Abstract
Circadian kaiBC expression in the cyanobacterium Synechococcus elongatus PCC 7942 is generated by temporal information transmission from the KaiABC-based circadian oscillator to RpaA, a putative transcriptional factor, via the SasA-dependent positive pathway and the LabA-dependent negative pathway which is responsible for feedback regulation of KaiC. However, the labA/sasA double mutant has a circadian kaiBC expression rhythm, suggesting that there is an additional circadian output pathway. Here we describe a third circadian output pathway, which is CikA-dependent. The cikA mutation attenuates KaiC overexpression-induced kaiBC repression and exacerbates the low-amplitude phenotype of the labA mutant, suggesting that cikA acts as a negative regulator of kaiBC expression independent of the LabA-dependent pathway. In the labA/sasA/cikA triple mutant, kaiBC promoter activity becomes almost arrhythmic, despite preservation of the circadian KaiC phosphorylation rhythm, suggesting that CikA largely accounts for the residual kaiBC expression rhythm observed in the labA/sasA double mutant. These results also strongly suggest that transcriptional regulation in the labA/sasA/cikA triple mutant is insulated from the circadian signals of the KaiABC-based oscillator. Based on these observations, we propose a model in which temporal information from the KaiABC-based circadian oscillator is transmitted to gene expression through three separate output pathways.
Keywords: CikA, circadian clock, LabA, SasA, transcriptional feedback
Circadian clocks are endogenous self-sustained oscillators with a period of ≈24 h. A variety of organisms from bacteria to mammals use circadian clocks to adapt to daily environmental changes. Autotrophic cyanobacteria are the simplest organisms with a circadian clock, and one of the cyanobacterial species, Synechococcus elongatus PCC 7942 (hereafter Synechococcus), is a model organism for studying the circadian clock system (1). Conceptually, the circadian clock system can be divided into three components: the oscillator that generates the basic timing loop, the input pathway that transmits signals from the environment to entrain the oscillator, and the output pathway that transmits temporal information from the oscillator to regulate various cellular activities, such as transcription. Three proteins encoded by the kaiABC gene cluster, which is essential for the circadian rhythm, generate the basic timing loop of the circadian clock in Synechococcus (2, 3). A temperature-compensated, self-oscillatory circadian clock can be reconstituted by mixing the three Kai proteins, KaiA, KaiB, and KaiC, with ATP in a test tube (3). KaiC, the central component of the circadian clock, is an autokinase and autophosphatase, and its activity is modulated by KaiA and KaiB through dynamic interactions (4–6). These Kai protein reactions including KaiC phosphorylation oscillate both in an in vitro oscillatory system and in Synechococcus in a circadian fashion (3, 4, 6).
Temporal information generated by the KaiABC-oscillator in Synechococcus is transmitted to the transcriptional regulatory system. Circadian gene expression is widely observed in Synechococcus; kaiBC transcription shows a robust circadian rhythm (2). The regulation of kaiBC expression forms a part of circadian transcriptional-translational feedback loop and thought to contribute the robustness of the entire circadian clock system in Synechococcus (7). The mechanism of circadian kaiBC expression has been gradually revealed, as well as the biochemical properties of the Kai proteins. A KaiC-interacting histidine kinase, SasA, and its cognate response regulator, RpaA, which has a putative DNA binding domain, are important output factors for transcription of clock-controlled genes including kaiBC (8). Disruption of either sasA or rpaA dramatically reduces circadian kaiBC expression and causes an almost arrhythmic phenotype, indicating that SasA and RpaA are circadian clock-controlled transcriptional activators (8, 9). This reduction of kaiBC expression results in reduction of KaiC accumulation in these mutants (8). SasA autophosphorylates in response to KaiC binding, and the phosphate is transferred to RpaA (8). It is likely that the KaiC-SasA-RpaA pathway activates kaiBC expression mainly during subjective day, depending on the status of the KaiABC-based oscillator (8).
Phosphorylated KaiC is capable of repressing its own gene expression in a dose-dependent manner (i.e., negative feedback regulation of KaiC) (2, 10, 11). labA was identified as a gene required for this negative feedback regulation of KaiC (12). kaiBC promoter activity is maintained when KaiC is overexpressed in labA mutants, suggesting that the negative feedback regulation is abolished by the labA mutation. The labA mutation increases the troughs of circadian kaiBC expression, resulting in a low-amplitude rhythm, suggesting that labA represses kaiBC expression mainly during subjective night in wild-type Synechococcus. The lack of the negative feedback regulation in the labA mutant increases KaiC accumulation. More importantly, the labA mutation suppresses the phenotypes of the sasA mutant; circadian kaiBC expression and KaiC accumulation are restored in the labA/sasA double mutant (12). These results strongly suggest that sasA and labA govern kaiBC expression through different pathways. In addition, bioluminescence from a PkaiBC::luc (firefly luciferase) reporter in the labA/rpaA double mutant is similar to that in the rpaA single mutant, indicating that rpaA is epistatic to labA in the bioluminescence assay (12). This suggests that rpaA and labA regulate kaiBC expression through the same pathway. LabA is likely involved in the repression of circadian kaiBC expression through RpaA, mainly during subjective early night when phosphorylated KaiC level is high. Thus, we propose a model in which the temporal information from the KaiABC-based oscillator diverges into two output pathways (the LabA-dependent negative pathway and the SasA-dependent positive pathway), which then converge onto RpaA to control the kaiBC expression in a circadian fashion (12).
Although SasA and LabA are involved in the two major output pathways for circadian regulation of transcription, an additional output pathway has been postulated because the labA/sasA double mutant in the PkaiBC::luc reporter strain background has a remarkable circadian bioluminescence rhythm (12). In this study, we report that CikA largely accounts for the circadian bioluminescence rhythm in the labA/sasA double mutant. CikA was originally identified as a sensor histidine kinase involved in the input pathway of the cyanobacterial circadian clock (13). The cikA mutation affects phosphorylation of KaiC and decreases phase shifts of the bioluminescence rhythm in response to 5-h dark pulses (13, 14). CikA is capable of binding to quinone molecules that sense the cellular redox state, and the stability of CikA protein in vivo is affected by the cellular redox state and environmental light intensity (14, 15). CikA is also likely to associate with the KaiABC-based protein complex (14, 15). Thus, it has been proposed that CikA senses changes in the cellular redox state, and through association with the KaiABC-based protein complex, CikA functions to reset the KaiABC-based circadian oscillator in harmony with the daily environmental cycle (14). However, it was unclear how the cikA mutation causes a low-amplitude phenotype under constant light (LL) conditions (13). Here, we analyzed the molecular and genetic relationship between CikA and other clock-related components to demonstrate a role for CikA in the transcriptional regulation.
Results
Identification of cikA as a Gene that Enhances Feedback Inhibition Induced by KaiC Overexpression.
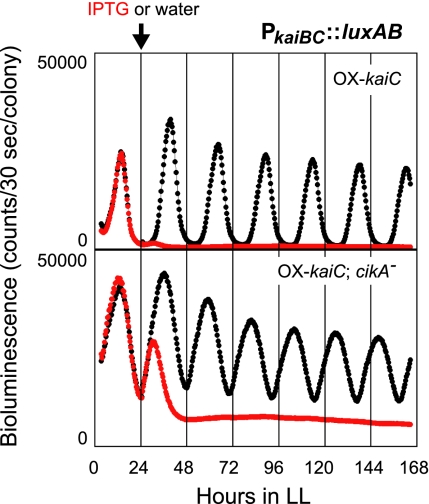
Previously, we identified labA as a gene indispensable for negative feedback regulation of KaiC by a genetic screening (12). We also identified a cikA mutant during the screen (Fig. 1). In this screen, we used a transgenic Synechococcus reporter strain [OX-kaiC(Ω)/NUC42] carrying additional loci in which the Vibrio harveyi luciferase gene (luxAB) was placed under the control of the kaiBC promoter (PkaiBC::luxAB) and kaiC was placed under the control of an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter (Ptrc::kaiC) (10). In the absence of IPTG, OX-kaiC(Ω)/NUC42 cells had a robust circadian bioluminescence rhythm that followed the circadian changes in kaiBC promoter activity. IPTG treatment induced KaiC overexpression and dramatically decreased bioluminescence by repressing luciferase expression via negative feedback inhibition of the kaiBC promoter (Fig. 1) (10). To identify mutants that lack this inhibition, we performed random transposon mutagenesis of OX-kaiC(Ω)/NUC42 and monitored the bioluminescence of individual colonies of the transformants in the presence of 1 mM IPTG. About 20,000 colonies were screened as described (12), and a cikA mutant [OX-kaiC(Ω)/cikA−(Km)/NUC42] that expresses the luxAB reporter gene even in the presence of IPTG was isolated (Fig. 1). Sequencing analysis of the genomic region flanking the transposon revealed that the transposon was inserted at nucleotide position 2218 (the first nucleotide of the translation initiation ATG was defined as “position 1”). In the absence of IPTG, OX-kaiC(Ω)/cikA-(Km)/NUC42 had a low-amplitude bioluminescence rhythm with higher troughs and a short period (Fig. 1), as reported in a previous work (13). Addition of 1 mM IPTG rapidly decreased the bioluminescence intensity of OX-kaiC(Ω)/cikA−(Km)/NUC42, suggesting that negative feedback of KaiC occurs in this mutant. However, the feedback inhibition halted incompletely and bioluminescence remained at an intermediate level without rhythmicity, suggesting that the feedback regulation is attenuated in this strain (Fig. 1).
Fig. 1.
Identification of cikA as a gene that supports negative feedback regulation of KaiC. Bioluminescence traces of the cikA mutant isolated from the screen [OX-kaiC(Ω)/cikA−(Km)/NUC42] is shown. Cells were grown on agar plates and their bioluminescence rhythms were measured under constant light (LL) conditions after entrainment with two 12-h light/12-h dark (LD) cycles. IPTG (1 mM) or water was added at hour 24 in LL (LL 24). (Upper) Bioluminescence rhythm of the parental reporter strain OX-kaiC(Ω)/NUC42; (Lower) bioluminescence rhythm of OX-kaiC(Ω)/cikA-(Km)/NUC42. (red trace) Bioluminescence of cells treated with 1 mM IPTG; (black trace) bioluminescence of cells treated with water. The arrow indicates the timing of IPTG (or water) addition.
Genetic Interaction Between cikA and labA.
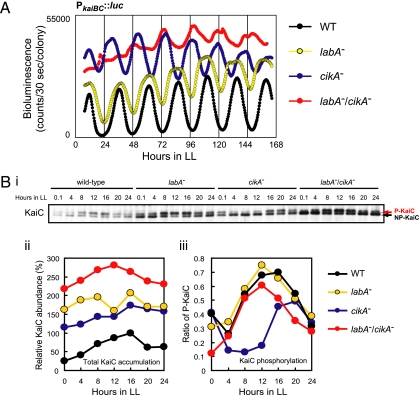
The phenotypes of the cikA mutant shown in Fig. 1 (increased trough levels of circadian kaiBC expression and attenuated negative feedback regulation of KaiC) strongly suggest that CikA negatively regulates kaiBC expression. LabA is another negative regulator for kaiBC expression (12). To clarify the roles of these two factors in the feedback regulation, we examined their genetic interaction. We generated a labA/cikA double mutant [labA−(Ω)/cikA−(Km)/AMC541] in a PkaiBC::luc reporter strain, AMC541, and evaluated the phenotype using a bioluminescence assay. We did not use luxAB reporter strains in this experiment because the labA mutation produces a “bright phenotype” in luxAB reporter strains (12). As reported (12, 13), the labA single mutant [labA−(Ω)/AMC541] had a low-amplitude rhythm with high trough levels, whereas the cikA single mutant [cikA−(Km)/AMC541] had a short period and low-amplitude rhythm with high trough levels (Fig. 2A). The labA/cikA double mutant had a much more severe phenotype: a very low-amplitude rhythm with a higher overall level (Fig. 2A; see also Fig. 3B), indicating an additive effect of these two mutations on the amplitude of the output oscillation.
Fig. 2.
Circadian phenotypes of the labA/cikA double mutant. (A) Bioluminescence rhythm of the labA/cikA double mutant. Wild-type AMC541 (WT), labA−(Ω)/AMC541 (labA−), cikA−(Km)/AMC541 (cikA−), and labA−(Ω)/cikA−(Km)/AMC541 (labA−/cikA−) were grown on BG-11M agar plates under LL conditions. After entrainment with two LD cycles, bioluminescence rhythms of the cells were measured in the presence of 0.5 mM luciferin. (B) Total abundance and phosphorylation profile of KaiC in the labA/cikA double mutant. AMC541, labA−(Ω)/AMC541, cikA−(Gm)/AMC541, and labA−(Ω)/cikA−(Gm)/AMC541 were cultured in BG-11M medium, and cells were collected every 4 h after entrainment with LD cycles. Total cellular proteins (2 μg per lane) were analyzed by immunoblotting using anti-KaiC antiserum. (Bi) One of blots from two independent experiments is shown. The upper and lower bands correspond to phosphorylated KaiC (P-KaiC; red arrow) and nonphosphorylated KaiC (NP-KaiC; black arrow), respectively. (Bii and Biii) Densitometric analysis of the blot shown in Bi; (Bii) total KaiC abundance; (Biii) KaiC phosphorylation rhythm.
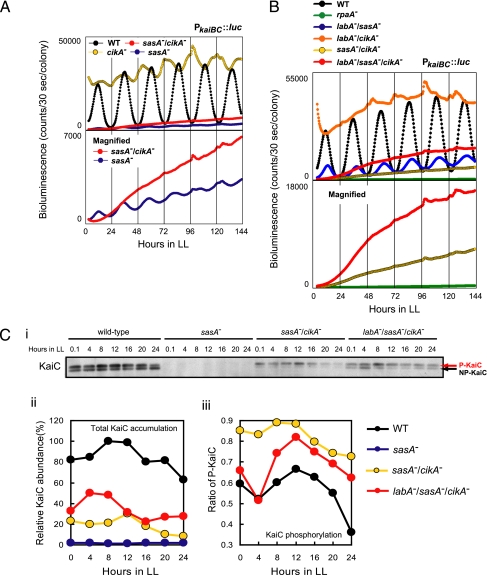
Fig. 3.
Circadian phenotypes of the sasA/cikA double mutant and the labA/sasA/cikA triple mutant. (A) Bioluminescence rhythm of the sasA/cikA double mutant. Wild-type AMC541 (WT), sasA−(Km)/AMC541 (sasA−), cikA−(Gm)/AMC541 (cikA−), and sasA−(Km)/cikA−(Gm)/AMC541 (sasA−/cikA−) were grown on BG-11M agar plates under LL conditions. After entrainment with an LD cycle, bioluminescence rhythms of the cells were measured in the presence of 0.5 mM luciferin. Bioluminescence traces for sasA−(Km)/AMC541 and sasA−(Km)/cikA−(Gm)/AMC541 are also shown on a magnified scale in the lower panel. (B) Bioluminescence rhythm of the labA/sasA/cikA triple mutant. Bioluminescence of AMC541, rpaA−(Km)/AMC541 (rpaA−), labA−(Ω)/sasA−(Km)/AMC541 (labA−/sasA−), labA−(Ω)/cikA−(Gm)/AMC541 (labA−/cikA−), sasA−(Km)/cikA−(Gm)/AMC541, and labA−(Ω)/sasA−(Km)/cikA−(Gm)/AMC541 (labA−/sasA−/cikA−) was measured as described in A. Bioluminescence traces for rpaA−(Km)/AMC541, sasA−(Km)/cikA(Gm)/AMC541, and labA−(Ω)/sasA−(Km)/cikA−(Gm)/AMC541 are also shown on a magnified scale in the lower panel. (C) Total KaiC levels and KaiC phosphorylation rhythm in the sasA/cikA double mutant and the labA/sasA/cikA triple mutant. KaiC levels of AMC541, sasA−(Km)/AMC541, sasA−(Km)/cikA−(Gm)/AMC541, and labA−(Ω)/sasA−(Km)/cikA−(Gm)/AMC541 were analyzed by immunoblotting as described in Fig. 2B. (Ci) One of blots from two independent experiments is shown. (red arrow) Phosphorylated KaiC (P-KaiC); (black arrow) nonphosphorylated KaiC (NP-KaiC). (Cii and Ciii) Densitometric analysis of the blot shown in Ci; (Cii) total KaiC abundance; (Ciii) KaiC phosphorylation rhythm. Note that KaiC level is extremely low in the sasA mutant, and thus we were unable to evaluate the phosphorylation rhythm.
Resetting of the circadian rhythm is dysfunctional in cikA mutants. Thus, desynchronization of circadian rhythms among cyanobacterial cells may produce normalized output signals with a severely low-amplitude rhythm in the labA/cikA double mutant. To assess this possibility, we examined KaiC accumulation and phosphorylation using immunoblotting. In the labA mutant, total KaiC was increased by ≈2- to 6-fold, and the circadian rhythm of KaiC phosphorylation was robustly sustained (Fig. 2B), as reported (12). The KaiC phosphorylation profile in the labA mutant was similar to that in the wild-type strain (Fig. 2Biii). Total KaiC level was also increased in the cikA mutant, similar to the labA mutant (Fig. 2Bi and Fig. 2Bii). As Ivleva et al. reported (14), the cikA mutation altered the phase of the phosphorylation rhythm after dark treatment (Fig. 2Bi and Fig. 2Biii). KaiC level in the labA/cikA double mutant was higher than that in labA or cikA mutant (Fig. 2Bi and Fig. 2Bii), suggesting that labA and cikA mutations have an additive effect on KaiC level. More importantly, the KaiC phosphorylation ratio in the labA/cikA double mutant fluctuated similar to the wild-type strain (Fig. 2Bi and Fig. 2Biii), suggesting that circadian oscillations of KaiC phosphorylation are synchronized among the labA/cikA double-mutant cells. Therefore, the severely low-amplitude rhythm of the bioluminescence reporter in the labA/cikA double mutant is likely caused by abnormalities in the cellular output from the oscillator, rather than desynchronization of individual oscillatory cells. These results suggest that cikA negatively regulates kaiBC expression independent of labA, and then it supports feedback regulation of KaiC.
Genetic Analyses of the sasA/cikA Double Mutant and the labA/sasA/cikA Triple Mutant.
Next, we evaluated genetic interaction between cikA and sasA using a sasA/cikA double mutant. We generated a sasA/cikA double mutant strain in the AMC541 background [sasA−(Km)/cikA−(Gm)/AMC541] and assayed its bioluminescence (Fig. 3A). The sasA single mutant in the AMC541 background [sasA−(Km)/AMC541] had a low-amplitude rhythm and dramatically reduced bioluminescence level (Fig. 3A), as described (12). The bioluminescence trace of the sasA/cikA double mutant was arrhythmic or almost arrhythmic, with an extremely low-amplitude rhythm that was more severe than that of the sasA mutant (Fig. 3A). These results suggest that cikA may generate the residual rhythmicity of the sasA mutant by regulating circadian gene expression independent of sasA.
We also evaluated the bioluminescence of the labA/sasA/cikA triple mutant in the AMC541 background [labA−(Ω)/sasA−(Km)/cikA−(Gm)/AMC541] (Fig. 3B). The labA/sasA double mutant in the AMC541 background [labA−(Ω)/sasA−(Km)/AMC541] had a low-amplitude, but remarkably robust, bioluminescence rhythm (Fig. 3B), as described (12). However, bioluminescence trace of the labA/sasA/cikA triple mutant was almost arrhythmic or had a severely low-amplitude rhythm (Fig. 3B), suggesting that cikA largely accounts for the bioluminescence rhythm in the labA/sasA double mutant. The bioluminescence level of the triple mutant was consistently higher than that of the sasA/cikA double mutant, indicating that the labA mutation has a positive effect on bioluminescence level irrespective of sasA and cikA. These results are consistent with those for the labA/sasA and the labA/cikA double mutants (Figs. 2A and 3B) (12). Bioluminescence level of the labA/sasA/cikA triple mutant was greater than that of the rpaA single mutant, rpaA−(Km)/AMC541 (Fig. 3B). These results suggest that RpaA can be activated by factors other than LabA, SasA, and CikA. Note that our attempts to isolate a cikA/rpaA double mutant for further genetic analyses resulted in failure, probably because of a synthetic lethal interaction between the two (Fig S1).
KaiC levels in these multiple mutants were examined by immunoblotting (Fig. 3C). KaiC decreased to undetectable levels in the sasA single mutant in this experiment (Fig. 3C). KaiC could be detected in the sasA/cikA double mutant, but the level was much lower than that in the wild-type strain (Fig. 3Ci and Fig. 3Cii). KaiC phosphorylation was persistently elevated in the sasA/cikA double mutant and its rhythmicity was obscure, probably because of low KaiC level (Fig. 3Ci and Fig. 3Ciii). Total KaiC level in the labA/sasA/cikA triple mutant was greater than that in the sasA/cikA double mutant, and the phosphorylation rhythm of KaiC was restored, likely a result of restoration of KaiC accumulation (Fig. 3C). In other words, like the in vitro KaiC phosphorylation rhythm, the cellular Kai-based oscillator is capable of working synchronously regardless of LabA, SasA, and CikA levels, if Kai accumulation in the cell reaches appropriate levels. Thus, the arrhythmicity of kaiBC expression in the labA/sasA/cikA triple mutant is likely caused by impairment of circadian output pathways governing gene expression, rather than the dysfunction of the oscillator per se or desynchronization among cells.
Discussion
In this study, we have genetically dissected the role of cikA in the regulation of kaiBC expression under LL conditions. The trough levels of circadian gene expression and KaiC accumulation were increased by the cikA mutation, suggesting that CikA is a negative regulator of kaiBC expression (Figs. 1, 2, and 3A) (13). Unlike LabA, which is critical for negative feedback regulation of KaiC, CikA was not essential for the feedback regulation in response to KaiC accumulation, rather CikA supports the feedback repression of kaiBC expression (Fig. 1). Increased KaiC accumulation in the cikA mutant is probably a result of insufficient feedback regulation of KaiC. This hypothesis is supported by epistasis analysis of labA and cikA mutations. The effects of labA and cikA mutations on kaiBC expression were additive (Fig. 2), suggesting that CikA and LabA act through different mechanisms to repress kaiBC expression. Double mutant analysis of cikA and sasA suggests that CikA also regulates kaiBC transcription independent of SasA (Fig. 3). Thus, the CikA-dependent transcriptional regulatory mechanism appears to be independent of LabA and SasA. Because LabA and SasA function independently to regulate kaiBC transcription (12), the regulatory system appears to consist of at least three separate pathways. It is interesting to note that the low-amplitude phenotype in the sasA single mutant becomes more severe under strong light conditions (8, 9). Because accumulation of CikA protein is inversely related to light intensity (15), it is possible that the decreased CikA level in sasA mutant cells under high-light conditions exacerbates the low-amplitude phenotype, similar to effect observed in the sasA/cikA double mutant.
The labA/cikA double mutant and the labA/sasA/cikA triple mutant clearly demonstrate the separation between transcriptional regulation and the circadian rhythm of the KaiABC-based oscillator in vivo (Figs. 2 and 3), strongly suggesting that the regulatory machinery for circadian transcription is mainly composed of three output pathways. In addition, kaiBC transcription is evident in strains that lack LabA, SasA, and CikA, although it is almost arrhythmic. These data suggest that RpaA can be activated by other factors through signaling pathways independent of the circadian clock. This finding seems to be related to the phenotype of the ΔkaiABC mutant: kaiBC promoter activity was evident in the ΔkaiABC mutant and similar to that in the triple mutant in bioluminescence assays (Fig. 3B) (4). The nullification of the oscillator appears to be equivalent to that of these three output pathways in terms of transcriptional regulation of kaiBC, implying that the regulatory machinery for basic kaiBC transcription is independent of the circadian system.
Previously, we proposed a model for feedback regulation of kaiBC in which the LabA-dependent negative pathway and the SasA-dependent positive pathway regulate the putative master transcriptional regulator, RpaA, to generate circadian gene expression (12). Based on our current results, we have added a third CikA-dependent circadian clock output pathway to our model. CikA is a negative factor and thus, CikA-dependent negative pathway could not function if the target promoters are not active. Because RpaA appears essential for activation of kaiBC expression, we speculate that the CikA-dependent negative output pathway requires the transcriptional activator RpaA as a downstream factor. Although biochemical interaction between SasA and RpaA (8) and genetic relationship between labA and rpaA (12) were demonstrated in previous studies, so far we have not established the role of RpaA in the CikA-dependent pathway under the KaiABC-based oscillator (Fig. S1). Further studies should be required for the elucidation of the CikA-dependent transcriptional regulatory network.
The bioluminescence trace of the labA/sasA/cikA triple mutant was not completely arrhythmic. Thus, there may be additional output pathways. Alternatively, the bioluminescence of the labA/sasA/cikA triple mutant may oscillate because of cellular activity, such as metabolism, translation, and protein degradation, which fluctuates in a circadian fashion under direct control of the KaiABC-based circadian oscillator.
When does CikA function during the circadian cycle? Under LL conditions, CikA accumulation increases and decreases in subjective night and day, respectively (14). CikA protein seems to fluctuate with a peak at CT 20 to 24 (subjective late night to dawn) (14). Thus, it is plausible that CikA represses kaiBC expression mainly during the subjective late night phase under constant conditions. The low-amplitude phenotype of cikA mutants suggests that CikA deepens the trough levels of circadian kaiBC expression (Figs. 1, 2, and 3A) (13). Furthermore, CikA proteins are unstable in the light (15), suggesting that CikA functions mainly during the dark phase under natural cyclic conditions. Combined regulation by the circadian clock and light must amplify the fluctuation in CikA level under natural conditions. CikA is also involved in phase-setting in response to a dark pulse (13), and the circadian profile of CikA level is reasonable for resetting the circadian clock in response to the dark-light transition at dawn under natural conditions.
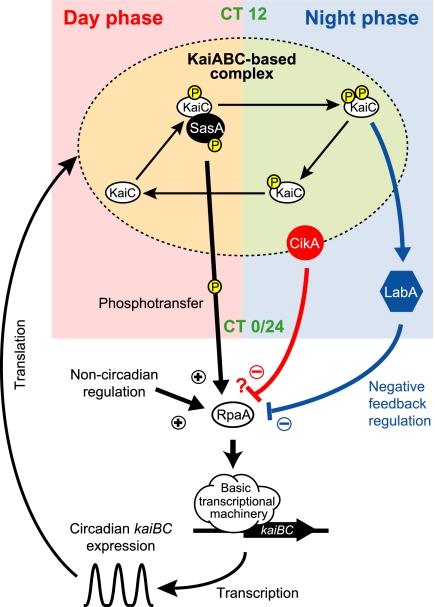
Considering all of the results, we propose a revised model for regulation of circadian kaiBC expression in Synechococcus (Fig. 4). The KaiABC-based complex is thought to serve as the basis for cyanobacterial circadian rhythm generation. KaiC has two phosphorylation sites and they are circadianly phosphorylated and dephosphorylated in sequence (16). KaiC phosphorylation oscillates with a peak around CT 16 (4). In vitro, SasA-RpaA phosphorylation occurs 4 to 8 h before the peak of KaiC phosphorylation (8). Therefore, we can extrapolate that SasA phosphorylation fluctuates with a peak around CT 8 to 12 (subjective day to dusk) in vivo and activation of the SasA-dependent pathway up-regulates kaiBC expression via RpaA mainly during the subjective day phase (8). Because phosphorylation of KaiC is indispensable for negative feedback regulation of KaiC (11), LabA appears to function downstream of phosphorylated KaiC. Both accumulation and phosphorylation of KaiC oscillate with a peak around CT 16 (subjective night) (4, 5). Thus, repression of kaiBC expression by LabA is strongest around CT 16 (12), when doubly phosphorylated KaiC is most prevalent (16). The LabA-dependent negative pathway appears to function by repressing RpaA activity. CikA abundance fluctuates with a peak around CT 20 to 24 (14). Thus, CikA may repress kaiBC expression during the subjective late night, possibly by associating with the KaiABC-based protein complex. Because CikA is involved in both input and output pathways of the circadian clock, the interaction of CikA with the KaiABC-based oscillator should function not only in the modulation of the phasing of the KaiABC-based oscillator but also in the activation of a negative circadian output pathway. We speculate that the CikA-dependent pathway also functions through RpaA to regulate transcription. It is likely that RpaA is also regulated by noncircadian mechanisms.
Fig. 4.
A possible model for the transcriptional-translational feedback loop of the cyanobacterial circadian clock. Details are described in the text.
The three major output pathways likely regulate kaiBC promoter activity according to different temporal information sources that are generated by a single circadian clock. SasA activates kaiBC promoter activity according to the reaction states of KaiC, whereas LabA represses kaiBC promoter activity in response to accumulation of phosphorylated KaiC. The function of CikA in transcriptional regulation likely depends on CikA level, which is under the control of the clock, as well as binding to the KaiABC-based complex. This scenario explains the phase of the kaiBC mRNA expression rhythm: peak at subjective dusk (CT 12) and trough at subjective dawn (CT 0/24) (12). Around subjective dawn, the SasA-dependent positive pathway is likely inactive and noncircadian regulatory mechanisms for RpaA activation may be diminished by the CikA-dependent negative pathway. After subjective dawn, the negative effect of the CikA-dependent pathway gradually decreases and the noncircadian regulatory mechanisms and the SasA-dependent positive pathway can activate RpaA. The positive effect of the SasA-dependent pathway peaks around subjective dusk. Later, the positive effect of the SasA-dependent pathway decreases as KaiC is dephosphorylated. The negative effect of the LabA-dependent pathway peaks in the early- to midsubjective night, and then the positive effects of noncircadian regulatory mechanisms and residual SasA-dependent activity are suppressed. The phase differences among these pathways should increase the amplitude of circadian kaiBC expression. Regulations by three pathways likely ensure robust circadian kaiBC expression against perturbations in cellular systems and the clock system. The robustness appears to be achieved by the mechanism of the Kai protein-based oscillation and oscillatory kaiBC expression, which can be uncoupled from the phosphorylation rhythm of the protein oscillator (7). The output pathways for kaiBC expression are likely required for the machinery to coordinate these rhythmic molecular events. Furthermore, in Synechococcus, the circadian clock regulates many promoters; they exhibit a range of waveforms, amplitudes, and phase angles (17, 18). The combination of three output pathways and noncircadian regulation may underlie the generation of a variety of circadian promoter activities.
In summary, we have highlighted the contribution of CikA on circadian kaiBC expression. Although this and previous studies revealed that CikA plays important roles in both phasing of the KaiABC-based oscillator and circadian gene expression, underlying mechanisms of these roles are still unclear. Further analysis is needed to reveal the functional relationship between CikA and the KaiABC-based oscillator in the cyanobacterial circadian-clock system.
Materials and Methods
Bacterial Strains and Media.
All cyanobacterial reporter strains used in this study were derivatives of Synechococcus elongatus PCC 7942. The PkaiBC::luxAB reporter strain NUC42 (19) and the PkaiBC::luc reporter strain AMC541 (1) were generated previously. OX-kaiC(Ω)/NUC42, labA−(Ω)/AMC541, sasA−(Km)/AMC541, labA−(Ω)/sasA−(Km)/AMC541, and rpaA−(Km)/AMC541 have been described (12). The other cyanobacterial strains were constructed in this study. Transformation of Synechococcus was performed as described (20). AMC541 and labA−(Ω)/AMC541 were transformed with pDcikA(Km) (described later) to generate cikA−(Km)/AMC541 and labA−(Ω)/cikA−(Km)/AMC541, respectively. AMC541, labA−(Ω)/AMC541, sasA−(Km)/AMC541, and labA−(Ω)/sasA−(Km)/AMC541 were transformed with pAM2152 (21) to generate cikA−(Gm)/AMC541, labA−(Ω)/cikA−(Gm)/AMC541, sasA−(Km)/cikA−(Gm)/AMC541, and labA−(Ω)/sasA−(Km)/cikA−(Gm)/AMC541, respectively. Synechococcus cells and Escherichia coli strains were cultured and maintained as described (12).
Genetic Screening Using Transposon Mutagenesis.
Genetic screening using transposon mutagenesis to identify the cikA mutant shown in Fig. 1 [OX-kaiC(Ω)/cikA−(Km)/NUC42] has been described (12). The genomic DNA of OX-kaiC(Ω)/cikA−(Km)/NUC42 was digested with EcoRI, subcloned into pBluescript II, introduced into DH5α, and kanamycin-resistant clones were selected. The plasmid obtained from the clone was pDcikA(Km). pDcikA(Km) was sequenced with KAN-2 FP-1 primer and KAN-2 RP-1 primer to determine the transposon insertion site.
Bioluminescence Assay.
Bioluminescence assays were performed as described (12). The results shown in the figures are representative of two or more independent experiments.
Immunoblotting.
Cell culture, sampling, KaiC immunoblotting, and densitometric analysis were performed as described (11). The results shown in the figures are representative of two independent experiments.
Supplementary Material
Acknowledgments
We thank R. Ito for collaborating in the screening, and H. Kondo and T. Nishikawa for technical support. We also thank Dr. S. S. Golden (University of California, San Diego) for providing pAMC2152 and pAM2477. This research was supported in part by Grants-in-Aid Grant 15GS0308 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T. K. and T.O.), and the Japan Society for the Promotion of Science Grant 17370088 (to T.O.). Analysis of DNA sequences was conducted in conjunction with the Life Research Support Center at Akita Prefectural University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909924107/DCSupplemental.
References
- 1.Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu Rev Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 2.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 3.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kageyama H, et al. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 7.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki H, et al. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 10.Nakahira Y, et al. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiwaki T, et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA. 2004;101:13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi Y, et al. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 14.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiwaki T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 18.Ito H, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimura H, et al. Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology. 2002;148:2903–2909. doi: 10.1099/00221287-148-9-2903. [DOI] [PubMed] [Google Scholar]
- 20.Porter RD. DNA transformation. Methods Enzymol. 1988;167:703–712. doi: 10.1016/0076-6879(88)67081-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol Microbiol. 2006;60:658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.