Abstract
Centrosomes have recently emerged as key regulators of the cell cycle. The G1/S transition requires a functional centrosome, and centrosomal localization of numerous proteins, including cyclin/Cdk complexes, is important for the G2/M transition. Here we identify a modular centrosomal localization signal (CLS) localizing cyclin A to centrosomes independently of Cdk binding. The cyclin A CLS is located in a distinct part of the molecule compared with the cyclin E CLS and includes the MRAIL hydrophobic patch involved in substrate recognition. The cyclin A CLS interacts with p27KIP1, and expression of p27KIP1 removes cyclin A but not cyclin E from centrosomes. Expression of the cyclin A CLS displaces both endogenous cyclin A and E from centrosomes and inhibits DNA replication, supporting an emerging concept that DNA replication is linked to centrosomal events. Structural analysis indicates that differences in surface charge and length of the C-terminal helix explain why the MRAIL region in cyclin E is not a functional CLS. These results indicate that the cyclin A CLS may contribute to targeting and recognition of centrosomal Cdk substrates and is required for specific effects of p27KIP1 on cyclin A-Cdk2.
Keywords: centrosomal localization signal, p27KIP1, cell cycle, hydrophobic patch
Centrosomes, the major microtubule organizing centers in animal cells, have been intensively studied for their role in building and organizing the mitotic spindle poles (1). Recent studies have shown they are also crucial for multiple steps throughout the entire cell cycle (2–4). Removal of centrosomes by microsurgery or laser ablation leads to G1 arrest of the acentriolar cell (5, 6), and G1 arrest was also observed after siRNA-mediated depletion of proteins from the pericentriolar material or the centrioles (7). Furthermore, centrosomal localization and activation of numerous regulatory proteins have been shown to be important for the G2/M transition (4), as well as the metaphase to anaphase transition (8–10). Altogether, these findings indicate that centrosomes regulate cell cycle progression by serving as a scaffold capable of concentrating, coordinating, and modulating more than 100 regulatory proteins (11). Thus, defining the mechanisms controlling centrosomal localization of proteins is of major importance in understanding cell cycle control.
The enzymes present on centrosomes and involved in controlling cell cycle transitions include the cyclin-Cdk complexes and regulators of Cdk activity (12–15). During the G1/S transition and S phase progression, Cdk2 activity is positively regulated through association with either cyclin E or cyclin A (16, 17). These cyclins have different functions during the cell cycle: cyclin E is largely required for the G1/S transition, whereas cyclin A is important for late S phase progression and the G2/M transition (18, 19). However, a certain redundancy exists between these cyclins: in absence of cyclin E-Cdk2, cyclin A-Cdk2 can promote the G1/S transition (20–22). In fact, it has been suggested that the differences in function between these cyclin-Cdk complexes are due less to their specificity toward particular substrates than to their presence or absence at the appropriate location and time during the cell cycle (23). Recently, we found that cyclin E centrosomal localization was mediated by a 20-aa motif termed the CLS (centrosomal localization signal) (14). The acceleration of S phase entry by cyclin E overexpression proved to be independent of Cdk2 binding but dependent on centrosomal localization and CLS integrity (14). Expression of the cyclin E CLS itself displaces both endogenous cyclins E and A from centrosomes (14), suggesting that cyclin A may also possess a CLS. However, the domain in cyclin A responsible for centrosomal localization has not been defined, nor has the potential requirement of Cdk binding for cyclin A centrosomal localization been examined. Comparing the sequences and crystal structures of cyclin E and cyclin A (24), it is evident that most of the cyclin E CLS domain is not present in the cognate region of cyclin A. Therefore, a systematic analysis of cyclin A localization was undertaken and led to the discovery of a distinct domain both necessary and sufficient for targeting the centrosome. We demonstrate that cyclin A centrosomal localization is independent of Cdk binding and is important for cell cycle progression through S phase. Finally, cyclin A, but not cyclin E, is displaced from centrosomes by the Cdk inhibitor p27KIP1, providing insight into the mechanism by which p27KIP1 may mediate cell cycle arrest.
Results
Cyclin A CLS Is Located in the N-Terminal Cyclin Box Fold Containing the Hydrophobic Pocket.
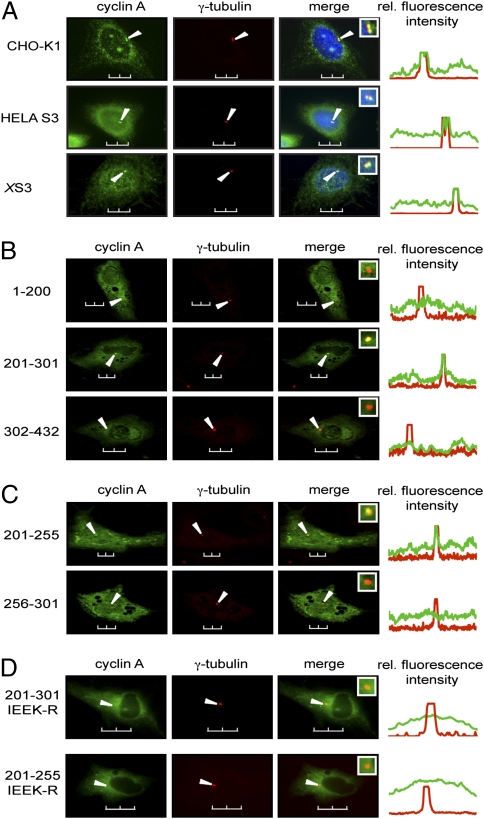
We first examined the localization of endogenous cyclin A in multiple different cell lines. As shown in Fig. 1A, mammalian and Xenopus cyclin A are present both in the nucleus and the cytoplasm of asynchronized cells, and a detectable fraction colocalizes with γ-tubulin, a specific centrosomal marker (25). The amino acid sequence alignment of human and Xenopus cyclin A shows a high level of conservation, with 71% similarity between the full-length sequences (Fig. S1). This similarity increases to 86% in the region containing the two cyclin box folds (CBOX 1 and 2) (26) that are shared by all cyclins, and it increases even further to 98% within CBOX1, which contains the MRAIL hydrophobic patch (Fig. S1). The region of cyclin A responsible for centrosomal localization was determined by transiently transfecting EGFP-tagged truncation constructs of cyclin A into Xenopus S3 cells and examining colocalization with γ-tubulin by confocal microscopy. For initial experiments, three cyclin A deletion constructs were investigated: the N-terminal domain containing the N-terminal helix (amino acids 1–200); the N-terminal cyclin box fold (CBOX1; amino acids 201–301); and the C-terminal cyclin box fold (CBOX2) with the C-terminal helix (amino acids 302–432) (Fig. S1). As shown in Fig. 1B, the N-terminal domain of cyclin A (1–200) does not localize to the centrosome. However, a construct containing CBOX1 (201–301) did localize to centrosomes with nearly the same efficiency as full-length cyclin A, whereas a construct containing CBOX2 (302–432), corresponding to the CLS-containing region of cyclin E (14), did not localize to centrosomes. Further truncation analysis revealed that amino acids 201–255 were sufficient for centrosomal localization, whereas amino acids 256–301 did not localize to centrosomes (Fig. 1C). Altogether, it is evident that the CLS motif of cyclin A is contained within the first 55 amino acids of the CBOX1 region (amino acids 201–255).
Fig. 1.
The MRAIL-containing sequence within CBOX1 localizes cyclin A to the centrosome. (A) Asynchronous CHO-K1, HeLa S3, and Xenopus S3 (XS3) cells were methanol-fixed and immunostained with antibodies to cyclin A (green) and γ-tubulin (red). (B and C) Plasmids encoding the indicated human cyclin A truncations fused to EGFP (amino acid numbers shown on the left) were transfected into XS3 cells. Cells were methanol-fixed and immunostained with antibodies to γ-tubulin. Expression and localization of the EGFP constructs were observed by direct fluorescence (green) and compared with immunolocalization of γ-tubulin (red). (D) Plasmids encoding human cyclin A truncations mutated on critical surface residues (IEEK-R) and fused to EGFP were transfected into XS3 cells. Their expression and localization were analyzed as in B. Arrows indicate the position of centrosomes. Insets: Magnification of the centrosomal region in the merged image. Line scans measuring centrosome-associated relative fluorescence intensity (rel. fluorescence intensity) are displayed on the right, with the green and red lines representing the GFP- and the γ-tubulin-associated fluorescence, respectively. (Scale bars, 10 μm.)
Within the cyclin A CLS lies the “MRAIL” sequence motif that is highly conserved in the α1 helix of CBOX1 among different cyclins (27, 28). Several conserved residues are on the surface of the α1 helix in a position accessible for interactions with substrates and regulators of the cyclin A-Cdk complexes (29–31). Therefore, point mutations were generated to determine whether these residues are important for cyclin A CLS functions. Mutation to arginine of I213 in the MRAIL sequence, as well as three other solvent-accessible residues (E220, E224, and K226), would not be expected to alter the structure of cyclin A because these substitutions retain the primarily hydrophilic character of the original amino acids. However, the arginines would protrude and potentially block binding and functionality of the cyclin A CLS. Indeed, Fig. 1D shows that the cyclin A CLS containing these four substitutions (IEEK-R) does not localize to centrosomes.
Cyclin A CLS Functions Independently of Cdk Binding.
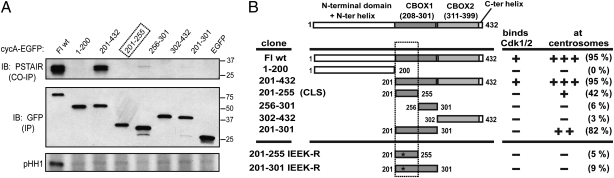
The region in cyclin A containing both cyclin box folds (amino acids 201–432) has been previously proposed to bind Cdk2 (24). To determine whether Cdk binding is required for cyclin A centrosomal localization, the cyclin A-EGFP constructs described above were expressed in Xenopus S3 cells and examined for Cdk binding by coimmunoprecipitation (Fig. 2A, Middle and Top, respectively). Moreover, the precipitates were analyzed for histone H1 kinase activity (Fig. 2A, Bottom). As shown in Fig. 2A, full-length cyclin A and a construct encoding amino acids 201–432 containing both cyclin box folds efficiently bound Cdk (Top), but kinase activity was evident only with the full-length construct (Fig. 2A, Bottom). These results are in agreement with a previous study predicting the importance of the N-terminal helix (amino acids 183–194) in reinforcing contacts between cyclins and Cdks, as well as stimulation of kinase activity (26). The other constructs, including the entire CBOX2 after the CLS (302–432), did not bind Cdk and had no detectable associated kinase activity. A relationship between the centrosomal localization and the Cdk-binding abilities of these truncated versions of cyclin A is summarized in Fig. 2B. Because fragments encoding the cyclin A CLS were sufficient for centrosomal localization but not for interaction with Cdk, it is evident that the cyclin A CLS functions independently of Cdk binding.
Fig. 2.
Cyclin A centrosomal localization is independent of Cdk binding. (A) Plasmids encoding cyclin A-EGFP truncations were transfected into XS3 cells, and the resulting proteins were immunoprecipitated with an antibody to GFP (Middle). Coimmunoprecipitation of Cdk1/2 was analyzed by Western blot using an anti-PSTAIR antibody (Top). (Bottom) Histone H1 kinase activity of the cyclin A-EGFP/Cdk immune complexes (pHH1, autoradiograph). (B) Graphic representation of the different cyclin A-EGFP constructs transfected into Xenopus S3 cells and tested for their ability to localize at the centrosomes and to bind Cdk1/2. For centrosomal localization, +, ++, and +++ represent 30–50%, 50–75%, and >75% of cells with centrosomal staining, respectively. The percentage of cells with centrosomally localized cyclin A from four independent experiments is in parentheses. The stars show the position of the four mutations. Boxes indicate the cyclin A CLS.
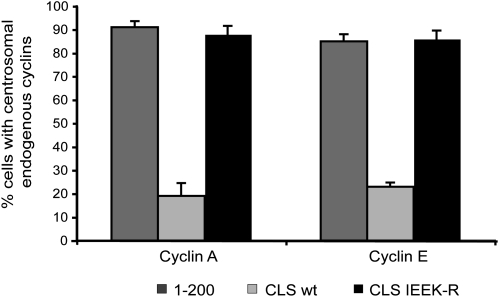
Fig. 3.
Overexpression of the cyclin A CLS displaces endogenous cyclins A and E from centrosomes. Bar graph showing the percentage of cells with endogenous cyclins A (Left) or E (Right) localized on centrosomes in the presence of cyclin A 1–200 (dark gray), cyclin A CLS wild type (light gray), or cyclin A CLS IEEK-R (black). Bars indicate mean ± SEM (n = 4). The corresponding immunofluorescence is displayed in Fig. S2.
Expression of the Cyclin A CLS Displaces Endogenous Cyclins A and E from Centrosomes.
It has been previously demonstrated that expression of the cyclin E modular CLS domain displaces both endogenous cyclin E and cyclin A from the centrosome (14). To assess whether the cyclin A CLS is capable of similar displacement, we transiently transfected the cyclin A CLS into CHO-K1 cells synchronized at the G1/S boundary and examined the centrosomal localization of endogenous cyclin E and cyclin A. As shown in Fig. S2A, with the cyclin A 1–200 construct that does not localize on centrosomes, both endogenous cyclin E and cyclin A (blue) colocalize with γ-tubulin (red), indicative of centrosomal localization (appearing pink in the enlarged box of the merged panels). However, after expression of the cyclin A CLS (201–255; Fig. S2B), neither endogenous cyclin A nor cyclin E were colocalized with γ-tubulin (yellow centrosomes in the merged picture), indicative of displacement from the centrosome. Furthermore, this displacement depended on the presence of a functional CLS, because expression of the mutant CLS (IEEK-R) that does not localize on centrosomes did not displace endogenous cyclins A or E (Fig. S2C). The percentage of transfected cells with centrosomal endogenous cyclins A and E under all these conditions is summarized in Fig. 3. Taken together, although the cyclin A CLS resides in a region of the molecule different from the cyclin E CLS, it is functionally homologous because it displaces both cyclins A and E from centrosomes.
Expression of the CLS Wild Type but Not the CLS IEEK-R Inhibits EdU Incorporation into DNA.
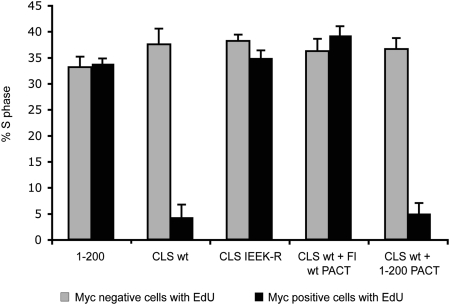
To investigate the effect of the cyclin A CLS on cell cycle progression, and particularly on DNA replication, various 6-Myc-tagged cyclin A constructs were transfected into unsynchronized CHO-K1 cells, followed by incubation with EdU (5-ethynyl-2’-deoxyuridine), a modified nucleoside incorporated during DNA synthesis (Fig. 4). Cells expressing a cyclin A construct unable to localize on centrosomes (1–200) showed EdU incorporation in ≈35% of the cells, similar to untransfected cells (Fig. S3A and Fig. 4, gray bars). In contrast, expression of the wild-type cyclin A CLS reduced incorporation to <5% (Fig. S3A and Fig. 4, black bars), whereas expression of the CLS construct carrying the four mutations (IEEK-R) that prevent its centrosomal localization did not reduce EdU incorporation. Because cyclin A CLS expression leads to displacement of both endogenous cyclins A and E and possibly other associated proteins from centrosomes (Fig. S2 and Fig. 3), inhibition of DNA synthesis could be due to other effects of cyclin A CLS expression besides displacement of cyclin A. To exclude this possibility, cyclin A was specifically targeted back to centrosomes by expressing the full-length cyclin A wild type fused to the PACT domain (Fl wt PACT), a motif responsible for targeting AKAP450 and pericentrin to centrosomes (32). As shown in Fig. S3B, both cyclin A Fl WT-PACT and cyclin A 1–200-PACT (1-200 PACT) localize on centrosomes, even in the presence of the cyclin A CLS. When coexpressed with the cyclin A CLS, cyclin A Fl WT-PACT but not cyclin A 1–200-PACT restored EdU incorporation into DNA, indicating that the centrosomal pool of cyclin A is required for DNA replication (Fig. S3A and Fig. 4).
Fig. 4.
Overexpression of the cyclin A CLS inhibits DNA replication. Unsynchronized CHO-K1 cells were transfected with the indicated cyclin A-6myc constructs and incubated with EdU to follow DNA replication. Results are represented as a bar graph showing the percentage of cells in S-phase after transfection with the indicated constructs. For each experiment, ≈100 cells were counted. Experiments were done in triplicate. Bars indicate mean ± SEM (n = 3). The corresponding EdU fluorescence is displayed in Fig. S3A.
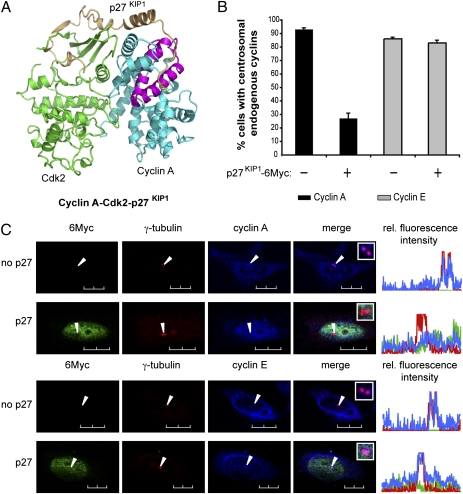
Cdk Inhibitor p27KIP1 Displaces Cyclin A but Not Cyclin E from Centrosomes.
Although the cyclin A CLS is not directly involved in the binding of Cdk2 (Fig. S4), it overlaps the MRAIL motif known for its important role in substrate/regulator selection (27). One of the most important regulators of cyclin A-Cdk2 activity is the Cdk inhibitor p27KIP1 (33, 34). The C terminus of p27KIP1 is thought to prevent the binding of substrates to cyclin-Cdk complexes and binds cyclin A in the region that includes the MRAIL and the cyclin A CLS (Fig. 5A) (27, 31). To test whether expression of p27KIP1 could interfere with CLS function, we transfected CHO-K1 cells with a construct encoding the full-length Myc-tagged human p27KIP1 and investigated the centrosomal localization of both endogenous cyclins A and E. The results summarized in Fig. 5B show that expression of p27KIP1 strongly displaces endogenous cyclin A from centrosomes (Fig. 5B, black bars, and Fig. 5C, Top). In the same experiment, the localization of endogenous cyclin E was not affected by the expression of p27KIP1 (Fig. 5B, gray bars, and Fig. 5C, Bottom), suggesting that centrosomal cyclin A-Cdk2 is differentially regulated by p27KIP1.
Fig. 5.
The Cdk inhibitor p27KIP1 displaces cyclin A but not cyclin E from centrosomes. (A) Ribbon diagram (adapted from ref. 31) showing that the interaction between p27KIP1 (gold) and the cyclin A-Cdk2 complex (blue and green, respectively) involves the cyclin A CLS region (purple). (B) Bar graph showing the percentage of cells with centrosomal endogenous cyclins A (black) or E (gray) after transfection with p27KIP1-6Myc, as illustrated in C. Bars indicate mean ± SEM (n = 3). (C) CHO-K1 cells were transfected with a plasmid encoding p27KIP1-6Myc. Cells were methanol-fixed, and expression of p27KIP1-6Myc was assessed by indirect immunofluorescence using an anti-Myc antibody (green). Localization of endogenous cyclin A (Top) and E (Bottom) was monitored by confocal microscopy (blue) and compared with the centrosomal localization of γ-tubulin (red). Arrows indicate the position of centrosomes. Insets: Magnification of the centrosomes from the merged image. Line scans measuring centrosome-associated relative fluorescence intensity (rel. fluorescence intensity) are displayed on the right, with the green, blue, and red lines representing the 6Myc, the endogenous cyclins, and the γ-tubulin fluorescence, respectively. (Scale bars, 10 μm.)
Discussion
The results in this study provide important information about a functional domain in cyclin A that is responsible for centrosomal localization. This domain resides between amino acids 201 and 255 in CBOX1 and is sufficient for localization on centrosomes. Like the cyclin E CLS, even though it is located in a different part of the molecule, the cyclin A CLS can function as a modular domain that has no contact with Cdk2 (Fig. S4) and that localizes to the centrosome independently of Cdk binding. Additionally, when expressed in cells, the cyclin A CLS, but not its mutated derivative (IEEK-R), displaces both endogenous cyclin A and cyclin E from centrosomes and greatly reduces DNA synthesis. This ability is consistent with the similar sequences and 3D structures of the two CLS domains, suggesting they may function through conserved mechanisms (Fig. 6). The mechanism by which both cyclin CLS domains prevent Edu/BrdU incorporation into DNA has not yet been identified, but it seems likely that displacement of endogenous cyclins from the centrosome by the expressed CLS is responsible for inducing this phenotype. Indeed, specifically targeting cyclin A back to centrosomes by expressing a PACT domain-fused cyclin A construct restored DNA replication to its normal level (Fig. 4). Furthermore, the inhibition of cell cycle progression around the G1/S transition by cyclin displacement from the centrosome is consistent with previous evidence that acceleration of DNA synthesis by cyclin E required its centrosomal localization (14). Altogether, these results suggest that the centrosomal pool of cyclins plays an important role in cell cycle progression through S phase and indicate the importance of communication between centrosomes and the nucleus.
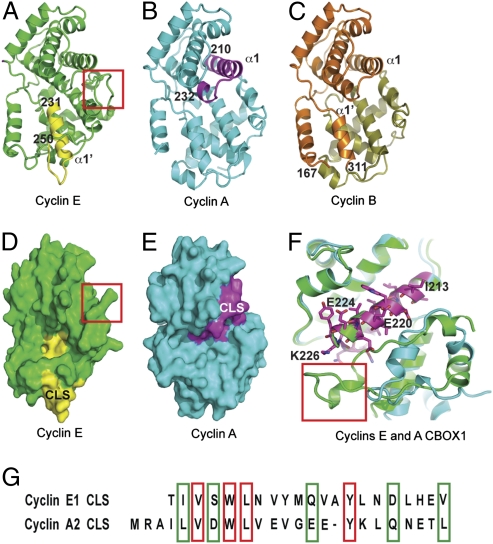
Fig. 6.
The CLSs of cyclins E and A are in distinct molecular regions. (A–C) Ribbon diagrams showing, respectively, the position of the cyclin E [Protein Data Bank (PDB) ID 1W98] and the cyclin A CLS (PDB ID 1VIN) within the 3D representations of the entire molecules, as well as the region of cyclin B (PDB ID 2JGZ) sufficient for centrosomal localization. The cyclin E CLS (A; yellow) belongs to the first helix of CBOX2 (231–251), whereas the cyclin A CLS (B; magenta) belongs to the first helix of CBOX1 (amino acids 210–232), and the cyclin B region sufficient for centrosomal localization (C; orange) belongs to CBOX1 and the beginning of CBOX2 (amino acids 166–311). (D and E) Space-filling models corresponding to A and B, respectively. D clearly shows that the C-terminal helix (red box) impinges on the “cyclin A CLS-like” region and may explain why this region is not a functional CLS in cyclin E. (F) Superimposition of cyclin A and E in the region containing the cyclin A CLS (magenta). The structure was rotated by ≈120° in the clockwise direction compared with Fig. 5 A–E. Residues exposed at the surface and substituted by arginines in IEEK-R mutant are annotated. This ribbon diagram also points out the differences between the C-terminal helices of cyclin A and E (bottom left corner). In cyclin E, the C-terminal helix (boxed in red) is longer than in cyclins A and B and may interfere with the accessibility of the “cyclin A CLS-like” region for protein–protein interaction. (G) Alignment of cyclin A and cyclin E CLS amino acid sequences, with the identical residues boxed in red and similar residues boxed in green.
It is of considerable interest that the CLS sequences of both cyclin E and cyclin A have similar effects on cyclin localization and cell cycle control despite residing in different parts of their respective molecules. Fig. 6 A–C shows the positions identified as the CLS in cyclin E (yellow) and cyclin A (magenta), as well as the region sufficient for localizing cyclin B (orange) to centrosomes. The cyclin A CLS is largely contained in the α1 helix of CBOX1, which is a subdomain within the larger region identified as both necessary and sufficient for cyclin B centrosomal localization (amino acids 167–311) (35). In contrast, the cyclin E CLS is in the α1’ helix in CBOX2 (14, 24). This location is clearly distinct from the location that we have defined as the CLS in cyclin A but is at least partially within the large region defined as sufficient to localize cyclin B to centrosomes (compare Fig. 6 A–C). The CLS in cyclin E comprises a loop domain (α1’–α2’ loop) that is greatly reduced in size in the cognate region of cyclins B and A, and the amino acid sequences from the cyclin E, the cyclin B, and the cyclin A α1’ helices share low similarity (24, 28). This may at least partially account for why this region of cyclin A does not function as a CLS.
The cyclin A CLS includes the MRAIL sequence, which is highly conserved among the cyclins. In addition, the α1 regions of cyclins A, B, and E are quite similar in structure and sequence (Fig. 6 A–C). Therefore it might be predicted that cyclin E would have CLS functions mediated by this region as well, but it does not (14). However, the individual cyclin structures and the superimposition of the cyclin A, B, and E structures (Fig. 6 A–F) show that cyclin E is an outlier in the cyclins in that it has an extended loop-helix-loop structure (C-terminal helix) that folds back near the α1 helix (Fig. 6 D and F, boxed in red). This additional polypeptide alters the structure, accessibility, and electrostatic properties of the cyclin E α1 helix compared with both cyclins B and A (see also Fig. S5). Thus, the protrusion of this loop-helix-loop at the C terminus of cyclin E potentially reduces the accessibility of the MRAIL-containing helix site corresponding to the cyclin A CLS (Fig. 6 D and F). There are also significant differences in surface area shape and charge distribution between cyclins E and A around the cyclin A CLS region (Fig. 6 D and E and Fig. S5). Cyclin E has a predominantly basic surface, whereas cyclin A has an acidic patch adjacent to the α1 helix. Altogether, these differences likely explain why the cyclin A CLS region defined here does not function as a CLS in the corresponding region of cyclin E. In contrast to the differences between cyclins A and E, the similarities of cyclins A and B in the region of the cyclin A CLS are quite striking. The surface shape and charge characteristics are remarkably similar, as shown in Fig. S5 B and C. They both have an acidic patch adjacent to the cyclin A CLS region, which is flanked by two basic surfaces. Furthermore, the structure of the C-terminal regions of cyclin A and B are highly similar and would not be predicted to block access to the α1 helix.
The cyclin A CLS includes the MRAIL sequence previously described as a “hydrophobic patch” that facilitates binding of the Cdk complexes to potential substrates (27). The importance of this sequence for the CLS is also evident in the loss-of-function phenotype observed with the IEEK-R mutant, which alters the MRAIL region (Figs. 1 and 3). Thus, our results suggest that the CLS motif in cyclin A may facilitate the recruitment and phosphorylation of centrosomal substrates and regulators of cyclin A-Cdks. Here we show that the Cdk inhibitor p27KIP1, a well-known regulator of cyclin-Cdk complexes, not only inhibits cyclin A-Cdk activity, but also specifically displaces the complex from centrosomes, where it may play a major role in progression through S phase (Fig. 3 and Fig. S2). This displacement may play a role in the G1 arrest usually observed when p27KIP1 expression is high or induced after stress (36). Despite the absence of a crystal structure for the cyclin E-Cdk2-p27 complex, it is likely that the binding of p27KIP1 would occur in the same region of cyclin E (containing the MRAIL motif) as in cyclin A, but the centrosomal localization of cyclin E is not affected by p27KIP1 expression, most likely because the cyclin E CLS is on the opposite side of the molecule.
In summary, our results define a domain in cyclin A promoting centrosomal localization that has important consequences for cell cycle progression into S phase. The identification of the centrosomal substrates/anchors responsible for localization through interaction with the CLS will be essential for uncovering the mechanism by which cyclin A regulates S phase. The concept of a link between the nuclear cycle and the centrosome cycle has also been suggested by recent evidence that centrosome duplication is regulated by centrosomal localization of proteins involved in DNA replication (37, 38), and that the G2/M transition involves centrosomal/nuclear communication (39).
Materials and Methods
Antibodies.
For Western blotting, polyclonal and monoclonal anti-GFP antibodies were from Clontech (#632460, and # 632381, respectively). To assess Cdk binding, a monoclonal anti-PSTAIRE antibody from Sigma-Aldrich (P7962) was used. For immunofluorescence, we used polyclonal anti-cyclin A antibodies from Abcam (Ab7956) and Santa Cruz Biotechnology (SC-751), a polyclonal anti-cyclin E antibody from Abcam (Ab7959), an anti-c-Myc (9E10)/FITC-conjugated monoclonal antibody from Santa Cruz Biotechnology (SC-40), and a monoclonal anti-γ tubulin [TU-30] antibody from Abcam (Ab27074). Secondary antibodies were conjugated with Alexa fluorochromes AF555 or AF594 and AF633 (Invitrogen).
Constructs.
The pCDNA5-FRT vector (Invitrogen) was modified by addition downstream of the multicloning site of a sequence encoding either EGFP, derived from the pEGFP vector (Clontech) between BamHI and ApaI, or a sequence encoding 6Myc, derived from the pCS2-6Myc vector between BamHI and XhoI (40). Sequences encoding full-length or truncated human cyclin A were produced by PCR using primers to introduce a HindIII restriction site at the 5′ end and a BamHI site at the 3′ end. Mutations in the cyclin A sequence were generated by two successive rounds of PCR using primers carrying the desired mutations. The human cDNA clone encoding p27KIP1 (Origene SC117607) was subcloned into the pCDNA5-FRT-6Myc vector. The cDNA encoding the PACT domain of AKAP450 (32) was a kind gift of Dr. Sean Munro (Medical Research Council, United Kingdom). It was subcloned into the pCDNA5-FRT-6Myc vector containing sequences encoding full-length or truncated human cyclin A. All products were sequenced in full to ensure that no mutations had been introduced by the PCR reactions.
Microscopy.
For immunolocalization, Xenopus S3 cells were seeded onto glass-bottom dishes (WillCo Wells) and then transfected with the indicated pCDNA5-FRT-cyclin A-EGFP constructs using Lipofectamine LTX (Invitrogen). Protein expression was carried out for 48 h, during which the cells were synchronized in G1/S by a double thymidine block (41), each followed by an 8-h release. After ice-cold methanol fixation and blocking of nonspecific epitopes with PBS containing 3% BSA, the EGFP signal was observed by direct fluorescence and compared with γ-tubulin staining of the centrosomes. Cells were selected for comparative analysis on the basis of similar total levels of EGFP expression. More than 100 cells were analyzed for each construct in each experiment. For investigating displacement of endogenous cyclin A and E, G1/S synchronized CHO-K1 cells were transfected with the pCDNA5-FRT-cyclin A-EGFP constructs or the pCDN5-FRT-p27KIP1-6Myc construct using Lipofectamin 2000 (Invitrogen) and fixed in ice-cold methanol after 18 h. The EGFP signal was observed by direct fluorescence and compared with γ-tubulin staining of the centrosomes. Endogenous cyclins A and E localization was monitored by indirect imunofluorescence using antibodies that do not recognize the cyclin A-EGFP proteins. Expression of p27KIP1 was monitored by indirect imunofluorescence with the anti-Myc antibody. Images were obtained with a Nikon Eclipse TE300, PCM2000 inverted confocal laserscan microscope with a 100× oil Plan Apo objective, NA 1.40. Images and line scans measuring centrosome-associated integrated intensity were generated using Simple PCI imaging software (Compix).
Immunoprecipitation.
Xenopus S3 cells were transfected with the pCDNA5-FRT-cyclin A-EGFP constructs, and protein expression was monitored for 24 h. Cell lysates were prepared as previously described (37) and precleared using 20 μL of anti-rabbit normal IgG-agarose beads for 2 h at 4 °C. Immunoprecipitation was performed by incubating the precleared lysates with an anti-GFP rabbit polyclonal antibody (Clontech) for 90 min at 4 °C and then with protein A-agarose beads for another 90 min at 4 °C. Beads were washed in lysis buffer and boiled in Laemmli Sample Buffer. Proteins were resolved by SDS/PAGE, and immunoprecipitates were subjected to SDS/PAGE followed by Western blot analysis using a mouse monoclonal anti-GFP antibody (Clontech), whereas coimmunoprecipitation of Cdk1/2 was detected with a monoclonal antibody to PSTAIR. A fraction corresponding to one fifth of the immunoprecipitation was also used to assay the kinase activity of the cyclin A-Cdk complexes toward histone H1, as previously described (42).
EdU Incorporation.
Unsynchronized CHO-K1 cells were transfected with the pCDNA5-FRT-cyclin A-6Myc constructs, and protein expression was monitored for 20 h. Then, EdU (Invitrogen) was added for 45 min to a final concentration of 10 μM, and cells were fixed in ice-cold methanol. Cells were stained for EdU according to the manufacturer’s directions, then with anti-c-Myc (9E10)/FITC-conjugated antibody, and counterstained with DAPI. Cells were observed using a Nikon Eclipse TE300 inverted microscope with a 40× air Plan Fluor objective, NA 0.75.
Supplementary Material
Acknowledgments
We thank David Morgan (University of California, San Francisco, CA) for a gift of human cyclin A2 cDNA; and Rebecca Ferguson for valuable discussions. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant GM79154 (to M.E.A.C.). G.P. and F.E. are Research Associates and J.L.M. an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914874107/DCSupplemental.
References
- 1.Wiese C, Zheng Y. Microtubule nucleation: Gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- 2.Cuschieri L, Nguyen T, Vogel J. Control at the cell center: The role of spindle poles in cytoskeletal organization and cell cycle regulation. Cell Cycle. 2007;6:2788–2794. doi: 10.4161/cc.6.22.4941. [DOI] [PubMed] [Google Scholar]
- 3.Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- 4.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 6.Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikule K, et al. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 8.Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol. 2002;157:1139–1149. doi: 10.1083/jcb.200203035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–2195. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakefield JG, Huang JY, Raff JW. Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr Biol. 2000;10:1367–1370. doi: 10.1016/s0960-9822(00)00776-4. [DOI] [PubMed] [Google Scholar]
- 11.Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 12.Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: Association with a detergent-resistant compartment and with the centrosome. J Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- 13.De Boer L, et al. Cyclin A/cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene. 2008;27:4261–4268. doi: 10.1038/onc.2008.74. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto Y, Maller JL. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- 15.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 16.Dulić V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 17.Koff A, et al. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 18.Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swenson KI, Farrell KM, Ruderman JV. The clam embryo protein cyclin A induces entry into M phase and the resumption of meiosis in Xenopus oocytes. Cell. 1986;47:861–870. doi: 10.1016/0092-8674(86)90801-9. [DOI] [PubMed] [Google Scholar]
- 20.Strausfeld UP, et al. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 21.Su TT, Stumpff J. Promiscuity rules? The dispensability of cyclin E and Cdk2. Sci STKE. 2004;2004:pe11. doi: 10.1126/stke.2242004pe11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg AR, et al. Overexpression of human cyclin A advances entry into S phase. Oncogene. 1995;10:1501–1509. [PubMed] [Google Scholar]
- 23.Moore JD, Kirk JA, Hunt T. Unmasking the S-phase-promoting potential of cyclin B1. Science. 2003;300:987–990. doi: 10.1126/science.1081418. [DOI] [PubMed] [Google Scholar]
- 24.Honda R, et al. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005;24:452–463. doi: 10.1038/sj.emboj.7600554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieder CL, Faruki S, Khodjakov A. The centrosome in vertebrates: More than a microtubule-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/s0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 26.Petri ET, Errico A, Escobedo L, Hunt T, Basavappa R. The crystal structure of human cyclin B. Cell Cycle. 2007;6:1342–1349. doi: 10.4161/cc.6.11.4297. [DOI] [PubMed] [Google Scholar]
- 27.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown NR, et al. The crystal structure of cyclin A. Structure. 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 29.Cheng KY, et al. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem. 2006;281:23167–23179. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
- 30.Lowe ED, et al. Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry. 2002;41:15625–15634. doi: 10.1021/bi0268910. [DOI] [PubMed] [Google Scholar]
- 31.Russo AA, Jeffrey PD, Patten AK, Massagué J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 32.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polyak K, et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 34.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 35.Bentley AM, Normand G, Hoyt J, King RW. Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis. Mol Biol Cell. 2007;18:4847–4858. doi: 10.1091/mbc.E06-06-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardner LB, et al. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 37.Ferguson RL, Maller JL. Cyclin E-dependent localization of MCM5 regulates centrosome duplication. J Cell Sci. 2008;121:3224–3232. doi: 10.1242/jcs.034702. [DOI] [PubMed] [Google Scholar]
- 38.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krämer A, Lukas J, Bartek J. Checking out the centrosome. Cell Cycle. 2004;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- 40.Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 41.Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol Biol. 2005;296:157–166. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 42.Peng A, Lewellyn AL, Maller JL. Undamaged DNA transmits and enhances DNA damage checkpoint signals in early embryos. Mol Cell Biol. 2007;27:6852–6862. doi: 10.1128/MCB.00195-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.