH2 recognizes a surface of a charged E2 not previously known to be important for interactions with E3-ligases.
The NMR mapping studies also demonstrate that resonances corresponding to residues in the β-sheet and C-terminus of the Ub subunit are significantly affected when SspH2 binds to UbcH5-O-Ub conjugate (Figs. 2E and F). The apparent interaction site on Ub is centered on a hydrophobic patch composed of residues Leu8, Ile44, and Val70. This surface of Ub is often involved in binding interactions with heterologous proteins (26). It remains to be determined if the interaction surfaces defined in this study are particular to SspH2 and, by homology, other bacterial effectors, or if these regions are also important for eukaryotic E3/E2 ∼ Ub interactions.
The surfaces of UbcH5-O-Ub defined by NMR mapping experiments are consistent with mutational studies (Fig. 3A). Mutation of UbcH5 Ala96 to Asp (A96D), a residue located in Loop L7 of the binding interface, does not interfere with formation of the UbcH5 ∼ Ub conjugate by E1 (27). However, no significant interactions of (A96D)-UbcH5 ∼ Ub with SspH2477–788 were observed by NMR (Fig. 2D), and activity was significantly reduced in SspH2 catalyzed reactions (Fig. 3B). In contrast, mutations outside the interaction surface defined by NMR mapping have little or no influence on activity. Mutations outside the SspH2 interaction surface do not appreciably alter the observed activity of UbcH5c with SspH2477–788 or other SspH/IpaH E3 family members (Fig. 3) (14, 15). Mutations in UbcH5c such as S22R UbcH5c, which affects the Ub-transfer activity of UbcH5 with a number of RING-domain E3-ligases (28), or R5A , K63A, and F62A in the canonical eukaryotic E3 interface, do not eliminate UbcH5 activity with these enzymes. The observation that SspH/IpaH E3s are not affected by mutations in the canonical eukaryotic E3 binding surface (i.e., R5 and F62) indicates that other E3s in this family may also recognize a noncanonical surface on the charged E2 ∼ Ub conjugate.
Fig. 3.
The A96D-UbcH5 ∼ Ub conjugate is impaired in reactivity with SspH2. (A) Ribbon diagram of UbcH5 highlighting the SspH2 contact region (Red) and locations of mutations described in the text. Ala 96 (Red Spheres) resides within the mapped contact region whereas R5, S22, F62, and K63 (Gray Spheres) lie outside this region. The UbcH5 active site residue C85 is shown in yellow. (B) (A96D)-UbcH5 ∼ Ub exhibits reduced activity with SspH2 in the synthesis of poly-Ub chains relative to wt-UbcH5, whereas (F62A)-UbcH5 ∼ Ub, like R5A, S22R, and K63A (14, 15), is only moderately affected. Reactions were conducted with 10 μM E2 ∼ UbT7 and 1 μM SspH2477–788 at 32 °C. Samples were collected at 0, 10, and 20 minutes after addition of 5 mM ATP and visualized by western analysis with α-T7 antibodies.
SspH2 Generates Poly-Ub Chains Conjugated to the Active Site of Wild-Type UbcH5.
Addition of UbcH5 ∼ Ub to SspH2477–788 leads to the rapid production of poly-Ub chains as shown on reducing SDS–PAGE gels (Fig. 1B). Unexpectedly, when reaction products are analyzed under nonreducing conditions, Ub-chains are found directly tethered to the active site of UbcH5 (Fig. 4). Reaction mixtures containing purified UbcH5 ∼ Ub and SspH2477–788 were separated by SDS–PAGE under nonreducing conditions and UbcH5 ∼ Ubn (where n denotes 2 or more Ubs) was visualized using UbcH5-specific antibodies. This product was lost upon addition of DTT (Fig. 4). Incubation of UbcH5 ∼ Ub at 32 °C for the same duration without SspH2477–788 in the mixture did not result in accumulation of UbcH5 ∼ Ubn species (Fig. S6), indicating that their production is dependent on SspH2477–788. UbcH5 is widely used in studies of Ub-transfer reactions and exhibits activity with most eukaryotic E3s, both HECT and RING/U-box E3s (29). To our knowledge, this is the only example of E3-dependent synthesis of poly-Ub chains directly linked to the active site of UbcH5.
Fig. 4.
SspH2 generates poly-Ub chains linked to the active site of UbcH5. Western analysis with α-UbcH5c antibodies performed on a reaction mixture containing purified 37 μM UbcH5-S ∼ Ubwt and 0.6 μM SspH2477–788. Samples were collected at 0, 1, 2, 3, and 4 minutes after addition of SspH2477–788 at 30 °C. Products were separated by SDS–PAGE under nonreducing (-DTT) and reducing (+DTT) conditions. The samples at 0 minutes were collected prior to the addition of SspH2477–788. Incubation at 30 °C of a similar control sample containing only UbcH5-S ∼ Ubwt and lacking SspH2 revealed no changes in the species detected (Fig. S6). Higher molecular weight species of UbcH5 ∼ Ub(n) are observed only in the presence of SspH2477–788 and under nonreducing conditions.
The rapid accumulation of UbcH5 ∼ Ubn in solution is consistent with the premise that poly-Ub chains tethered to the E2 are synthesized as part of the normal SspH2 catalytic cycle. An alternative possibility is that in vitro the sequential transfer of Ub from E2 ∼ Ub to E3 to target lysine is blocked when a bona fide substrate is not present. Possible ways to restore Ub transfer via SspH2 is 1) by simple hydrolysis of the charged SspH2 ∼ Ub conjugate to free E3 and Ub or 2) via the transfer of Ub to an available proxy target (Fig. 5A). In this case, a suitable target substrate is one that can bind to the E3 (or E3 ∼ Ub) and has appropriately positioned Lys residues that can accept activated Ub. Under the experimental conditions described in Fig. 4, the only available candidate for a target protein that can lead to the synthesis of UbcH5 ∼ Ubn is the Ub subunit of a separate UbcH5 ∼ Ub conjugate.
Fig. 5.
UbcH5-O-Ub is not used as a platform for the synthesis of poly-Ub chains. (A) Reaction scheme depicting how UbcH5c ∼ Ub could conceivably be used, in the absence of a bona fide substrate, to yield UbcH5c ∼ Ubn via the sequential addition of Ub. (B) Test of the hypothesis. Lanes 1–3: Selective visualization of UbcH5-O-UbT7 and its derivatives in the reaction mixture. Equimolar amounts (5 μM) of UbcH5-O-UbT7 and wt UbcH5 ∼ Ub were present at the start of the reaction. Samples were collected at 0, 3, and 10 minutes after the addition of SspH2477–788. The primary products detected are UbcH5-O-Ub2 and UbcH5-O-Ub3. Lanes 4–6: Identical reaction mixture except all Ub in the reaction mixture has a T7-epitope tag. The predominant reaction products are poly-Ub chains. A cross-reacting species that is present prior to the addition of SspH2477–788 is indicated by an asterisk.
This latter scenario suggests that UbcH5 ∼ Ub could play a dual role serving as both a source of activated Ub for SspH2 (i.e., substrate) and also as the final acceptor of Ub from SspH2 (i.e., target). This would require that there exist multiple sites on SspH2 capable of binding UbcH5 ∼ Ub—one site that normally binds UbcH5 ∼ Ub as the substrate and a second distinct site that can maintain the interaction through multiple rounds of Ub addition (Fig. 5A). Chains would then be synthesized by the sequential addition of Ub from SspH2 ∼ Ub to the end of a growing Ub chain that remains tethered to the UbcH5 active site. If true, then UbcH5-O-Ub should also serve as a platform for the synthesis of Ub chains. A reaction mixture containing SspH2 and both UbcH5-O-Ub and wtUbcH5 ∼ Ub should result in the generation of both DTT-sensitive wtUbcH5 ∼ Ubn and DTT-resistant UbcH5-O-Ubn. To test this scenario, UbcH5-O-Ub was purified with the Ub subunit containing an N-terminal T7-epitope tag to allow for selective visualization of UbcH5-O-UbT7 and products containing UbcH5-O-UbT7. As shown in Fig. 5B (lanes 1–3), when present with an equivalent concentration of wtUbcH5 ∼ Ub under conditions that lead to chain synthesis, UbcH5-O-UbT7 is primarily modified with a single Ub. The addition of a second Ub is apparent at longer reaction times. However, in an identical reaction mixture with all Ub labeled with a T7-epitope tag, it is clear that the primary products in this reaction are high molecular weight poly-Ub chains (Fig. 5B, lanes 4–6). These results demonstrate that UbcH5-O-Ub can be modified but is involved in only one or two catalytic cycles and is not an optimal target for chain synthesis. Thus SspH2 does not appear to catalyze the progressive addition of Ub to the distal end of a UbcH5c-conjugated Ub chain. Rather, chain growth appears to require exchange of the growing Ub chain between E2 and E3 catalytic cysteines.
Discussion
Interaction of the bacterial E3 ligase SspH2 with E2 ∼ Ub.
Bacterial Ub-ligases are a recently recognized addition to the panoply of enzymes that catalyze Ub-transfer. All these enzymes must recognize and function with components of the eukaryotic ubiquitylation machinery, yet the structures of the bacterial enzymes differ markedly from their eukaryotic counterparts (Fig. S7). Although SspH/IpaH family members have been classified as HECT-type E3s, NMR mapping studies of protein interacting surfaces reveal significant differences in the binding and interaction with cognate E2s. Previous structural studies of E3/E2 interactions examined binding of a free E2 to an E3. Examples like the structure of E6-AP in complex with the E2 UbcH7 provide an important foundation for our understanding of E2/E3 interactions among HECT E3-ligases (20). However, experiments with free UbcH5 and free Ub failed to detect any direct interaction with SspH2477–788 or SspH2171–788, even at protein concentrations in excess of 300 μM. This contrasts somewhat with a previous report of weak interactions between IpaH9.8 and free UbcH5 (Kd of 317 μM) (14) and may reflect individual variations among SspH/IpaH E3 ligases.
Interaction of UbcH5c or Ub with SspH2477–788 was only observed for the UbcH5-O-Ub conjugate and involves both the UbcH5 and Ub subunits. The interaction surface is distinct from that defined for free UbcH7 binding to E6-AP and has not been previously recognized as important for E3 recognition. It is not likely that the binding of UbcH5 ∼ Ub to SspH2 relies on protein conformational changes induced in either the UbcH5 or Ub subunits of the conjugate because no large changes in 13Cα chemical shifts were observed in either subunit upon activation (Fig. S4). Thus, binding of UbcH5 ∼ Ub to SspH2 is consistent with a paradigm in which affinity and selectivity arise from a combination of multiple weak but coordinated interactions.
Possible Mechanisms for SspH2-Directed Synthesis of UbcH5 ∼ Ubn.
An unexpected feature of SspH2-directed poly-Ub chain synthesis is the rapid accumulation of poly-Ub chains directly tethered via a thioester linkage to the active site of UbcH5. Hochstrasser (30) described several mechanisms relevant to HECT-type poly-Ub chain synthesis: see-saw, sequential, indexation, and hybrid mechanisms (Fig. S8). In the sequential mechanism, the E3 is able to bind both an E2 ∼ Ub conjugate and a target protein. Ub is then transferred from E2 ∼ Ub to the E3 (to form E3 ∼ Ub) followed by modification of the protein target. Synthesis of a poly-Ub chain is accomplished by successive addition of Ub from the E3 (i.e., E3 ∼ Ub) to the distal end of a growing Ub chain tethered to a substrate. Fig. 5 demonstrates that UbcH5-O ∼ Ub does not serve as a platform for the sequential addition of Ub, a finding that argues against the sequential model for the synthesis of UbcH5 ∼ Ubn. Additional experiments testing free Ub as a platform for chain synthesis or proteins directly fused to Ub yielded similar results (Fig. S9). The indexation model differs from the sequential mechanism in that Ub is successively added to the distal end of a growing chain linked to the E3 active site. This mechanism predicts the accumulation of poly-Ub chains tethered to the E3 but not the E2 active site. The hybrid mechanism (not depicted) requires that free Ub noncovalently bind to the E3 and serve as a platform for the synthesis of poly-Ub chains. Once a chain is synthesized, it is activated by an E1, transferred to an E2, to an E3, and finally to a protein target. Because poly-Ub chains tethered to the UbcH5 active site Cys can be generated from purified UbcH5 ∼ Ub conjugates in the absence of an E1, and because SspH2 has little affinity for free Ub (Fig. 2A), the hybrid mechanism is considered the least plausible explanation. Other hybrid or “mixed” mechanisms are possible, but each makes additional assumptions and adds additional complexity to reaction schemes.
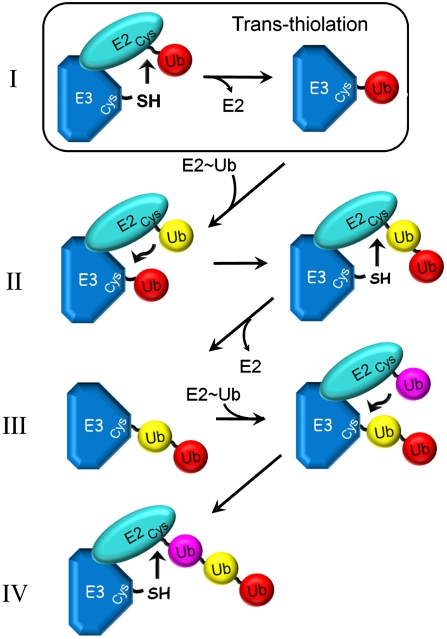
Of the mechanisms outlined above, the see-saw mechanism is most consistent with our data (Fig. 6). In this mechanism the growing chain is reciprocally transferred between E2 and E3 active sites. During each round of catalysis a new molecule of UbcH5 ∼ Ub reacts with SspH2 ∼ Ubn to yield UbcH5 ∼ Ub1+n and SspH2. The elongated chain is then transferred back to yield SspH2 ∼ Ub1+n. This mechanism predicts that only UbcH5 ∼ Ub (i.e., UbcH5 charged with a single Ub) is preferred as a substrate for charged SspH2 ∼ Ubn. As shown in Fig. 5, once UbcH5-O-diUb is formed it is no longer a preferred substrate for SspH2 ∼ Ubn, a finding consistent with this model. Furthermore, reciprocal transfer of the growing Ub chain between UbcH5 and SspH2 active sites directly results in the synthesis of the observed product, UbcH5 ∼ Ubn.
Fig. 6.
Reaction Scheme depicting the see-saw mechanism. In the see-saw mechanism the growing Ub chain is reciprocally transferred between E2 and E3 active sites. After the initial transthiolation step to yield charged E3 ∼ Ub and free E2 (I), a new E2 ∼ Ub conjugate binds to the ligase. A Ub Lys residue on this E2 ∼ Ub conjugate would react with the E3 ∼ Ub thioester to form charged E2 ∼ Ub-Ub (II). The nascent Ub chain is then transferred back to the E3, in a manner analogous to the initial trans-thiolation step (I), to give E3 ∼ Ub-Ub and free E2 (III). Multiple cycles extend the poly-Ub chain and result in the synthesis of charged E2 ∼ Ubn.
Part of the see-saw mechanism requires that the growing chain reside, at least temporarily, tethered to the E3 active site. There is, however, no fundamental requirement that all intermediates accumulate to the same extent in the reaction mixture. The ability to purify wtUbcH5 ∼ Ub conjugates and to visualize wtUbcH5 ∼ Ubn species (Fig. 4) suggests that E2 ∼ Ub adducts are relatively stable allowing for detection under nonreducing conditions. SspH2 ∼ Ubn is harder to isolate. However, under conditions that favor formation of charged E3 ∼ Ub and reduce the probability for the reciprocal transfer of Ub back to UbcH5 ∼ Ub ([SspH2] > [UbcH5 ∼ Ub]), we can observe SspH2 ∼ Ub conjugates (Fig. S1). Close inspection of these results suggest that higher molecular weight DTT-sensitive poly-Ub chains are detected linked to the active site of SspH2477–788. Though not definitive, this result favors the see-saw mechanism over an indexation type of mechanism where synthesis of a poly-Ub chain tethered to the E3 actives site requires some stability of the E3 ∼ Ubn conjugate through multiple cycles of elongation.
Other Cases of E3-Generated E2 ∼ Ubn.
There are few examples of E3-generated E2 ∼ poly-Ub conjugates in the literature. Ravid and Hochstrasser (31) demonstrated that the yeast HECT E3 Ufd4 can assemble a poly-Ub chain onto the active site Cys of its cognate E2, Ubc7, in vivo. The authors also showed that a preassembled poly-Ub chain can be transferred to a target Lys residue, suggesting that chain synthesis occurs before substrate modification. Recently, Li et al. (32) demonstrated that the mammalian RING E3 gp78c can facilitate assembly of a K48-linked chain on the active site cysteine of the E2 Ube2g2, and that this chain can be transferred en bloc to the protein substrate HERPc in a gp78c-dependent reaction (32). gp78c is a RING E3 that does not have an active site cysteine. A critical factor for the synthesis of preassembled chains is the interplay between two suitably positioned thiol groups that can each carry thiolester-linked Ub. Therefore, the authors suggest that gp78c facilitates the proper juxtaposition or dimerization of two charged E2s to promote chain building and elongation (33). A similar mechanism is unlikely for SspH2-directed synthesis as mutation of SspH2 Cys580 to Ala, which abrogates all SspH2 activity (Fig. S2), should not affect transfer between two bound and charged E2s. C580 is located in a flexible loop connecting two antiparallel α-helices and mutation to Ala does not affect binding of UbcH5-O-Ub as judged by NMR (Fig. 2C). For SspH2-directed chain synthesis, the interplay between E3 and E2 active sites combined with a preference for the association of SspH2 with the UbcH5 ∼ Ub conjugate is critical for facilitating rapid poly-Ub chain synthesis.
Finally, we note that not all structurally related E3s are expected to operate via the same mechanism. For instance, recent studies of E6-AP directed synthesis of poly-Ub chains are consistent with a sequential mechanism (34). However, homologous HECT domains may employ distinct mechanisms for the synthesis of Ub polymers (35). It is possible that the SspH/IpaH family of E3 ligases exhibits similar variation in mechanism for the synthesis of poly-Ub chains that depends on the functional role of each effector within its eukaryotic host.
Functional Implications SspH2-Directed Synthesis of UbcH5 ∼ Ubn.
SspH2 synthesizes K48-linked poly-Ub chains, implying that its targets are destined for proteasomal destruction. Upon introduction into the cytosol of a eukaryotic host, it must compete with endogenous E3s for E2 ∼ Ub conjugates and target proteins. This may be achieved by increased catalytic activity relative to their eukaryotic counterparts. Additionally, synthesis and accumulation of UbcH5 ∼ Ubn may provide a mechanism for rapid transfer of a preassembled K48-linked poly-Ub chain to a protein target. Such a mechanism may increase the likelihood that encounters with a target would result in a productive modification of that target with an intact poly-Ub signal. Furthermore, synthesis and accumulation of UbcH5 ∼ Ubn may increase the local pool of E2 ∼ Ubn that is preferentially used by bacterial effector Ub-ligases. Further testing of these possibilities must await identification of the cellular substrates for the bacterial E3 ligases and analysis of eukaryotic E3 interactions with E2 ∼ Ub species.
Materials and Methods
Expression and Purification of Proteins.
All expression plasmids were generated using standard methods (see SI Text). Protein constructs were expressed and purified as previously described (27). UbcH5 ∼ Ub and UbcH5-O-Ub was prepared by incubation of UbcH5c with E1, ATP, and isolated by gel filtration over a Superdex 75 column (25 mL) equilibrated with 25 mM NaPi, 150 mM NaCl, pH 7 (standard buffer).
Biochemical Assays.
SspH2-catalyzed reactions were performed in standard buffer with either purified UbcH5 ∼ Ub conjugates or in reaction mixtures at 30 °C containing, 1 mM MgCl2, 0.5 μM E1, 2 μM UbcH5c, 20 μM Ubwt or UbT7, and 1 μM E3 and initiated with 5 mM ATP. Samples were collected at intervals and quenched by the addition of SDS–PAGE loading buffer. Samples were separated by SDS–PAGE and visualized by Coomassie staining or western analysis.
NMR Spectroscopy.
All NMR samples were prepared in standard buffer at concentrations of 0.3–0.5 mM and data were collected at 25 °C. NMR data were collected on Bruker 500 MHz DMX (University of Washington) or Varian INOVA 600, 800, and 900 MHz spectrometers (Pacific Northwest National Labs). Collection of data for backbone resonance assignments utilized standard three-dimensional NMR techniques (22). NMR titration experiments were performed by sequential addition of unlabeled SspH2477–788 or SspH2171–788 to either ∼0.35 mM uniformly 15N-labeled free UbcH5 and free Ub or to 0.35 mM 15N-labeled UbcH5-O-Ub.
Supplementary Material
Acknowledgments.
We thank D. Wenzel, J. Pruneda, R. Pfeutzner, M. Christen, and L. R. Hoffman for critical reading of the manuscript and helpful discussions. NMR experiments were performed, in part, in the Environmental Molecular Sciences Laboratory located at Pacific Northwest National Laboratory and operated by Battelle. Mass spectrometry experiments were performed with the help of M. MacCoss, D.Tomazela, and P.Gafken. I.L. was supported by the Interdisciplinary Training Program in Bacterial Pathogenesis at the University of Washington. This work was supported by National Science Foundation Grant MCB-0615632 (R.E.K) and National Institutes of Health Grant RO1 AI048683 (S.I.M).
Note Added in Proof.
During review of this manuscript a structure of a UbcH5B ~ Ub/HECTNEDD4L complex was published revealing similar E2/E3 interactions to those previously observed 3081in a E6AP/UbcH7 complex. Ub is also involved in binding the C-terminal lobe of NEDD4L, resulting in the assembly of an overall compact structure (36).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914821107/DCSupplemental.
References
- 1.Pickart CM, Eddins MJ. Ubiquitin: Structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1-3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6(8):599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 3.Hochrainer K, Lipp J. Ubiquitylation within signaling pathways in- and outside of inflammation. Thromb Haemostasis. 2007;97(3):370–377. [PubMed] [Google Scholar]
- 4.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2(3):195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 5.Joazeiro CA, Weissman AM. RING finger proteins: Mediators of ubiquitin ligase activity. Cell. 2000;102(5):549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 7.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311(5758):222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 8.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15(1):65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 9.Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67(6):1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 10.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1(1):77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Rytkonen A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104(9):3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444(7119):567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 13.Quezada CM, Hicks SW, Galan JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci USA. 2009;106(12):4864–4869. doi: 10.1073/pnas.0811058106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer AU, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15(12):1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol. 2008;15(12):1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 16.Walters KJ, et al. Characterizing protein-protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods Enzymol. 2001;339:238–258. doi: 10.1016/s0076-6879(01)39316-3. [DOI] [PubMed] [Google Scholar]
- 17.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41(1):1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 18.Brzovic PS, Klevit RE. Ubiquitin transfer from the E2 perspective: why is UbcH5 so promiscuous? Cell Cycle. 2006;5(24):2867–2873. doi: 10.4161/cc.5.24.3592. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12(4):633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, et al. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286(5443):1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 21.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102(4):533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 22.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 1999;34(2):93–158. [Google Scholar]
- 23.Brzovic PS, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA. 2003;100(10):5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13(10):915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 25.Verdecia MA, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11(1):249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 26.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124(6):1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14(10):941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 28.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21(6):873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Lorick KL, Jensen JP, Weissman AM. Expression, purification, and properties of the Ubc4/5 family of E2 enzymes. Methods Enzymol. 2005;398:54–68. doi: 10.1016/S0076-6879(05)98006-3. [DOI] [PubMed] [Google Scholar]
- 30.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124(1):27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 31.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9(4):422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446(7133):333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 33.Li W, et al. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106(10):3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29(12):3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Pickart CM. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. Embo J. 2005;24(24):4324–4333. doi: 10.1038/sj.emboj.7600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B ~ ubiquitin-HECTNEDD4L complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.