Abstract
Cholesterol metabolism has been implicated in the pathogenesis of several neurodegenerative diseases, including the abnormal accumulation of amyloid-β, one of the pathological hallmarks of Alzheimer disease (AD). Acyl-CoA:cholesterol acyltransferases (ACAT1 and ACAT2) are two enzymes that convert free cholesterol to cholesteryl esters. ACAT inhibitors have recently emerged as promising drug candidates for AD therapy. However, how ACAT inhibitors act in the brain has so far remained unclear. Here we show that ACAT1 is the major functional isoenzyme in the mouse brain. ACAT1 gene ablation (A1−) in triple transgenic (i.e., 3XTg-AD) mice leads to more than 60% reduction in full-length human APPswe as well as its proteolytic fragments, and ameliorates cognitive deficits. At 4 months of age, A1− causes a 32% content increase in 24-hydroxycholesterol (24SOH), the major oxysterol in the brain. It also causes a 65% protein content decrease in HMG-CoA reductase (HMGR) and a 28% decrease in sterol synthesis rate in AD mouse brains. In hippocampal neurons, A1− causes an increase in the 24SOH synthesis rate; treating hippocampal neuronal cells with 24SOH causes rapid declines in hAPP and in HMGR protein levels. A model is provided to explain our findings: in neurons, A1− causes increases in cholesterol and 24SOH contents in the endoplasmic reticulum, which cause reductions in hAPP and HMGR protein contents and lead to amelioration of amyloid pathology. Our study supports the potential of ACAT1 as a therapeutic target for treating certain forms of AD.
Keywords: Alzheimer disease, cholesterol esterification, lipid metabolism, oxysterols
Alzheimer disease (AD) is characterized by extracellular accumulation of plaques, which are aggregates of amyloid-β (Aβ) peptides derived from proteolytic cleavages of amyloid precursor protein (APP), and intracellular accumulation of hyperphosphorylated tau (1). APP can be cleaved via the α- or the β-secretase pathways (2). The β-secretase pathway, but not the α-secretase pathway, generates Aβ from APP. Cholesterol content in cells can affect the production of Aβ, in part by its ability to modulate the enzyme activities of various secretases in cell membranes (3). Cholesterol has also been implicated in the pathogenesis of AD via other mechanisms (4–6).
In the brain, cholesterol is almost exclusively derived from endogenous biosynthesis (7). The transcription factor SREBP2 controls the expression of enzymes involved in cholesterol biosynthesis, including the rate-limiting enzyme HMG-CoA reductase (HMGR) (8). In addition, the nuclear receptors LXRs control the gene expression of proteins involved in cholesterol transport (9, 10), including apoE, ABCA1, ABCG1, and ABCG4 (11, 12). Cholesterol can be enzymatically converted by a brain-specific enzyme, 24-hydroxylase (CYP46A1) (13), to the oxysterol 24(S)-hydroxycholesterol (24SOH); the concentration of 24SOH far exceeds those of other oxysterols in the brain (14). In intact cells and in vitro, 24SOH can down-regulate sterol synthesis (15, 16). When provided to neurons, 24SOH decreases the secretion of Aβ (17). 24SOH levels were decreased in brain samples from patients with AD (18).
Acyl-CoA:cholesterol acyltransferase (ACAT) converts free cholesterol to cholesteryl esters (CEs). There are two ACAT genes, Acat1 and Acat2 (also known as Soat1 and Soat2), with different tissue expression patterns (19). Both are considered drug targets for treating cardiovascular diseases. Hutter-Paier et al. reported that treating a mouse model for AD with isotype-nonselective ACAT inhibitors substantially diminished amyloid plaque density (20, 21), suggesting that ACAT inhibitors may serve as therapeutic agents for AD; however, it was not clear whether the effects of the ACAT inhibitors observed were caused by inhibition of ACAT activity and/or effects on other biological process(es) in the mouse brain. In the current work, we undertake a combined mouse genetic and biochemical approach to evaluate the potential role of ACAT in AD.
Results
ACAT Expression in Mouse Brains.
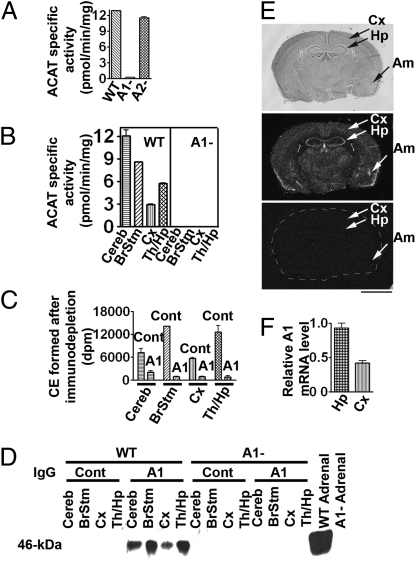
Whether the brain has active ACAT enzyme was previously unknown. To examine this issue, we prepared brain homogenates from WT, Acat1−/− (A1−) and Acat2−/− (A2−) mice and found that WT and A2− mouse brains contained comparable ACAT enzyme activity, whereas A1− mouse brains contained negligible activity (Fig. 1A). Various regions prepared from WT mouse brains, but not A1− mouse brains, all contained active ACAT (Fig. 1B). Mouse ACAT1 is a 46-kDa protein (22). Immunoblot analysis showed that, in homogenates prepared from mouse brain (but not from other mouse tissues), a non-ACAT1 protein band appeared in the 46-kDa region; the presence of this nonspecific band precluded us from using immunoblotting or histochemical staining to identify ACAT1 in the mouse brain. To unambiguously identify the ACAT1 protein, we first performed immunoprecipitation (IP) experiments with detergent-solubilized WT mouse brain extracts. The results showed that ACAT activity could be efficiently immunodepleted by ACAT1-specific antibodies (i.e., A1), but not by control antibodies (Fig. 1C). We then performed immunoblotting on the immunoprecipitates; the result showed that, in homogenates from WT mouse brain regions, the ACAT1 antibodies specifically identified a 46-kDa protein band; this band is absent in homogenates prepared from the adrenals and brains of A1− mice (Fig. 1D). These results indicate that mouse brains do express ACAT1 as the major ACAT isoenzyme.
Fig. 1.
ACAT protein, enzyme activity, and RNA expression in mouse brains. (A) ACAT activity in 53-d-old mouse brain homogenates and (B) ACAT activities in various regions. Cereb, cerebellum; BrStm, brainstem; Cx, cortex; Th, thalamus; Hp, hippocampus. (C) Immunodepletion of ACAT activity in the WT mouse brain homogenates. IP with nonspecific rabbit IgG or with ACAT1-specific (A1) IgG. The ACAT activities in the supernatants were measured. (D) Identification of A1 protein. After IP, pellets were resolved by SDS/PAGE; A1 protein (46 kDa) was detected with polyclonal A1 antibodies. Lysates from WT and A1− mouse adrenals were used as controls. (E) A1 mRNA distribution. Upper: Nissl staining from a 2-month-old WT mouse. Cx, cortex; Hp, hippocampus; Am, amygdale. (Middle and Bottom) In situ hybridizations using [32P]-ACAT1 antisense riboprobe or sense riboprobe (as negative control). For bottom panel, brain periphery was outlined artificially. (Scale bar: 250 mm.) (F) A1 mRNA levels in WT mouse hippocampus and cortex as measured by real-time PCR and normalized against neurofilament polypeptide chain (NF120) mRNA. Data in A–D and F represent mean ± SEM; n = 2.
To determine the ACAT1 mRNA distribution in mouse brains, we performed in situ hybridization experiments. Both hippocampus and cortex contain ACAT1 mRNA; hippocampus expresses a stronger signal (Fig. 1E Middle ). Other ACAT1-positive regions included choroids plexus, medial habenular nucleus, amygdala, and rostral extension of the olfactory peduncle. We next isolated hippocampus-rich regions and cortex-rich regions from WT mice and compared their ACAT1 mRNA levels by real-time PCR (primer sequences are listed in Table S1). The result showed that ACAT1 mRNA is approximately twofold higher in hippocampus than in cortex (Fig. 1F). A separate RT-PCR experiment using ACAT2-specific primers showed that only the thalamus-rich region, and no other brain regions, expresses low but detectable ACAT2 mRNA levels (Fig. S1A), confirming an early report by Anderson et al. (23), who showed in monkey brains that the ACAT2 mRNA level was nearly undetectable.
Effect of A1− on Aβ Deposition/hAPPswe Processing and on hTau.
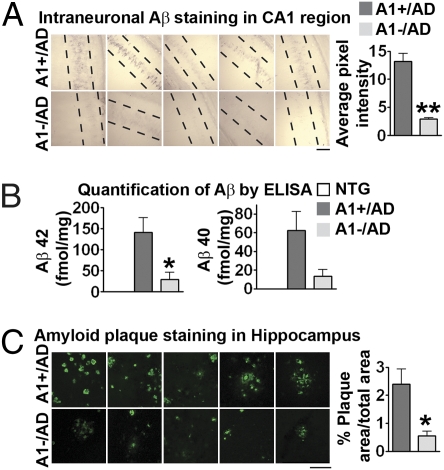
To investigate the effect of inactivating ACAT1 on amyloid and tau pathologies in the triple transgenic (3XTg)-AD mice (24), we produced Acat1−/−/AD (A1−/AD) mice by crossing the Acat1−/− mice with the 3XTg-AD mice. The breeding scheme is described in Fig. S2. To examine the effect of A1− on amyloid pathology, we first used the human specific anti-Aβ antibody 6E10 to perform intraneuronal immunostaining in the CA1 region of hippocampi of 4-month-old mice. Results showed that the staining was significantly diminished (by approximately 78%) in the A1−/AD mice (Fig. 2A Bottom). We next used ELISA to measure the total Aβ40 and Aβ42 levels in mouse brain homogenates at 17 months of age. Results showed that the Aβ42 levels were significantly decreased (by approximately 78%) in A1−/AD mice; the Aβ40 levels were also decreased, but the difference observed was not statistically significant. The brains of nontransgenic (NTG) mice did not contain measurable Aβ (Fig. 2B). We next used thioflavin S to stain amyloid plaques in AD mouse brains at 17 months of age. The results showed that, in A1−/AD mice, the amyloid plaque load in the hippocampi was significantly reduced (by approximately 77%; Fig. 2C); in the cortex, the amyloid plaque load in these mice showed a trend toward decreasing (P = 0.17; Fig. S1B).
Fig. 2.
Effect of A1− on Aβ pathology in AD mice. (A) Intraneuronal Aβ (using human Aβ-specific antibody 6E10) in the hippocampal CA1 region of male mice at 4 months (P = 0.0059; n = 4 or 5). (B) Aβ42 and Aβ40 levels analyzed by ELISA in the forebrains of mice at 17 months. For Aβ42, P = 0.035; for Aβ40, P = 0.084; n = 5. (C) Amyloid plaque load (using thioflavin S staining) in the hippocampus of 17-month-old mice; P = 0.031; n = 5. (Scale bars: 100 μm in A and C.) Data represent mean ± SEM. *P < 0.05; **P < 0.01.
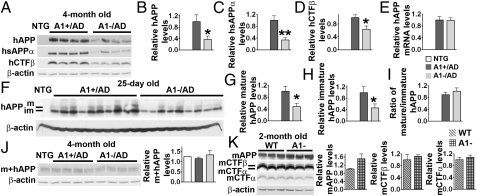
We next studied the effect of A1− on human APP processing in 4-month-old AD mice. We used the antibody 6E10 to detect full-length APP [human APP harboring the Swedish mutation (hAPPswe)] and its proteolytic fragments sAPPα [soluble APP fragment produced by α-secretase cleavage (hsAPPα)] and CTFβ [C-terminal APP fragment produced by β-secretase cleavage (hCTFβ)]. The results showed that, in A1−/AD mice, hsAPPα and hCTFβ levels were decreased (by approximately 67% and 37%, respectively; Fig. 3 A, C, and D). To our surprise, the hAPP level was also significantly reduced (by approximately 62%; Fig. 3 A and B). In contrast to the hAPP protein levels, there was no difference in hAPP mRNA levels between the A1+/AD mice and the A1−/AD mice (Fig. 3E) (primer sequences are listed in Table S1). hAPP is synthesized in the ER in its immature form (with a molecular weight of approximately 105 kDa); the immature form moves from the ER to the Golgi via the secretory pathway (25) and becomes highly glycosylated (mature form has a molecular weight of approximately 115 kDa) (26, 27). We examined the effects of A1− on the levels of immature and mature forms of hAPP in young AD mice (at 25 d of age). The results showed that A1− led to decrease in both forms to approximately the same extent (by approximately 52%–54%; Fig. 3 F–I), suggesting that the effect(s) of A1− act on newly synthesized hAPP. The AD mice express both hAPP and endogenous (mouse) APP (mAPP). To test the possibility that A1− may affect both the hAPP and the mAPP levels, we used a different antibody (antiserum 369), which recognizes the C-terminal fragments of both hAPP and mAPP (28) to investigate the total APP levels in AD mice. The results showed that there was no detectable difference in the total APP levels among NTG, A1+/AD, and A1−/AD mice (Fig. 3J), indicating that, in our AD mouse strain, the hAPP is not overexpressed compared with the endogenous mouse APP protein level. We also examined the mAPP processing in mice that do not contain the hAPP gene. In these mice, A1− also did not affect the levels of mAPP and its homologue APLP2 (29), or any of the proteolytic fragments derived from mAPP (Fig. 3K). These results led us to conclude that A1− leads to reduction in only the hAPP level, not the mAPP level. Subtle sequence differences exist between hAPP and mAPP, and these differences may play important roles in causing differential fates of hAPP and mAPP (30, 31). We investigated the effect of A1− on mutant human tau (htau) in 3XTg-AD mice. The results showed that, at 17 months of age, no significant change was observed in the number of hippocampal neurofibrillary tangles between the A1+/AD and the A1−/AD mice, suggesting that A1− may not lead to tau pathology attenuation in AD mice.
Fig. 3.
Effect of A1− on human and mouse APPs (mAPP) and their cleavage products in AD and NTG mouse brains. (A) Immunoblot analysis and (B–D) quantification. For hAPP, P = 0.042; for hCTFβ, P = 0.017; for hsAPPα, P = 0.004. Forebrains of 4-month-old mice were used; n = 7. (E) mRNA analysis of hAPP gene by real-time PCR; n = 6. (F) Immunoblot analysis and (G and H) quantification of mature and immature forms of hAPP from 25-d-old AD mouse forebrains; P = 0.019 (G) and P = 0.046 (H); n = 9. (I) For the ratio of mature to immature hAPP, P = 0.368; n = 9. (J) Immunoblot analysis and quantitation of mouse and human APP (m+hAPP) from 4-month-old NTG and A1/AD mouse forebrains. Full-length m+hAPP is detected by using antiserum 369, which recognizes both mouse and human APP (n = 7). (K) Immunoblot analysis and quantitation of mouse endogenous APP (mAPP) and its cleavage fragments from 2-month-old C57BL/6 mouse forebrains. Antiserum 369 was used to detect mAPP and the CTF fragments (mCTFβ and mCTFα). P = 0.168 for mAPP, P = 0.605 for mCTFβ, and P = 0.504 for mCTFα; n = 3. Data represent mean ± SEM. *P < 0.05; **P < 0.01.
A1− Ameliorates Cognitive Deficits of AD Mice.
(SI Results, Fig. S3).
Effects of A1− on sterol metabolism in AD mouse brains.
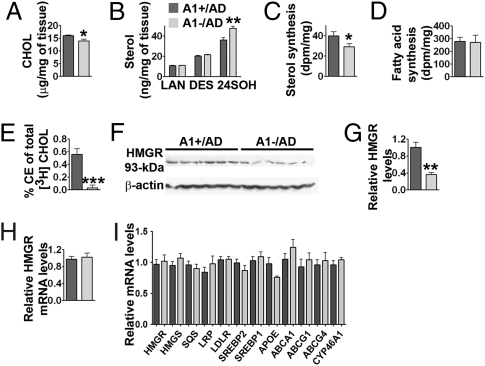
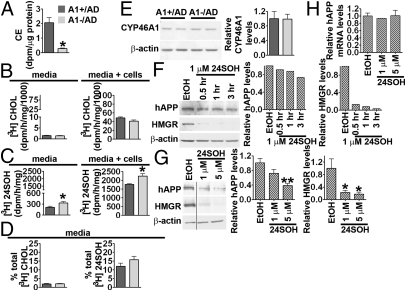
ACAT1 is involved in cellular cholesterol homeostasis. We hypothesized that A1− may cause a decrease in hAPP content by affecting sterol metabolism in AD mouse brains. To test this possibility, we isolated the sterol fractions from A1+/AD and A1−/AD mouse brains and analyzed them by GC/MS. The results showed that, at 4 months of age, lack of A1 caused an approximate 13% decrease in cholesterol content (Fig. 4A; P = 0.04) and an approximate 32% increase in 24SOH content per wet weight tissue (Fig. 4B; P = 0.007). The decrease in cholesterol content of the A1−/AD mouse brains was confirmed when a colorimetric enzyme assay kit (Wako) was employed to determine free cholesterol. The lanosterol or desmosterol contents per milligram tissue in the A1+/AD and A1−/AD mouse brains did not change significantly (Fig. 4B). Additional results showed that a lack of A1 caused an approximately 10% decrease in cholesterol content and an approximate 23% increase in 24SOH content in the 2-month-old AD mouse brains. We next compared the relative sterol synthesis and fatty acid synthesis rates in the brains of these mice in vivo. The results showed that A1− caused an approximate 28% decrease in the sterol synthesis rate (Fig. 4C; P = 0.04) without significantly changing the fatty acid synthesis rate (Fig. 4D). In mouse brains, CE contents are reported to be very low (32). We attempted to measure CE in A1+ mouse brains by separating the CE fraction from the free cholesterol fraction using column chromatography and determining the cholesterol content in CE by GC/MS after CE is saponified. The result suggested that CE might be present at no more than 1% of the total cholesterol mass in mouse brains. The low level of CE prevented us from reliably measuring a value. We used a similar procedure to determine the 24SOH ester content, and estimated that no more than 1% of total 24SOH is esterified in the brain. These results are consistent with the finding that ACAT prefers to use cholesterol to various oxysterols as its enzymatic substrate (33).
Fig. 4.
Sterol metabolism in A1+/AD and A1−/AD mouse forebrains; 4-month-old male mice were used. (A) GC-MS analysis of cholesterol (CHOL); P = 0.04; n = 5. (B) GC-MS analysis of lanosterol (LAN), desmosterol (DES), and 24SOH. For 24SOH, P = 0.007; n = 5. (C) Sterol synthesis in vivo (P = 0.04); n = 9. (D) Fatty acid synthesis in vivo; n = 9. (E) Esterification of [3H]cholesterol in mouse brains; P = 0.0009; n = 6. (F) Immunoblot analysis and (G) quantification of 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGR); P = 0.001; n = 6. (H) Relative expression of HMGR mRNA analyzed by real-time PCR; P = 0.71; n = 6. (I) Relative expressions of mRNAs of SRE response genes or LXR response genes analyzed by real-time PCR. HMGR, P = 0.71; 3-hydroxy-3-methylglutaryl CoA synthase (HMGS), P = 0.24; squalene synthase (SQS), P = 0.48; lipoprotein receptor-related protein-1 (LRP), P = 0.35; LDL receptor (LDLR), P = 0.91; sterol regulatory element–binding protein 2 (SREBP2), P = 0.35; sterol regulatory element–binding protein 1 (SREBP1), P = 0.54; apolipoprotein E (APOE), P = 0.07 (i.e., the difference approached but did not reach statistical significance); ATP-binding cassette transporter subfamily A member 1 (ABCA1), P = 0.27; ABCG1, P = 0.058; ABCG4, P = 0.63; cytochrome P450 46A1 (CYP46A1), P = 0.3; n = 6. Data represent mean ± SEM. *P < 0.05; **P < 0.01; and ***P < 0.001.
To test the functionality of ACAT1 in the intact mouse brain, we developed a procedure to measure CE synthesis in vivo by injecting [3H]-labeled cholesterol (as a cyclodextrin complex) into intact mouse brains. We monitored the 3H-CE produced in A1+ and A1− mice 3 h after injection. The result showed that, in A1+/AD mice, a small percentage of [3H] cholesterol was converted to [3H] CE (0.56% in 3 h); in contrast, such conversion was not detectable in the A1−/AD mouse brains (Fig. 4E). This result demonstrates that ACAT1 in intact mouse brains does biosynthesize CE, although at a low rate.
The data in Fig. 4 A–C suggest that, in AD mouse brains, A1− leads to an increased 24SOH level, which in turn leads to a down-regulation of the sterol synthesis rate. Studies in cell culture have suggested that 24SOH may down-regulate sterol synthesis by two mechanisms: by (i) blocking transcriptional activations of SREBP2 target genes and/or (ii) increasing the degradation rate of HMGR protein (8). To test the first possibility, we compared the mRNA levels of various SREBP2 target genes in the A1+/AD and A1−/AD mouse brains, but we failed to detect statistically significant alterations in the expression levels of these genes (Fig. 4I) (primer sequences are listed in Table S2). Additional results showed that no statistically significant alterations in the mRNA levels of various LXR target genes occurred in the brains of mice with or without A1 (Fig. 4I) (primer sequences are listed in Table S2). To test the second possibility, we performed immunoblot analysis in brain homogenates prepared from the AD mice with or without A1. The result showed that the HMGR protein content is decreased by approximately 65% in A1−/AD mouse brains (Fig. 4 F and G; P = 0.0009), whereas the HMGR mRNA in A1− mouse brains was not changed (Fig. 4H). Additional results showed that, in AD mice at 25 d of age, A1− caused an approximate 62% decrease in HMGR protein content, demonstrating that the effect of A1− on HMGR content occurs in mice at a young age.
Biosynthesis of 24SOH in Hippocampal Neuronal Cell Cultures.
The results described here show that A1−/AD mouse brains exhibit elevated 24SOH levels, suggesting that, in mouse neurons, A1− may cause an increase in the biosynthesis of 24SOH. Cultured neurons isolated from human and mouse brains synthesize and secrete 24SOH (13, 34). Based on these reports, we established a hippocampal-rich neuronal cell culture system from A1+/AD and A1−/AD mice to test this possibility. We first monitored CE biosynthesis in these neurons by incubating them with labeled [3H]oleic acid. Upon entering cells, [3H]oleic acid is rapidly converted to [3H]CE by ACAT. Both the A1+ cells and the A1− cells biosynthesize CE; however, A1− cells synthesize [3H]CE at much reduced capacity than A1+ cells (Fig. 5A). We next examined the effect of A1− on 24SOH biosynthesis by feeding neurons with the sterol precursor [3H]acetate for 3 h, then isolated and analyzed the labeled sterols present in the cells and in the media. The results showed that A1− cells exhibited a reduced trend in cholesterol synthesis rate; the difference observed between A1+ cells and A1− cells approached but did not reach statistical significance (P = 0.05; Fig. 5B Right). The 24SOH synthesis rate in A1− cells was significantly increased (by approximately 27%; Fig. 5C Right). We also analyzed the [3H]sterols in the media of A1+ and A1− cells. The result showed that the [3H]cholesterol contents were not significantly different (Fig. 5B Left); in contrast, the [3H]24SOH content in the media of A1− cells was significantly (approximately 56%) higher than in A1+ cells (Fig. 5C Left). We calculated the percentage of total [3H]sterols secreted into the media, and found that neurons secreted only approximately 2% of total [3H]cholesterol (Fig. 5D Left), but secreted 13% to 15% of total [3H]24SOH into the media (Fig. 5D Right).
Fig. 5.
Biosynthesis and regulatory activities of 24SOH in primary hippocampal neurons. For A–E, hippocampal neurons from A1+/AD and A1−/AD mice were employed. (A) Cholesterol esterification in intact cells. Cells were cultured for 7 d; lipids in cells were extracted and analyzed by TLC; P = 0.037. (B and C) Biosynthesis of [3H]sterols from [3H]acetate. Cells were cultured for 14 d. The lipids were analyzed by TLC. (B) [3H]Cholesterol (CHOL), P = 0.4 for media and P = 0.05 for cells and media. (C) [3H]24(S)-hydroxycholesterol (24SOH), P = 0.04 for media and P = 0.01 for cells and media. (D) Secretion of newly synthesized CHOL (P = 0.38) and 24SOH (P = 0.19); n = 2. (E) Immunoblot analysis of CYP46A1. Cells were cultured for 20 d; n = 2. (F) Immunoblot analysis of hAPP and HMGR in A1+/AD hippocampal neurons incubated with 1 μM 24SOH [delivered in ethanol (EtOH) at 0.1%] for 0.5 to 3 h. Cells were cultured for 35 d; (G) Effects of treating A1+/AD hippocampal neurons with 1 μM or 5 μM of 24SOH for 3 h on hAPP and HMGR levels. For 5 μM 24SOH, P = 0.0003 for hAPP and P = 0.03 for HMGR. Cells were cultured for 4 or 8 weeks; n = 3. For E–G, values were normalized against the β-actin signal in each lane. (H) Relative expression of hAPP mRNA by real-time PCR; n = 3. Data represent mean ± SEM. *P < 0.05; **P < 0.01.
The results described earlier (Fig. 5C) demonstrate that A1− causes an increased 24SOH biosynthesis rate in cultured neurons. Mouse neurons maintained in culture express CYP46A1 as a single 53-kDa protein, which can be identified by immunoblotting (13). It is possible that the increased synthesis of 24SOH observed in A1− neurons may be a result of an increase in CYP46A1 protein content in these neurons. To test this possibility, we examined CYP46A1 protein content in A1+ and A1− neurons by immunoblotting. The results showed that the intensities of the single 53-kDa protein band were comparable between these two cell types (Fig. 5E). This result suggests that, in hippocampal neurons, the mechanism(s) involved in A1−dependent increase in 24SOH synthesis may not require an increase in CYP46A1 protein content.
24SOH Provided to AD Mouse Neurons Decreases hAPP Protein Content.
The observations made in intact A1−/AD mouse brains [i.e., an increase in 24SOH content (Fig. 4B) and a decrease in hAPP content (Fig. 3A, B, and F–I)] suggest that 24SOH may decrease hAPP content in neurons. To test this possibility, we treated hippocampal neuronal culture from A1+/AD mice with 24SOH, and monitored the hAPP protein content and the HMGR protein content in parallel. We found that 1 μM 24SOH rapidly decreased the protein contents of both hAPP and HMGR (within 3 h; Fig. 5F). A separate experiment showed that 1 to 5 μM 24SOH causes a rapid decrease in hAPP protein content (Fig. 5G) without affecting its mRNA level (Fig. 5H) (primer sequences are listed in Table S1). This result supports the interpretation that accumulation of 24SOH in neurons may down-regulate hAPP protein content in vivo.
Discussion
Earlier work showed that when the ACAT inhibitor CP113818 or CI-1011 was administered to mice with AD, it significantly reduced amyloid plaques and rescued cognitive deficits, suggesting that inhibiting ACAT may prevent and/or slow the progression of AD (20, 35). The present work supports this hypothesis. However, close comparison revealed that several important differences exist between the effects of the ACAT inhibitors and the effects of A1−. CP113818 inhibited the processing of both human APP and mouse APP; CI-1011 decreased the mature/immature ratio of hAPP. In contrast, A1− causes a decrease in only the full-length human APP protein content; it does not affect the mouse APP at any level, and it does not alter the mature/immature ratio of hAPP. In addition, unlike the effect of A1−, CP113818 did not cause a reduction in the full-length hAPP content (20). The differences in results raise questions about the specificity of the ACAT inhibitors employed. ACAT is a member of the membrane bound O-acyltransferase enzyme family (36), which comprises 16 enzymes with similar substrate specificity and catalytic mechanisms, but with diverse biological functions. In addition, many ACAT inhibitors are hydrophobic, membrane-active molecules (37). When administrated to cells, they may partition into membranes at high concentration and perturb membrane properties nonspecifically. Although CP113818 and CI-1011 are designated as ACAT inhibitors, they may also inhibit other enzymes in the membrane bound O-acyltransferase enzyme family and/or interfere with other biological processes. Our present work shows that inactivating the ACAT1 gene alone is sufficient to ameliorate amyloid pathology, at least in the 3XTg-AD mouse model. In this mouse model, A1− acts to reduce Aβ load mainly by reducing the hAPP protein content. The action of A1− is similar to that of cerebrolysin, which reduces Aβ in an AD mouse model mainly by decreasing the hAPP protein content (38, 39). To explain how A1− leads to hAPP content reduction, we show that the brains of A1−/AD mice contain a significantly greater amount of 24SOH. We then demonstrate that, in neuron-rich cultures, 24SOH added to the medium leads to rapid decrease in hAPP protein content. How 24SOH acts on hAPP is currently unknown. APP may be a sterol-sensing protein (40); APP contains three CRAC motifs, a consensus motif known to bind cholesterol (41). It is possible that cholesterol and/or oxysterol may directly interact with the hAPP protein to accelerate its rate of degradation. Other possibilities cannot be excluded. The AD mice used in our current study express a mutant form of hAPP. Further investigations are required to determine whether A1− also leads to decreases in nonmutated hAPP. We also show that, in mouse brains, A1− causes a decrease in HMGR protein and a decrease in cholesterol biosynthesis. Earlier, Tabas et al. (42) and Scheek et al. (43), showed that inhibiting ACAT in macrophages or in CHO cells increases the ER “regulatory sterol pool” that mediates down-regulation of HMGR levels and SREBP processing. The “regulatory sterol” could be cholesterol itself and/or an oxysterol derived from cholesterol; however, whether oxysterol(s) play(s) important roles in regulating sterol biosynthesis in the brain in vivo is currently debated, as reviewed by Björkhem et al. (44). This issue can be addressed in the context of recent results from three different research groups: Russell and coworkers (45–47) showed that knocking out the 24-hydroxylase gene Cyp46a1 caused a near elimination in the 24SOH content and a decrease in cholesterol turnover in the mouse brains; Cyp46a1 −/− did not affect the amyloid pathology in an AD mouse model. In contrast, Hudry et al. (48) showed that over-expressing Cyp46a1 in mouse brains caused a twofold increase in 24SOH content and significantly ameliorated amyloid pathology in their AD mice. Hudry et al. (48) did not observe a reduction in the hAPP protein content; instead, they demonstrated a decrease in hAPP processing, an increase in SREBP2 mRNA, and no change in brain cholesterol content. Our present results show that, in A1−/AD mice, a 32% increase in 24SOH content and significant reductions in hAPP content and amyloid pathology occurred. The Cyp46a1 gene knockout or Cyp46a1 overexpression in mice might have produced compensatory effects that did not occur in the A1− mice, and vice versa; thus it is difficult to directly compare the results. Conversely, these (apparently conflicting) results together suggest that 24SOH may play an auxiliary but not an obligatory role in affecting cholesterol metabolism and amyloid biology. Based on other evidence, Brown and Jessup (49) have independently proposed that a given oxysterol may play auxiliary but not obligatory roles in regulating cellular cholesterol homeostasis.
We propose a mechanistic model that links cellular cholesterol trafficking with ACAT1, CYP46A1, 24SOH synthesis, hAPP, and HMGR at the ER (Fig. S4): in neurons, cholesterol trafficking in and out of the ER occurs. The unnecessary buildup of unesterified cholesterol at the ER (and other membranes) is toxic (50, 51). To minimize cholesterol accumulation, A1 located at the ER removes a portion of ER cholesterol by converting it to CE. A1− leads to an increase in the ER cholesterol pool and raises the substrate level for CYP46A1 at the ER (13), and leads to an increase in 24SOH biosynthesis in neurons. A similar scenario had previously been suggested by Sun et al. (52). The increased 24SOH and/or cholesterol concentration in the ER leads to rapid down-regulation of hAPP protein content, perhaps by accelerating its rate of degradation at the ER, thereby limiting its capacity to produce Aβ. 24SOH secreted by neurons can enter astrocytes and other cell types and lead to efficient down-regulation of HMGR and cholesterol biosynthesis in these cells. In summary, we attribute the beneficial effects of A1− on amyloid pathology in AD mouse brains to increase(s) in ER cholesterol and/or 24SOH level in the neurons. Barring the possible side effects caused by altering cholesterol metabolism in the brain, our work suggests agents that inhibit ACAT1 enzyme activity or decrease ACAT1 gene expression may have therapeutic value for treating AD in humans.
Materials and Methods
Generation of Acat1−/−/AD (A1−/AD) and Acat2−/− /AD (A2−/AD) Mice.
The Acat1−/− and Acat2−/− mice (53, 54) in C57BL/6 background were received from Sergio Fazio (Nashville, TN) and Shailesh Patel (Charleston, SC), respectively. The 3XTg-AD mice (AD mice) in hybrid 129/C57BL/6 background contain two mutant human transgenes, hAPP harboring Swedish mutation (hAPPswe), and mutant htau (htauP301L), and contain the knock-in mutant presenilin 1 (PS1M146V) (55). Breeding strategy is described in Fig. S2. Detailed methods are described in SI Text.
Statistical Analysis.
Statistical comparisons were made by using a two-tailed, unpaired Student t test. The difference was considered significant when the P value was less than 0.05.
Supplementary Material
Acknowledgments
We thank Drs. Robert Leaton, Yasuomi Urano, Gregory Holmes, Lorenzo Sempere, Scott Howell, Yohei Shibuya, and Paul Huang at Dartmouth University; Barbara Tate and Jim Harwood at Pfizer (New York, NY); Gregory Brewer at Southern Illinois University; Chunjiang Yu at University of Illinois Medical Center; and Irene Cheng at National Yang-Ming University (Taiwan, ROC) for advice during the course of this work. We also thank Stephanie Murphy for careful editing of the manuscript. The laboratory of T.Y.C. is supported by National Institutes of Health grant HL060306. The laboratory of N.G.S. is funded by Canadian Institutes of Health Research grants MOP 44363 and MOP 36496.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913828107/DCSupplemental.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolozin B. Cholesterol and the biology of Alzheimer’s disease. Neuron. 2004;41:7–10. doi: 10.1016/s0896-6273(03)00840-7. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Q, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wellington CL. Cholesterol at the crossroads: Alzheimer’s disease and lipid metabolism. Clin Genet. 2004;66:1–16. doi: 10.1111/j.0009-9163.2004.00280.x. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann T. Cholesterol, A beta and Alzheimer’s disease. Trends Neurosci. 2001;24(11) Suppl:S45–S48. doi: 10.1016/s0166-2236(00)01990-1. [DOI] [PubMed] [Google Scholar]
- 7.Dietschy JM, Turley SD. Thematic review series: brain lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- 10.Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 11.Wang N, et al. ATP-binding cassette transporters G1 and G4 mediate cholesterol and desmosterol efflux to HDL and regulate sterol accumulation in the brain. FASEB J. 2008;22:1073–1082. doi: 10.1096/fj.07-9944com. [DOI] [PubMed] [Google Scholar]
- 12.Tarr PT, Edwards PA. ABCG1 and ABCG4 are coexpressed in neurons and astrocytes of the CNS and regulate cholesterol homeostasis through SREBP-2. J Lipid Res. 2008;49:169–182. doi: 10.1194/jlr.M700364-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Russell DW, Halford RW, Ramirez DM, Shah R, Kotti T. Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem. 2009;78:1017–1040. doi: 10.1146/annurev.biochem.78.072407.103859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lütjohann D, et al. Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci USA. 1996;93:9799–9804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Muneton S, Sjövall J, Jovanovic JN, Griffiths WJ. The effect of 24S-hydroxycholesterol on cholesterol homeostasis in neurons: quantitative changes to the cortical neuron proteome. J Proteome Res. 2008;7:1606–1614. doi: 10.1021/pr7006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown J, 3rd, et al. Differential expression of cholesterol hydroxylases in Alzheimer’s disease. J Biol Chem. 2004;279:34674–34681. doi: 10.1074/jbc.M402324200. [DOI] [PubMed] [Google Scholar]
- 18.Heverin M, et al. Changes in the levels of cerebral and extracerebral sterols in the brain of patients with Alzheimer’s disease. J Lipid Res. 2004;45:186–193. doi: 10.1194/jlr.M300320-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Chang TY, Li BL, Chang CC, Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab. 2009;297:E1–E9. doi: 10.1152/ajpendo.90926.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutter-Paier B, et al. The ACAT inhibitor CP-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer’s disease. Neuron. 2004;44:227–238. doi: 10.1016/j.neuron.2004.08.043. [DOI] [PubMed] [Google Scholar]
- 21.Huttunen HJ, Kovacs DM. ACAT as a drug target for Alzheimer’s disease. Neurodegener Dis. 2008;5:212–214. doi: 10.1159/000113705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiner V, et al. Tissue expression studies on the mouse acyl-CoA: cholesterol acyltransferase gene (Acact): findings supporting the existence of multiple cholesterol esterification enzymes in mice. J Lipid Res. 1997;38:1928–1933. [PubMed] [Google Scholar]
- 23.Anderson RA, et al. Identification of a form of acyl-CoA:cholesterol acyltransferase specific to liver and intestine in nonhuman primates. J Biol Chem. 1998;273:26747–26754. doi: 10.1074/jbc.273.41.26747. [DOI] [PubMed] [Google Scholar]
- 24.Morrissette DA, Parachikova A, Green KN, LaFerla FM. Relevance of transgenic mouse models to human Alzheimer disease. J Biol Chem. 2009;284:6033–6037. doi: 10.1074/jbc.R800030200. [DOI] [PubMed] [Google Scholar]
- 25.Cai D, et al. Presenilin-1 regulates intracellular trafficking and cell surface delivery of beta-amyloid precursor protein. J Biol Chem. 2003;278:3446–3454. doi: 10.1074/jbc.M209065200. [DOI] [PubMed] [Google Scholar]
- 26.Weidemann A, et al. Identification, biogenesis, and localization of precursors of Alzheimer’s disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 27.Oltersdorf T, et al. The Alzheimer amyloid precursor protein. Identification of a stable intermediate in the biosynthetic/degradative pathway. J Biol Chem. 1990;265:4492–4497. [PubMed] [Google Scholar]
- 28.Buxbaum JD, et al. Processing of Alzheimer beta/A4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc Natl Acad Sci USA. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slunt HH, et al. Expression of a ubiquitous, cross-reactive homologue of the mouse beta-amyloid precursor protein (APP) J Biol Chem. 1994;269:2637–2644. [PubMed] [Google Scholar]
- 30.Du P, et al. Dominance of amyloid precursor protein sequence over host cell secretases in determining beta-amyloid profiles studies of interspecies variation and drug action by internally standardized immunoprecipitation/mass spectrometry. J Pharmacol Exp Ther. 2007;320:1144–1152. doi: 10.1124/jpet.106.114561. [DOI] [PubMed] [Google Scholar]
- 31.Muhammad A, et al. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci USA. 2008;105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B, et al. Reversal of defective lysosomal transport in NPC disease ameliorates liver dysfunction and neurodegeneration in the npc1-/- mouse. Proc Natl Acad Sci USA. 2009;106:2377–2382. doi: 10.1073/pnas.0810895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Chang CC, Westover EJ, Covey DF, Chang TY. Investigating the allosterism of acyl-CoA:cholesterol acyltransferase (ACAT) by using various sterols: in vitro and intact cell studies. Biochem J. 2005;391:389–397. doi: 10.1042/BJ20050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim WS, et al. Role of ABCG1 and ABCA1 in regulation of neuronal cholesterol efflux to apolipoprotein E discs and suppression of amyloid-beta peptide generation. J Biol Chem. 2007;282:2851–2861. doi: 10.1074/jbc.M607831200. [DOI] [PubMed] [Google Scholar]
- 35.Huttunen HJ, et al. Inhibition of acyl-coenzyme A: cholesterol acyl transferase modulates amyloid precursor protein trafficking in the early secretory pathway. FASEB J. 2009;23:3819–3828. doi: 10.1096/fj.09-134999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 37.Homan R, Hamelehle KL. Influence of membrane partitioning on inhibitors of membrane-bound enzymes. J Pharm Sci. 2001;90:1859–1867. doi: 10.1002/jps.1135. [DOI] [PubMed] [Google Scholar]
- 38.Rockenstein E, et al. Cerebrolysin decreases amyloid-beta production by regulating amyloid protein precursor maturation in a transgenic model of Alzheimer’s disease. J Neurosci Res. 2006;83:1252–1261. doi: 10.1002/jnr.20818. [DOI] [PubMed] [Google Scholar]
- 39.Rockenstein E, et al. Effects of Cerebrolysin on neurogenesis in an APP transgenic model of Alzheimer’s disease. Acta Neuropathol. 2007;113:265–275. doi: 10.1007/s00401-006-0166-5. [DOI] [PubMed] [Google Scholar]
- 40.Beel AJ, et al. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): does APP function as a cholesterol sensor? Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epand RM. Proteins and cholesterol-rich domains. Biochim Biophys Acta. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Tabas I, Weiland DA, Tall AR. Inhibition of acyl coenzyme A:cholesterol acyl transferase in J774 macrophages enhances down-regulation of the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase and prevents low density lipoprotein-induced cholesterol accumulation. J Biol Chem. 1986;261:3147–3155. [PubMed] [Google Scholar]
- 43.Scheek S, Brown MS, Goldstein JL. Sphingomyelin depletion in cultured cells blocks proteolysis of sterol regulatory element binding proteins at site 1. Proc Natl Acad Sci USA. 1997;94:11179–11183. doi: 10.1073/pnas.94.21.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Björkhem I. Are side-chain oxidized oxysterols regulators also in vivo? J Lipid Res. 2009;50(Suppl):S213–S218. doi: 10.1194/jlr.R800025-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lund EG, et al. Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem. 2003;278:22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- 46.Kotti TJ, Ramirez DM, Pfeiffer BE, Huber KM, Russell DW. Brain cholesterol turnover required for geranylgeraniol production and learning in mice. Proc Natl Acad Sci USA. 2006;103:3869–3874. doi: 10.1073/pnas.0600316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halford RW, Russell DW. Reduction of cholesterol synthesis in the mouse brain does not affect amyloid formation in Alzheimer’s disease, but does extend lifespan. Proc Natl Acad Sci USA. 2009;106:3502–3506. doi: 10.1073/pnas.0813349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudry E, et al. Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer’s disease. Mol Ther. 2009 doi: 10.1038/mt.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown AJ, Jessup W. Oxysterols: sources, cellular storage and metabolism, and new insights into their roles in cholesterol homeostasis. Mol Aspects Med. 2009;30:111–122. doi: 10.1016/j.mam.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Warner GJ, Stoudt G, Bamberger M, Johnson WJ, Rothblat GH. Cell toxicity induced by inhibition of acyl coenzyme A:cholesterol acyltransferase and accumulation of unesterified cholesterol. J Biol Chem. 1995;270:5772–5778. doi: 10.1074/jbc.270.11.5772. [DOI] [PubMed] [Google Scholar]
- 51.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J Clin Invest. 2002;110:905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Yao J, Kim TW, Tall AR. Expression of liver X receptor target genes decreases cellular amyloid beta peptide secretion. J Biol Chem. 2003;278:27688–27694. doi: 10.1074/jbc.M300760200. [DOI] [PubMed] [Google Scholar]
- 53.Meiner VL, et al. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc Natl Acad Sci USA. 1996;93:14041–14046. doi: 10.1073/pnas.93.24.14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buhman KK, et al. Resistance to diet-induced hypercholesterolemia and gallstone formation in ACAT2-deficient mice. Nat Med. 2000;6:1341–1347. doi: 10.1038/82153. [DOI] [PubMed] [Google Scholar]
- 55.Oddo S, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.