Retroviruses replicate by integrating a DNA copy of their genome into cellular DNA (1). The integrated DNA is replicated along with cellular DNA during each cell-division cycle and is the template for transcription of RNAs required for production of progeny virus. DNA integration is mediated by the virally encoded integrase enzyme and occurs at essentially any location in the host DNA, but each group of retroviruses exhibits distinct regional preferences (2). For example, HIV-1 and closely related retroviruses called lentiviruses preferentially integrate in active transcription units. The mechanistic basis of the target-site preference of HIV-1 DNA integration is not well understood, but studies suggest that a cellular protein called lens epithelium-derived growth factor (LEDGF) plays a key role (3). LEDGF binds to both HIV-1 integrase and chromatin, and in a popular model, it tethers the viral integration machinery to chromatin (4). In this issue of PNAS, Ferris et al. (5) provide strong support for this model and show that the targeting preference of HIV-1 can be changed simply by swapping the chromatin-binding domain (CBD) of LEDGF.
Retroviral DNA is made by reverse transcription in the cytoplasm after infection and forms part of a large nucleoprotein complex termed the preintegration complex (PIC). Integrase, which catalyzes the key DNA-cutting and -joining steps of integration, is tightly associated with the viral DNA within the PIC. In addition to integrase, many other viral and cellular proteins have been reported to be components of the PIC, although in most cases their functional role, if any, is unclear. Integrase is an obvious candidate for determining target specificity because the target site must juxtapose the active site of the enzyme during the catalytic steps. Indeed, it has been long established that retroviral integrases do not catalyze random integration into DNA but rather use some sites in DNA preferentially as targets (see ref. 6 and references therein). However, the specificity directed by integrase and target DNA alone is local, accounting for nonrandom distribution of integration within a few hundred base pair stretches of DNA; it cannot account for the regional preferences of integration into chromatin discussed here.
A paradigm for thinking about how integration can be targeted comes from studies of other mobile genetic elements that exhibit a very high degree of specificity for their sites of integration. Well-understood examples include the yeast Ty3 retrotransposon, which integrates almost exclusively into transcription initiation sites of genes transcribed by RNA polymerase III, and Ty5, which integrates into heterochromatin. This specificity is directed by tethering the integrase proteins to chromatin via different transcription factors (7, 8). HIV-1 integrase binds LEDGF (9), making this protein a prime candidate for a role in target-site selection. A domain in the C-terminal region of LEDGF, the integrase-binding domain (IBD), binds integrase, and the interface has been determined by x-ray crystallography (10, 11). The N-terminal part of LEDGF contains a proline-tryptophan-tryptophan-proline (PWWP) domain and adenine-thymine (AT) hooks that bind chromatin (12). Importantly, a viable mouse embryo fibroblast LEDGF knockout (MEF-KO) cell line exists that supports HIV-1 replication, albeit at a reduced level, enabling the potential role of this protein in directing integration specificity to be explored (13). Deletion of the PWWP chromatin-binding region of LEDGF reduces HIV-1 replication to the level observed in the absence of this cofactor, but replacing the PWWP domain with other chromatin-binding peptides restores efficient replication (14).
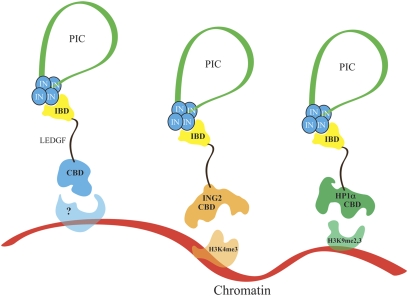
In the simplest model, LEDGF is an adaptor that enhances integration by tethering integrase within the PIC to chromatin. The study by Ferris et al. (5) tests this hypothesis by replacing the PWWP and AT hooks of LEDGF with other CBDs with known specificity (Fig. 1): the plant homeodomain finger from inhibitor of growth protein 2 (ING2), which binds histone 3 lysine 4 trimethylations (H3K4me3), and the chromodomain of heterochromatin protein 1-α (HP1α), which binds histone 3 that is methylated on lysine 9 (H3K9me2,3). The H3K4me3 modification is common in the 5′ regions of active genes (15), whereas H3K9me2,3 is widely distributed throughout chromatin (16). Not only do these LEDGF fusion proteins restore HIV-1 replication when expressed in MEF-KO cells; they also direct integration to the target sites of their respective CBDs.
Fig. 1.
Tethering of PICs to chromatin by LEDGF fusion proteins. The viral DNA within the PIC is shown in green with the ends associated with a tetramer of integrase (IN). The IBD of LEDGF (yellow) binds to integrase within the PIC. The CBD of LEDGF tethers the PIC to its binding sites in chromatin; the binding partner in chromatin is unknown. PICs can be tethered to different regions of chromatin by substituting other CBDs that bind different partners. The tether spatially restrains the PIC, and integration occurs in the vicinity of attachment.
How does LEDGF exert such a strong influence on the targeting of HIV-1 DNA integration? The stimulation of integration observed upon expression of LEDGF in MEF-KO cells and the redirection of integration sites directed by fusions with heterologous CBDs suggest that LEDGF normally plays a dominant role in anchoring PICs to chromatin before integrase catalyzes integration. Is the targeting in MEF-KO cells directed only by the affinity of integrase for chromatin, or do other cellular proteins substitute for LEDGF in its absence? The answer to this question must await the results of further studies. It also remains to be determined whether the functional interaction of integrase with LEDGF is through integrase protomers tightly associated with the viral DNA ends or through more loosely bound integrase protomers within the PIC. The interaction of the N-terminal region of LEDGF with chromatin is not well understood.
Why does HIV-1 recruit LEDGF to direct targeting? The most obvious consequence of targeting for a retrovirus is modulating the expression of the integrated provirus. However, the ING2-IBD and HP1α-IBD fusions increased HIV-1 titer to similar levels in MEF-KO cells, suggesting that, although these two domains target integration to different regions of chromatin, they do not result in significantly different expression of the integrated proviruses. HIV-1 simply may recruit LEDGF to boost the association of PICs with chromatin, and the resultant targeting may be an indirect consequence that is not functionally important for the virus. However, under different selection
This finding has potential importance in the application of retroviral vectors for gene therapy.
conditions there may be advantages that are not revealed in these studies. Because LEDGF binds only lentiviral integrases, another unanswered question is whether other retroviral integrases employ different cellular proteins in the same manner that HIV-1 exploits LEDGF.
It would come as no surprise if the repertoire of known cellular targeting proteins were to expand further.
Regardless of the functional significance of target-site selection for the virus, the study by Ferris et al. (5) shows that targeting can be redirected by substituting the CBD of the LEDGF with a different anchor. This finding has potential importance in the application of retroviral vectors for gene therapy. Such vectors can stably integrate the desired gene into the cellular genome with high efficiency, but integration at certain sites can lead to cancers (17). Biasing integration toward “safe” regions of the genome can mitigate this problem. Swapping the chromatin-binding anchor of exogenously expressed LEDGF alone is insufficient, because such proteins would need to compete with endogenous LEDGF. However, integrase and the IBD can be mutated in parallel so that they still bind one another, but integrase no longer binds endogenous LEDGF (11). In this study Ferris et al. (5) show that the half of LEDGF that binds to chromatin can be swapped to essentially any CBD of desired specificity. Although the extent to which retroviral target specificity can be directed remains to be determined, manipulating the dual-anchor LEDGF, which bridges the PIC and chromatin before integration, is the most promising strategy to date.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases and by the National Institutes of Health AIDS Targeted Antiviral Program.
Footnotes
The author declares no conflict of interest.
See companion article on page 3135.
References
- 1.Brown PO. Integration. In: Coffin JM, Hughes SH, Varums HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. pp. 161–203. [Google Scholar]
- 2.Bushman F, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 3.Ciuffi A, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 4.Ciuffi A. Mechanisms governing lentivirus integration site selection. Curr Gene Ther. 2008;8:419–429. doi: 10.2174/156652308786848021. [DOI] [PubMed] [Google Scholar]
- 5.Ferris AL. Lens epithelium-derived growth factor (LEDGF) fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci USA. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holman AG, Coffin JM. Symmetrical base prefer-ences surrounding HIV-1 and avian sarcoma/leukosis virus but not murine leukemia virus integration sites. Proc Natl Acad Sci USA. 2005;102:6238–6238. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchner J, Connolly CM, Sandmeyer SB. Require-ment of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 8.Zhu YX, Dai JB, Fuerst PG, Voytas DF. Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov P, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 10.Cherepanov P, Ambrosio ALB, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hare S, et al. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mu-tants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llano M, et al. Identification and characteriza-tion of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 13.Shun MC, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meehan AM, et al. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009;5:e1000522. doi: 10.1371/journal.ppat.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Nair V. Retrovirus-induced oncogenesis and safety of retroviral vectors. Curr Opin Mol Ther. 2008;10:431–438. [PubMed] [Google Scholar]