Abstract
Bacterial group II introns encode maturase proteins required for splicing. In organelles of photosynthetic land plants, most of the group II introns have lost the reading frames for maturases. Here, we show that the plastidial maturase MatK not only interacts with its encoding intron within trnK-UUU, but also with six additional group II introns, all belonging to intron subclass IIA. Mapping analyses of RNA binding sites revealed MatK to recognize multiple regions within the trnK intron. Organellar group II introns are considered to be the ancestors of nuclear spliceosomal introns. That MatK associates with multiple intron ligands makes it an attractive model for an early trans-acting nuclear splicing activity.
Keywords: chloroplast, RNA-binding protein, splicing, RNA-processing, plant
Group II introns are highly structured ribozymes, found in bacteria and organellar genomes of plants, fungi, protists, and some animals. They catalyze their own excision from precursor RNAs (1). In addition, bacterial group II introns are mobile genetic elements. These features, as well as structural similarities with nuclear introns, have led to the proposition that ancestors of modern group II introns have crossed kingdoms from prokaryotes to eukaryotes via eubacterial endosymbionts, and eventually evolved into nuclear introns (2). Whereas nuclear introns are removed by a large ribonucleoprotein complex called the spliceosome, each bacterial intron encodes a distinct maturase protein facilitating its splicing. The evolutionary transition from the highly specific, maturase-driven splicing toward the complex transacting spliceosomal machinery is unclear. Without any “living fossils” with a protospliceosome, a key question is how the emerging intron diversification was accompanied by a splicing machinery with the ability to act on multiple targets. At least initially, the already existing splicing factors encoded by the invading group II introns, i.e., modified maturases, may have fulfilled this task.
In present day organelles, maturase genes can be found in fungal and plant mitochondria as well as in plant chloroplasts. Although mitochondria are not directly amenable to transgenic studies, there are a number of plant species, foremost tobacco, for which directed mutagenesis of chloroplast genomes is possible (3). All land plant chloroplasts with the exception of some parasitic species in the genus Cuscuta (4, 5) contain a single maturase gene called matK in the intron of the lysine tRNA-K(UUU) gene. matK is expressed at least in green tissue (6–8). Sequence and structural homologies are found to the RNA binding motif (domain X) and the reverse transcriptase domain typically found in bacterial maturases (9, 10). Both domains are involved in splicing. By contrast, domains required for the mobility of group II introns are missing in MatK.
The function of matK in chloroplasts is so far unknown, but its position inside the trnK gene suggests a role for splicing the trnK precursor. Importantly, it has been suggested that other chloroplast introns are targeted by MatK as well: firstly, several chloroplast genomes of parasitic plants and ferns have lost trnK, but have retained a free-standing matK reading frame, suggesting additional tasks for MatK, presumably in splicing other introns (4, 5, 11–13); and secondly, loss of chloroplast translation leads to loss of splicing for some, but not all chloroplast introns, suggesting that a chloroplast reading frame is required for splicing these multiple introns (7, 15, 16). As there are no other candidates for RNA processing factors in the well-described and small chloroplast genome, it was suggested that matK is responsible for splicing these introns. If true, MatK would be an example of a group II intron maturase that has left the strict association with just a single target intron, joined by the group I intron maturase BI4 from yeast mitochondria, which supports splicing of two group I introns (17).
Attempts to generate knock-out alleles of matK to more directly assess a function of MatK in splicing failed: no homoplastomic mutant tissue could be obtained, a clear sign for matK being essential for cell survival (18). We therefore decided to investigate on a genomewide scale, with which RNAs MatK is associated in vivo and whether specific target sites can be identified. We found that MatK associates with seven intron-containing transcripts and provides evidence that the association takes place in a manner different from canonical bacterial maturase proteins.
Results and Discussion
MatK Is a Soluble Stroma Protein Expressed Predominantly in Young Leaf Tissue.
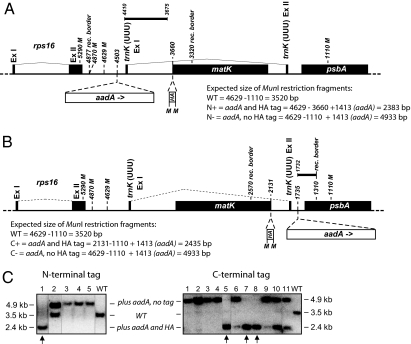
Taking advantage of homologous recombination in tobacco chloroplast (3), we appended an extension encoding the hemagglutinin (HA) antigen to the 5′- or 3′-end of the matK gene, respectively (Fig. 1 A and B). Control transformants without extensions but with selection markers were also generated. Correct integration and homoplastomy of the transgenes were verified by PCR, Southern hybridization, and segregation analysis (Fig. 1C, Fig. S1). Phenotypes of transplastomic plants were indistinguishable from WT under standard growth chamber conditions (Fig. S1), which indicates that neither the N-terminal nor the C-terminal HA-tag interferes with MatK function.
Fig. 1.
Tagging of chloroplast matK. (A) Map of the matK genomic region and description of transformation vectors for N-terminal tagging of matK (vectors pRZN+/pRZN−). Black boxes correspond to exons. Inserts are shown as open boxes. Introns are shown as thin dotted lines connecting exons. aadA, selection marker; HA, hemagglutinin tag. Bold lines indicate probe used in Southern blot experiment. Numbers refer to positions on the plastid chromosome of wild-type (WT) Nicotiana tabacum (acc. no. NC_001879). rec. border, border of chloroplast DNA fragment inserted in the transformation vector. M, MunI restriction sites relevant for Southern analysis (see C). Note that sites MunI 4629 and MunI 4870 were eliminated in pRZN+ and pRZN− for cloning strategy reasons. (B) Same as in A for C-terminal tagging of matK (vectors pRZC+/pRZC−). (C) Test for homoplastomy of transformed tobacco lines by Southern hybridization. Total DNA was digested with MunI, separated by agarose gel electrophoresis, blotted, and hybridized to a matK gene-specific probe. (Left) 5′ probe for the detection of the N-terminal sequence extension of matK. (Right) 3′ probe for the detection of the C-terminal sequence extension (probe position and restriction sites are shown in A and B). Identified MunI restriction fragments correlate with calculated sizes (note that Mun1 site 4629 is present in transplastomic plants, i.e., has not been removed during recombination with vectors pRZN+ or pRZN−). On the basis of these fragment sizes, it can be inferred that line 1 is homoplastomic for both the selective marker and the HA tag (arrow). Of the lines generated with the pRZC+ construct, numbers 5, 7, and 8 are homoplastomic for tag and cassette (arrows).
We verified expression of the tagged matK genes by Western blotting analysis and demonstrated that MatK is localized in the soluble fraction, and not the membrane fraction, of chloroplasts (Fig. 2A). We quantified matK expression during tobacco development (Fig. 2B). Intriguingly, strongest MatK accumulation was observed at day 7 postimbibition in very young tissue. In older plants, MatK is barely detectable. This expression pattern is reminiscent of activity patterns of other factors involved in chloroplast gene expression-like plastid RNA polymerases (19). It suggests that MatK is instrumental in staging early chloroplast development, whereas minute levels of MatK suffice for later maintenance of gene expression.
Fig. 2.
Expression Analysis of tagged MatK (A) Immunological detection of HA-tagged MatK protein. Chloroplasts isolated from transplastomic or WT lines were lysed and fractionated into membranes and soluble stroma. Fractions were separated by SDS-PAGE on a 12% gel, blotted and probed using an antiserum against the HA-epitope (Sigma). Arrow heads point to where an irrelevant marker lane has been removed from the images. RbcL = large subunit of RuBisCo detected by Ponceau staining. (B) Tagged MatK accumulation during tobacco development. Total protein was extracted from tobacco seeds or plants at different ages (41). Protein concentrations were determined and equal amounts of protein were subject to Western analysis. From top to bottom: a. Phenotypes of plants at different ages. b. Example Ponceau stain after transfer of proteins to a nylon membrane showing RbcL in green tissue, while this protein is absent in seeds and germinating seedlings. c. Exemplary chemolimunogram after detection of tagged MatK with an anti-HA antiserum in the eight tissues analyzed. d. Chemoluminescence signals from three independent Western analyses were quantified using a Lumi-Imager (Roche), analyzed with Quantity One software (Biorad) and plotted relative to the maximum signal obtained for 7d old plants (set to 1). Standard deviations are given as vertical bars.
MatK Associates with Seven Chloroplast Introns.
To identify MatK-bound RNAs, we immunoprecipitated N- and C-terminally tagged MatK proteins (Fig. 3A), labeled the coprecipitated RNA as well as the unbound RNA with fluorescent dyes, and hybridized both fractions competitively to a tobacco chloroplast genome tiling microarray (RNA coimmunoprecipitation and Chip analysis, RIP-Chip, refs. 20, 21). Comparison of bound/unbound ratios for samples and controls enabled calculation of a differential enrichment ratio, +HA/−HA, which was plotted against the chromosomal position of each probe on the microarray (Fig. 3B and Table S1). Seven of the eight most strongly enriched RNAs carry introns from the structurally defined group IIA intron subclass. Four tRNAs (V-UAC, I-GAU, A-UGC, and K-UUU) and two RNAs coding for ribosomal proteins (rpl2 and rps12) were enriched, as was an RNA encoding a subunit of chloroplast ATPase (atpF). The eighth signal corresponds to 23S rRNA, but scored much lower in t test analyses than the group IIA intron containing RNAs (Table S1), suggesting that the differences observed between +HA and –HA immunoprecipitations for 23S rRNA is not biologically significant. The 23S rRNA is cotranscribed with trnA-UGC and trnI-GAU. Therefore, low amounts of 23S rRNA found in the pellet of tagged MatK IPs are likely coenriched because it is part of the same precursor.
Fig. 3.
MatK specifically coimmunoprecipitates with seven group IIA introns. (A) Immunological analysis of aliquots from pellet and supernatant fractions of tagged MatK immunoprecipitations from transplastomic plants using a peroxidase-linked HA antiserum (Sigma). A 35th of the total supernatant and a 10th of the pellet fraction were loaded on the gel. P, pellet; S, supernatant; RbcL, large subunit of RuBisCo. (B) Identification of RNAs associated with HA-tagged MatK in chloroplast stroma by RNA coimmunoprecipitation and microarray analysis (RIP-Chip). The enrichment ratios (F635:F532) were normalized between four assays with C+ lines, three assays with N+ lines, three control experiments with C− lines, and three control experiments with N− lines. The background-corrected, median-normalized values for replicate spots from the tagged MatK assays were divided by those from the control assays and plotted according to their chromosomal position. Scale for y-axis is logarithmic. Corresponding data can be found in Table S1. (C) Validation of RIP-Chip data by dot-blot hybridization of radiolabeled RNAs that coimmunoprecipitated with HA-tagged MatK. RNA that coimmunoprecipitated with HA-tagged MatK was reverse transcribed in the presence of labeled deoxynucleotides and random primers, and hybridized to DNA probes on a nylon filter. For each experiment, signals were normalized to the psbE signal. (Normalization to other probes including psbA, rrn16, and rrn23 yielded similar results.) Normalized values from two C+ replicates were compared to two C− replicates to yield the relative enrichment ratios shown here (with standard deviations). The same analysis was done for one pair of N+/N− experiments. Scale for y-axis is logarithmic.
To validate our RIP-Chip findings, we analyzed coimmunoprecipitated, radiolabeled RNAs by dot-blot hybridization using membrane-immobilized probes corresponding to the eight most prominent peaks from RIP-Chip experiments, as well as several controls including all plastid introns. Signals from replicate experiments were quantified and normalized enrichment ratios, +HA/−HA, were calculated. Only the seven RNAs containing group IIA introns were significantly enriched (Fig. 3C). Neither group IIB introns nor any of the other RNAs scoring low in t tests of RIP-Chip experiments including 23S rRNA were enriched in HA-tagged MatK immunoprecipitations. Our data demonstrate that MatK interacts specifically with intron-containing RNAs, a result that is in agreement with the phylogenetically supported claim that MatK is an intron maturase. The only plastid group IIA intron not identified in either type of analysis as a MatK target was the second intron in the clpP mRNA. Further attempts to detect clpP-intron 2 in slot-blot assays were negative (Fig. S2). The clpP intron 2 is a structural outsider among group IIA introns (22) and is retained in parasitic plants that have lost matK (4, 5), suggesting that it does not require MatK for splicing. Together, our data show that MatK is a multivalent RNA-associated protein, which contrasts with the monospecific, bacterial group II intron maturases characterized to date (1).
Fine Mapping of the MatK Target Site in the trnK Precursor RNA: Association with Intronic Domains Excluding the matK Coding Region.
To narrow down the sequences associated with MatK in its target transcripts, we prepared an array that covers the tobacco trnK gene with oligonucleotides (Table S2). Similar approaches using oligos in slot-blot experiments were successfully applied to identify sequence areas of peak enrichment in Co-IP experiments of chloroplast-localized RNA-binding proteins (e.g., 21, 23). The oligos used here are with three exceptions all 50 nucleotides long. RNA isolated from stroma and incubated during immunoprecipitation is degraded due to the action of endogenous ribonucleases. This was controlled by RNA gel blot hybridization, which demonstrated that trnK precursor transcripts are indeed degraded (Fig. S3). The bulk of the degraded RNA is in a range of 200 nt to 500 nt. No intact precursor RNAs beyond that length were detectable. This suggests that fine mapping of RNAs associated with tagged MatK is possible. Like all group II introns, the trnK intron can be represented as six secondary structure domains (DI–VI) emanating from a central wheel (24). The positions of the oligos relative to these structural elements are given in Table S2. As controls, 12 oligos were chosen for rrn16, rbcL, psbA, the ndhB intron, the rpl16 intron, and the petD intron, none of which associate with MatK. This array was used in RIP-Chip experiments of N-terminally as well as C-terminally tagged MatK plus nontagged controls (three experiments for each of the four genotypes). The differential enrichment ratio (+HA/−HA) for each oligo was calculated and plotted against the oligo’s position within the trnK gene (Fig. 4A). The nonenriched sequences include the entire matK ORF, domain V, parts of DVI, and exon sequences. By contrast, all of the other oligos show enrichment values above control levels (Fig. 4). Enrichment is not a function of different GC content and thus different labeling or hybridization efficiencies (Fig. S4).
Fig. 4.
Mapping of tagged MatK binding sites on the trnK precursor. (A) Identification of trnK RNA segments associated with HA-tagged MatK in chloroplast stroma by RIP-Chip analysis. The enrichment ratios (F635:F532) were normalized between three assays for each genotype (C+, N+, C−, and N−). The background-corrected, median-normalized values for replicate spots from the tagged MatK assays were divided by those from the control assays and plotted according to their position within the trnK gene. Intron domains are marked below. (B) Enrichment values for control oligos corresponding to genes not identified in intial RIP-Chip experiments. B is based on the same set of experiments as described in A.
These data suggest that MatK makes contacts with intronic sequences upstream and downstream of the matK ORF, but does not interact with the ORF itself. These results are also in accordance with data from the initial RIP-Chip experiment, which showed two enrichment peaks corresponding to intron sequences, but no enrichment of the intervening matK sequences (Fig. 3B). Importantly, the binding surface of LtrA includes DII and regions of DI (25, 26). Both are also targets for MatK according to our RIP-Chip results, suggesting that the general positioning of MatK on the folded intron has been maintained from bacteria to chloroplasts.
Peak Enrichment of RNA Coprecipitated with MatK Is Found in Sequence Elements Inside trnK Intron Domains II and IV.
In the Lactococcus lactis L1.LtrB intron, the DIVa arm contains the maturase start codon and Shine-Dalgarno sequence and has been identified as a high-affinity binding site for LtrA (27). Binding of LtrA to DIVa is considered to be the first contact of LtrA with the intron and impacts splicing efficiency (28). In addition, this interaction represses translation of the LtrA reading frame (29) and has been suggested to be more important for intron mobility than for splicing (30). We see no strong association of the area surrounding the matK start codon or of any sequences between DIII and DIV with tagged MatK (Fig. 4A; oligos trnK-16 through -19). Thus, if MatK autoregulation is maintained in chloroplasts it is not likely to occur via docking to the translational start of MatK. Possibly, loss of binding in this area was tolerable, because the DIVa–maturase interaction was more important for intron mobility than for splicing (30), a feature definitely lost in chloroplasts.
Instead of enrichment of RNA close to the start codon, we find enrichment for an area corresponding to the 3′-end of DIV, downstream of the matK ORF (oligo trnK-39 and -40). Possibly, this area downstream of the matK ORF is a binding site for MatK. Speculatively, this binding could influence matK gene expression, as it is known that 3′-UTRs of plastid mRNAs are important determinants of RNA stability and often associate with regulatory RNA binding proteins (31).
The prime peak for enrichment of RNA in tagged MatK RIP-Chip experiments using the trnK oligo array corresponds to the oligo trnK-11. trnK-11 covers the 5′-part and the terminal loop of DII as well as D1(i) and D1(ii) (Table S2). In vitro binding studies will be needed in the future to decide on the exact binding site at a per-nucleotide resolution. Also, such in vitro work would clarify whether the associated RNA regions determined here are due to a direct interaction with tagged MatK or come about because of additional associated factors. Indeed, the nuclear-encoded RNA binding proteins WTF/RNC1 and OTP51 are known to be involved in trnK splicing (32, 33) and thus might interact with MatK. RNA recovered in pellets of tagged MatK immunoprecipitations could thus potentially be coprecipitated via a protein bridge. However, as other maturases bind RNA directly, it is more parsimonious to assume that the observed enrichments in tagged MatK RIP-Chip experiments are the result of direct MatK–RNA interactions. Another caveat of the analysis presented here is that it is not time resolved: Possibly the MatK–RNPs enriched during immunoprecipitation represent a collection of different MatK–RNA interactions caught at different stages of splicing. The contacts uncovered must therefore not necessarily take place at the same time, but could be consecutive. A dynamic role for bacterial maturases during splicing has indeed been proposed (25).
Among the intron elements with highest affinity for tagged MatK were sequences that are considered defining features of group IIA introns versus group IIB introns (24, 34), namely DI(i), the DI–II junction nucleotides as well as the DIV–DV junction. Specific contact with these areas could explain how MatK distinguishes between group IIA and group IIB introns. The exclusive association with group IIA introns is in agreement with earlier studies that reported a specific block in group IIA intron splicing upon disruption of chloroplast translation in monocotyledonous plants and suggested MatK as the responsible plastid-encoded factor (7,15,16). In dicotyledonous tobacco, plastid translation is essential for cell development, and targeted disruption of matK is not possible (18). Also, our own attempts to introduce point mutations into the matK reading frame to generate hypomorphic, viable mutants resulted exclusively in plants that retained the selective marker, but had lost the point mutations (Fig. S5). The MatK–RNA association data presented here suggest that this is attributable to the fact that MatK is required for splicing of essential plastid RNAs (four tRNA and two mRNA precursors coding for ribosomal subunits). Plastid tRNA genes and ribosomal protein genes are known to be essential in tobacco (35–37).
Our data support a model where MatK makes multiple contacts with its target intron, which suggests a large interacting surface. Such a large interacting protein–RNA interface could be instrumental for accommodating multiple introns by using different attachment points for each individual intron (28). The common and specific ways MatK associates with diverse group IIA introns can be addressed by using smaller tiling arrays for other MatK targets like those shown here for the trnK precursor or by in vitro binding studies. Our finding that a bacterial-type maturase is capable of diversifying its target intron range has intriguing ramifications for the evolution of nuclear introns. They show that a single organellar maturase could have served a multitude of introns after transfer of group II introns from organelles into the nuclear genome. Thus, MatK is a model for an early protospliceosomal activity during the arrival and initial spread of group II introns through the eukaryotic genome. Therefore, understanding how MatK accommodates multiple introns has the potential to help understand the mode of action of trans-acting proteins in the spliceosome.
Methods
Cloning and Generation of Transplastomic Lines.
Vector pRZN+: The target region for homologous recombination was amplified from purified chloroplast DNA using primers 5′matKAu1 and 5′matKAu2 and cloned into pBluescript II SK+ (Stratagene) via SalI linkers. A point mutation in primer 5′matKAu1 eliminated a first MunI site. A second MunI site in the intergenic matK-rps16 spacer was eliminated by restriction of the site, subsequent blunting with Pfu polymerase (Phusion; Finnzymes) and religation. The aadA-selective marker was introduced blunt-end into a blunted Bsp119I site. The matK start codon was changed to a leucine codon by site-directed mutagenesis and replaced with a MunI site, which required replacement of an A by a T using primers 5′matKIn1 and 5′matKIn2. A DNA fragment with MunI linkers encoding the hemagglutinin epitope was introduced into this MunI site. Vector pRZN− is identical to pRZN+, but lacks the HA tag and carries instead the wild-type matK 5′ terminus.
Vector pRZC+: The target region for homologous recombination was amplified from purified chloroplast DNA using primers 3′matKAu1 and 3′matKAu2, and cloned into pBluescript II SK+ (Stratagene) via SalI linkers and sequenced. The aadA-selective marker cassette was inserted into a blunted BglII site (38). The matK stop codon was eliminated by site-directed mutagenesis and replaced with a MunI site, which required addition of a single T using primers 3′matKIn1 and 3′matKIn2. A DNA fragment with MunI linkers coding for the hemagglutinin epitope was introduced into this MunI site. Vector pRZC− is identical to pRZC+, but lacks the HA tag and carries instead the wild-type matK 3′ terminus. The sequences of all four final transformation vectors have been deposited in GenBank (FN396957-FN396960).
Tobacco plastids were transformed by particle bombardment as described previously (3). Plastid transformants were analyzed for correct integration of the aadA cassette by PCR using the primer pairs, N+psbArev2 and aadA-RB-for or 5′matK-test-rps16 and aadA-LB-rev. Presence of the HA tag was verified by Southern blot, PCR, and immunologically.
Analysis of Coprecipitated RNA.
RIP-Chip analysis of HA-tagged MatK was performed essentially as described previously (21) using a full-genome tobacco chloroplast microarray (20) or a custom-made oligonucleotide array. Five microliters of anti-HA antiserum (Sigma) were used for each immunoprecipitation. Full-plastome RIP-Chip: Hybridization was carried out with the hybridization buffer III (Perkin-Elmer ASAP micromax labeling kit) at 50 °C overnight. Washes were done for 15 min in 0.01% SDS/0.5× SSC; 15 min 0.01% SDS and 0.06× SSC; and 15 min 0.06× SSC (all at room temperature, RT). After scanning in a Scanarray Gx unit (Perkin-Elmer) and extraction of raw data with the GenePix 6.0 software (Axon), enrichment ratios of signals from pellet fractions to supernatant fractions were determined for seven biological replicates—three from N-terminally tagged plants and four from C-terminally tagged plants. For each tag, 3 control RIP-Chip experiments were analyzed as well. All 13 experiments were normalized and cleaned for low signals or poor spots (21). Only probes for which more than half of all spots passed these citeria were further considered. trnK-oligo RIP-Chip: the analysis was carried out similarly to the full-genome array with the following modifications: Hybridization was done at 40 °C. Washes were done each for 8 min in 0.02% SDS/1× SSC (40 °C), 1× SSC (RT), 0.2× SSC (RT), and 0.05× SSC (RT). Altogether 12 experiments form the basis for the analysis, 3 for each genotype (C+, C−, N+, and N−).
Dot-blot analysis of RNA coimmunoprecipitated with HA-tagged MatK: Chloroplast genes were amplified by PCR. Amplification products (1 μg) were denatured with 0.5 M NaOH before spotting on Hybond-N+ membranes (Amersham) using a vacuum manifold. The DNA was fixed to the membrane by UV-crosslinking (250 mJ/cm2). RNA coprecipitated with HA-tagged MatK was DNase digested and reverse transcribed using the Quantitect reverse transcriptase kit (Qiagen) in the presence of α32P-dCTP, α32P-dGTP, and α32P-dATP. cDNA products were purified by gel filtration using Microspin G50 columns (Amersham), then denatured for 5 min at 96 °C and hybridized to spotted probes at 48 °C overnight in Church buffer (39). Blots were washed (10 min/wash) at 48 °C as follows: twice with 1× SSC/0.5% SDS, once with 0.5× SSC/0.1% SDS, and twice with 0.2× SSC/0.1% SDS. Signals were detected using a phosphorimaging system (Personal Molecular Imager FX; Bio-Rad) and quantified using the Quantity One software (Bio-Rad). Fig. 3C is based on IPs from a total of six independent experiments: two C+ extracts, two C− (negative) controls, one N+ extract and one N− (negative) control. For each blot, all spots were normalized to the psbE signal.
Slot-blot analysis of RNA coimmunoprecipitated with tagged MatK was carried out as described previously (21).
Analysis of Proteins.
Chloroplasts were prepared as previously described (40). Chloroplasts were lysed by incubation 10 min on ice in hypotonic lysis buffer (30 mM hepes–KOH pH 7.7, 10 mM Mg acetate, 200 mM K acetate, 2 mM DTT, and a mixture of protease inhibitors) and pulled 30 times through a 21G 1.5 gauge needle. Membranes were pelleted by centrifugation for 30 min at 30,000 g and 4 °C in a Sorvall RC6 centrifuge (SS34 rotor). Aliquots of the membrane fraction resuspended in lysis buffer were used directly in standard Western blot analysis. The supernatant constitutes the stromal fraction used for RIP-Chip experiments.
Supplementary Material
Acknowledgments
We gratefully acknowledge the excellent technical assistance provided by Simone Hardel, Janne Saint-Paul, and Reik Modrozynski and also thank Michi Tillich and Alice Barkan for critical discussions. This work was funded by a Deutsche Forschungsgemeinschaft (DFG) Emmy-Noether fellowship (to C.S.) and a Deutscher Akademischer Austausch Dienst (DAAD) short-term fellowship to (R.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences of all four final transformation vectors have been deposited in GenBank (FN396957–FN396960).
This article contains supporting information online at www.pnas.org/cgi/content/full/0909400107/DCSupplemental.
References
- 1.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. Five easy pieces. Science. 1991;254:663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- 3.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS One. 2009;4:e5982. doi: 10.1371/journal.pone.0005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barthet MM, Hilu KW. Expression of matK: Functional and evolutionary implications. Am J Bot. 2007;94:1402–1412. doi: 10.3732/ajb.94.8.1402. [DOI] [PubMed] [Google Scholar]
- 7.Vogel J, Börner T, Hess WR. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.du Jardin P, Portetelle D, Harvengt L, Dumont M, Wathelet B. Expression of intron-encoded maturase-like polypeptides in potato chloroplasts. Curr Genet. 1994;25:158–163. doi: 10.1007/BF00309542. [DOI] [PubMed] [Google Scholar]
- 9.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barthet MM, Hilu KW. Evaluating evolutionary constraint on the rapidly evolving gene matK using protein composition. J Mol Evol. 2008;66:85–97. doi: 10.1007/s00239-007-9060-6. [DOI] [PubMed] [Google Scholar]
- 11.Duffy AM, Kelchner SA, Wolf PG. Conservation of selection on matK following an ancient loss of its flanking intron. Gene. 2009;438:17–25. doi: 10.1016/j.gene.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Gao L, Yi X, Yang YX, Su YJ, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol. 2009;9:130. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel J, Hübschmann T, Börner T, Hess WR. Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: Support for MatK as an essential splice factor. J Mol Biol. 1997;270:179–187. doi: 10.1006/jmbi.1997.1115. [DOI] [PubMed] [Google Scholar]
- 15.Hübschmann T, Hess WR, Börner T. Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol Biol. 1996;30:109–123. doi: 10.1007/BF00017806. [DOI] [PubMed] [Google Scholar]
- 16.Hess WR, et al. Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell. 1994;6:1455–1465. doi: 10.1105/tpc.6.10.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labouesse M, Netter P, Schroeder R. Molecular basis of the ‘box effect’, a maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. Eur J Biochem. 1984;144:85–93. doi: 10.1111/j.1432-1033.1984.tb08434.x. [DOI] [PubMed] [Google Scholar]
- 18.Drescher A. München: LMU München; 2003. ycf1, ycf14 und RNA-Edierung: Untersuchungen an im Lauf der Plastidenevolution neu hinzu gewonnenen Genen und Eigenschaften. PhD thesis. [Google Scholar]
- 19.Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S. Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol. 2006;142:993–1003. doi: 10.1104/pp.106.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura T, et al. Array-based analysis on tobacco plastid transcripts: preparation of a genomic microarray containing all genes and all intergenic regions. Plant Cell Physiol. 2003;44:861–867. doi: 10.1093/pcp/pcg101. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz-Linneweber C, et al. A pentatricopeptide repeat protein binds to and facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 25.Dai L, et al. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell. 2008;30:472–485. doi: 10.1016/j.molcel.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- 28.Matsuura M, Noah JW, Lambowitz AM. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh RN, Saldanha RJ, D’Souza LM, Lambowitz AM. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol. 2002;318:287–303. doi: 10.1016/S0022-2836(02)00054-2. [DOI] [PubMed] [Google Scholar]
- 30.Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol. 2004;340:211–231. doi: 10.1016/j.jmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Bollenbach TJ, Schuster G, Portnoy V, Stern D. Processing, degradation, and polyadenylation of chloroplast transcripts. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol. 19. Berlin, Heidelberg: Springer; 2007. pp. 175–211. [Google Scholar]
- 32.de Longevialle AF, et al. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008;56:157–168. doi: 10.1111/j.1365-313X.2008.03581.x. [DOI] [PubMed] [Google Scholar]
- 33.Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci USA. 2009;106:4537–4542. doi: 10.1073/pnas.0812503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 35.Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol. 2008;15:192–198. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- 36.Rogalski M, Ruf S, Bock R. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 2006;34:4537–4545. doi: 10.1093/nar/gkl634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legen J, Wanner G, Herrmann RG, Small I, Schmitz-Linneweber C. Plastid tRNA genes trnC-GCA and trnN-GUU are essential for plant cell development. Plant J. 2007;51:751–762. doi: 10.1111/j.1365-313X.2007.03177.x. [DOI] [PubMed] [Google Scholar]
- 38.Koop HU, et al. Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta. 1996;199:193–201. doi: 10.1007/BF00196559. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Voelker R, Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.