Abstract
In Drosophila, Pumilio (Pum) is important for neuronal homeostasis as well as learning and memory. We have recently characterized a mammalian homolog of Pum, Pum2, which is found in discrete RNA-containing particles in the somatodendritic compartment of polarized neurons. In this study, we investigated the role of Pum2 in developing and mature neurons by RNA interference. In immature neurons, loss of Pum2 led to enhanced dendritic outgrowth and arborization. In mature neurons, Pum2 down-regulation resulted in a significant reduction in dendritic spines and an increase in elongated dendritic filopodia. Furthermore, we observed an increase in excitatory synapse markers along dendritic shafts. Electrophysiological analysis of synaptic function of neurons lacking Pum2 revealed an increased miniature excitatory postsynaptic current frequency. We then identified two specific mRNAs coding for a known translational regulator, eIF4E, and for a voltage-gated sodium channel, Scn1a, which interacts with Pum2 in immunoprecipitations from brain lysates. Finally, we show that Pum2 regulates translation of the eIF4E mRNA. Taken together, our data reveal a previously undescribed role for Pum2 in dendrite morphogenesis, synapse function, and translational control.
Keywords: dendritic spines, eIF4E, ribonucleoparticles, neuronal development, translational control
In Drosophila, Pumilio (Pum), a member of the Pum and FBF (PuF) family of proteins, plays an important role in the nervous system. First, Pum controls dendrite morphogenesis in Drosophila peripheral neurons (1). Second, Pum regulates synaptic growth and function by controlling the expression of eIF4E mRNA at the Drosophila neuromuscular junction (2, 3). Third, Pum inhibits neuronal excitability by repressing the translation of a voltage-gated sodium channel (4, 5). Finally, loss of pumilio impairs long-term memory (6). A homolog of Pum, mammalian Pum2, is expressed in hippocampal neurons and is found in ribonucleoparticles (RNPs) in the somatodendritic compartment (7). Its punctate microtubule-associated expression pattern in dendrites mimics that of RNA-binding proteins (RBPs) known to be involved in RNA transport and translational control (8, 9). Recently, Pum2 has also been linked to micro-RNA-regulated activity-dependent dendritogenesis in mammalian neurons (10). In this study, we set out to study the role of Pum2 in the activity-independent morphogenesis of dendrites, dendritic spines, and excitatory synapses of rat hippocampal neurons.
Results and Discussion
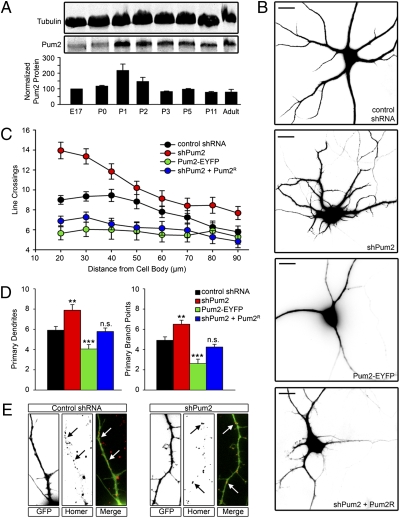
We first investigated whether Pum2 expression in the brain is developmentally regulated. Western blot analysis of protein extracts from whole brain at different points of development using a monospecific, affinity-purified, anti-Pum2 antibody revealed that Pum2 is expressed at all stages of neuronal differentiation (11) (Fig. 1A). Expression increases significantly at postnatal day 1 when dendritic outgrowth begins in the hippocampus, suggesting a potential role for Pum2 in dendrite morphogenesis. We tested this hypothesis using shRNA to down-regulate and a Pum2-EYFP fusion protein to overexpress Pum2, respectively. The efficiency of shRNA-mediated Pum2 (shPum2) down-regulation in dissociated hippocampal neurons has been previously demonstrated (7, 12). Hippocampal neurons were transiently transfected at 7 days in vitro (DIV), fixed 3 days later, and subjected to Sholl analysis (13). Neurons lacking Pum2 show a significant increase in dendritic complexity (Fig. 1 B, Top Middle, and C, red circles). Conversely, neurons overexpressing Pum2-EYFP demonstrate a simplified dendritic arbor (Fig. 1 B, Bottom Middle, and C, green circles) compared with neurons lacking Pum2 and control neurons (mismatch control shRNA; Fig. 1 B, Top, and C, black circles). In neurons lacking Pum2, we observed a significant increase in the number of dendritic line crossings between 20 and 36 μm (P < 0.001). In contrast, neurons overexpressing Pum2-EYFP displayed a significant reduction in dendritic line crossings between 20 and 60 μm (P < 0.001 between 20 and 36 μm, P < 0.01 between 44 and 60 μm) as compared with control shRNA-transfected neurons.
Fig. 1.
Pum2 is a negative regulator of dendrite development. (A) Whole-brain extracts were probed for Pum2 and tubulin at selected time points of neuronal morphogenesis. Pum2 protein levels were expressed as a ratio of tubulin and subsequently normalized to the ratio at embryonic day 17 (E17). P, postnatal day. (B) Representative photographs of neurons transfected with control pSUPERIOR shRNA (Top), shPum2 (Top Middle), or Pum2-EYFP (Bottom Middle) and cotransfected with shPum2 and an RNAi Pum2R (Bottom). E17 rat hippocampal neurons were transfected at 7 DIV, followed by fixation and staining for MAP2 and analysis at 10 DIV. (Scale bars: 10 μm.) (C) Sholl quantification of dendritic arbor complexity. Neurons lacking Pum (shPum2, red circles) show significantly more line crossings between 20 and 36 μm than control (control shRNA, black circles; P < 0.001). Conversely, neurons overexpressing Pum2-EYFP (green circles) demonstrate significantly fewer (P < 0.001 between 20 and 36 μm and P < 0.01 between 44 and 60 μm) line crossings between 20 and 60 μm than control neurons. Neurons cotransfected with shPum2 and Pum2R also demonstrated significantly fewer line crossings (P < 0.001 between 20 and 36 μm and P < 0.01 between 44 and 60 μm). Three independent experiments were performed, and at least 30 neurons were analyzed for each group. (D) Quantification of primary dendrites and primary branch points. (Left) Neurons lacking Pum2 demonstrate significantly more for shPum2-treated neurons (red box) compared with control (black box). Overexpression of Pum2 resulted in a significant decrease of primary dendrites (green box). Cotransfection with shPum2 and Pum2R yielded no significant changes in primary dendrite numbers (blue box). (Right) Quantitative analysis of primary branch points reveals a significant increase in shPum2-treated neurons (red box) and a significant decrease in Pum2-EYFP-overexpressing neurons (green box) when compared with control (black box). Neurons cotransfected with shPum2 and Pum2R (blue box) had a comparable number of primary branch points as control. n.s., not statistically significant. **P < 0.01; ***P < 0.001. (E) Protrusions from neurons lacking Pum2 express dendritic markers. Control neurons and neurons transfected with shPum2 were stained for the dendritic postsynaptic protein homer. Dendrites from both groups expressed this marker (white arrows), indicating that these structures are functionally comparable to control dendrites. All boxes are 50 μm in height.
To determine whether the shPum2 effects are specific, a shRNA cleavage-resistant Pum2 mRNA (Pum2R; Fig. S1) was cotransfected together with the shPum2 vector. This yielded comparable results to Pum2-EYFP overexpression in that the dendritic arbor was simplified (Fig. 1 B, Bottom, and C, blue circles). shPum2- and Pum2R-coexpressing cells displayed a significant reduction in dendritic line crossings between 20 and 60 μm (P < 0.001 between 20 and 36 μm, P < 0.01 between 44 and 60 μm) as compared with control. This indicates that the effects of the shRNA targeting Pum2 are specific, because the resistant form of Pum2 was able to reverse the observed phenotype. The fact that we see overcompensation suggests that the Pum2R construct is expressed at higher levels than the shPum2, causing a similar phenotype, as observed on Pum2–EYFP overexpression.
We next asked if these changes in dendritic complexity were the result of changes in primary dendritic outgrowth from the soma and/or changes in dendritic branching. We quantified primary dendrites (Fig. 1D, Left) and primary branch points (Fig. 1D, Right). Control neurons had an average of 5.9 (±0.38 SE) primary dendrites emerging from the soma, whereas neurons lacking Pum2 had significantly more (7.9 ± 0.55 SE; P < 0.01). In contrast, Pum2-EYFP overexpressing had significantly fewer (4.1 ± 0.44 SE; P < 0.001). Neurons coexpressing shPum2 and Pum2R had no significant change in primary dendritic outgrowth (5.8 ± 0.35 SE) as compared with control. When primary branch points were quantified (Fig. 1D, Right), neurons lacking Pum2 had significantly more (6.5 ± 0.40 SE; P < 0.01) than control neurons (4.9 ± 0.34 SE). Pum2-EYFP overexpressing neurons had significantly fewer branch points (2.6 ± 0.43 SE; P < 0.001), whereas neurons coexpressing shPum2 and Pum2R did not show significant changes compared with control (4.3 ± 0.27 SE). If primary dendrites branch at the same frequency in all conditions, one would expect that branching would change in relation to primary dendrite outgrowth. Control neurons had a ratio of 0.67 (±0.03 SE) primary branch points compared with primary dendrites. Although this is the trend in our data, neurons lacking Pum2 or overexpressing Pum2-EYFP have a slightly lower ratio of primary branch points to primary dendrites as compared with control (0.61 ± 0.03 SE vs. 0.57 ± 0.07 SE), respectively. Taken together, this suggests that Pum2 negatively regulates dendritic outgrowth. Loss of Pum2 leads to enhanced dendritic arborization, and overexpression of Pum2 causes the reverse effect. We were interested in determining if these additional dendrites forming on neurons lacking Pum2 were functionally comparable to normal or control dendrites. They were MAP2-positive (Fig. 1B). We also stained shPum2-treated neurons with an antibody directed against the postsynaptic protein homer (Fig. 1E). The majority of dendrites on control and shPum2-treated neurons expressed homer puncta along the shaft (Fig. 1E, white arrows), indicating that they are functionally comparable to control dendrites. These findings on dendrite morphogenesis are reminiscent of Drosophila, wherein Pum is essential for proper development of the dendritic arbor of peripheral neurons (1). Furthermore, miR-134 has been recently discovered to promote dendritic outgrowth by inhibiting translation of Pum2 mRNA (10).
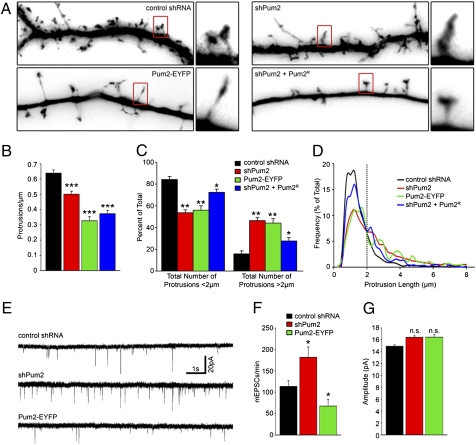
Because other dendritically localized RBPs have been demonstrated to influence dendritic spine morphogenesis (8, 14), we asked whether loss of Pum2 would have similar properties. Mature 15 DIV neurons were transfected with control shRNA, shPum2, or Pum2-EYFP or were cotransfected with shPum2 and Pum2R and fixed 3 days later (Fig. 2A). On visualization of GFP-filled dendrites, a convenient marker for transfection, control dendrites formed stereotypical mushroom-shaped protrusions, typically less than 2 μm in length (15), whereas neurons lacking Pum2 or overexpressing Pum2-EYFP had predominantly long, thin, filopodia-like structures (>2 μm in length). Although both types of structures were found in all experimental conditions, the ratio was significantly altered (Fig. 2 C and D). When measured, 84.67% of all protrusions were under 2 μm in length in control shRNA-treated neurons. In neurons lacking Pum2, 53.55% of all protrusions were less than 2 μm in length. Similar results were obtained on Pum2-EYFP overexpression, wherein 55.92% of all protrusions were <2 μm in length. When expressed as a percentage of the total number of quantified protrusions, the shift toward longer filopodia-like structures greater than 2 μm in length was significant for both shPum2 and Pum2-EYFP (Fig. 2C; P < 0.01). We also noticed a significant reduction in protrusion density in both treatment groups (Fig. 2B): Control neurons had 0.64 (±0.02 SE) protrusions per micrometer, whereas neurons lacking Pum2 had 0.50 (±0.02 SE; P < 0.001) and Pum2-EYFP-expressing neurons had 0.33 (±0.03 SE; P < 0.001) protrusions per micrometer, respectively. Coexpression of shPum2 and Pum2R resulted in partial rescue of protrusion morphology. Although these neurons had significantly fewer protrusions less than 2 μm compared with control (72.32% ± 2.98 SE; P < 0.05), there was significant recovery of the phenotype produced by shPum2 treatment. Coexpression of shPum2 and Pum2R did not result in rescue of protrusion density (0.37 ± 0.02 SE protrusions per micrometer; P < 0.001). However, while attempting to rescue the dendritic arbor phenotype, we observed that expression of Pum2R was stronger than that of shPum2, and therefore produced a phenotype reminiscent of Pum2-EYFP overexpression. Interestingly, both shPum2 knock-down and Pum2-EYFP overexpression lead to a similar phenotype of longer protrusions with a lower density. It may not be possible to observe a rescue phenotype in this instance, because Pum2R expression will presumably induce the same phenotype as Pum2-EYFP and mask any potential rescue.
Fig. 2.
Pum2 regulates dendritic spine morphology and mEPSC frequency. (A) Representative dendrites from neurons treated with control shRNA (Top Left), shPum2 (Top Right), or Pum2-EYFP (Bottom Left) and cotransfected with shPum2 and Pum2R (Bottom Right) at 15 DIV and fixed at 18 DIV. Representative examples for each group are boxed in red and shown as enlargements to the right. The length of each dendrite panel is 50 μm. (B) shPum2-treated neurons (red bars), Pum2-EYFP-expressing neurons (green bars), and cotransfected neurons with shPum2 and Pum2R (blue bars) all had a significant (***P < 0.001) reduction in GFP-positive protrusion density. (C) Quantification of GFP-filled dendritic protrusion length. Control shRNA-treated neurons (black bars) had significantly more protrusions that were less than 2 μm in length compared with neurons lacking Pum2 (red bars; P < 0.01), neurons expressing Pum2-EYFP (green bars; P < 0.01), and neurons expressing shPum2 and Pum2R (blue bars; P < 0.05). (D) Frequency distribution of protrusion length. Pum2 down-regulation (red line) increased the average length of protrusions compared with control shRNA-treated neurons (black line), Pum2-EYFP-expressing neurons (green line) and shPum2- and Pum2R-cotransfected neurons (blue line). The dotted line indicates a length of 2 μm. (E) Representative recordings of mEPSCs from shPum2-treated neurons (Middle), Pum2-EYFP-overexpressing neurons (Bottom), and control shRNA-treated neurons (Top). (F) Neurons lacking Pum2 (red bar) show a significant increase in mEPSC frequency (*P < 0.05). Pum2-EYFP-expressing neurons (green bar) demonstrated a significant (*P < 0.05) reduction in mEPSC frequency. (G) shPum2-treated and Pum2-EYFP-expressing neurons (red and green bars, respectively) do not display significant changes in mEPSC amplitude. n.s., not statistically significant.
These changes in protrusion morphology and density prompted us to investigate the electrophysiological properties of neurons misexpressing Pum2. We therefore measured the frequency and amplitude of miniature excitatory postsynaptic currents (mEPSCs) of control, shPum2-treated, and Pum2-EYFP-overexpressing neurons (Fig. 2E). Surprisingly, loss of Pum2 led to an increase in the mean number of mEPSCs, whereas overexpression of Pum2-EYFP led to a decrease (Fig. 2 E and F). Control neurons produced a mEPSC frequency of 118.39 (±14.27 SE) per minute. Loss of Pum2 led to a significant increase (204.33 ± 27.29 SE; P < 0.05) and neurons overexpressing Pum2-EYFP displayed a significant reduction (73.79 ± 13.12 SE; P < 0.05). Interestingly, the mEPSC amplitude was not significantly affected (Fig. 2G). These changes in mEPSC frequency but not amplitude were unexpected. Loss of Stau2 in cultured hippocampal neurons resulted in a similar dendritic protrusion phenotype. However, these neurons demonstrated no changes in mEPSC frequency but significant reductions in amplitude (8).
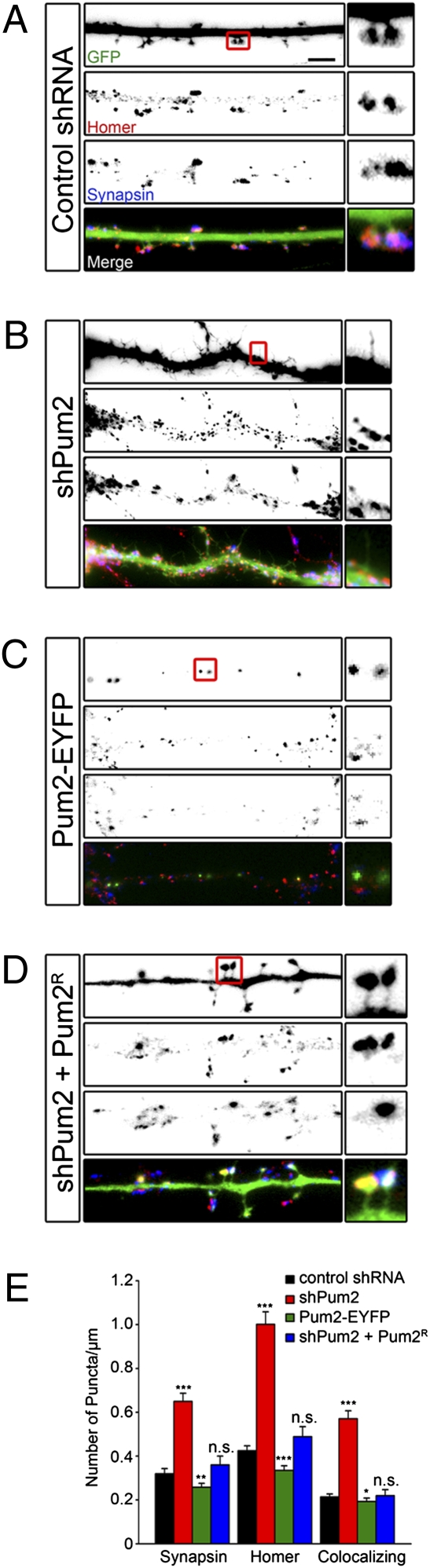
These unexpected results led us to explore whether there are concomitant changes in dendritic excitatory synapse numbers in neurons with altered Pum2 expression levels. We stained neurons lacking Pum2 or overexpressing Pum2-EYFP with synapsin and homer, pre- and postsynaptic marker proteins, respectively. Both have been previously used to quantify excitatory synapse number (16). In control neurons, GFP-filled dendritic spines and homer puncta very often colocalized and were in close proximity to synapsin antibody puncta, suggesting that they represent functional excitatory synapses (Fig. 3A). We quantified both synapsin and homer puncta along control dendrites and found an average of 0.32 (±0.02 SE) and 0.42 (±0.02 SE) puncta per micrometer, respectively (Fig. 3E, black bars). Neurons lacking Pum2 predominantly formed the long thin filopodia previously described in Fig. 2. However, when stained with the synapsin and homer antibodies, there were noticeable increases in puncta for both markers along the dendritic shafts (0.62 ± 0.04 SE and 0.99 ± 0.05 SE for synapsin and homer, respectively; P < 0.001; Fig. 3 B and E, red bars). Conversely, neurons overexpressing Pum2-EYFP demonstrated significantly fewer synapsin and homer puncta as compared with control (0.23 ± 0.02 SE, P < 0.01 and 0.28 ± 0.02 SE, P < 0.001, respectively; Fig. 3 C and E, green bars). Furthermore, the homer puncta found along the dendritic shafts of Pum2-EYFP-overexpressing neurons were weaker in intensity and smaller in size (Fig. 3C). Critically, we were able to rescue this effect by cotransfecting shPum2 and Pum2R. These neurons displayed no significant changes in synapsin or homer puncta (0.36 ± 0.04 SE vs. 0.49 ± 0.05 SE, respectively; Fig. 3 D and E, blue bars). Because colocalization of these two markers is considered to represent a functional synapse (16), we quantified the frequency of synapsin and homer colocalization along the dendrites of the neurons, which was 0.21 (±0.01 SE) per micrometer (Fig. 3E) in control neurons. Neurons lacking Pum2 expressed significantly more colocalizing puncta (0.52 ± 0.03 SE; P < 0.001) per micrometer, and neurons overexpressing Pum2-EYFP expressed significantly fewer (0.15 ± 0.02 SE; P < 0.05) per micrometer, respectively. Neurons coexpressing shPum2 and Pum2R did not demonstrate any significant changes in colocalizing puncta (0.22 ± 0.03 SE; Fig. 3E) as compared with control. Taken together, our data suggest that loss of Pum2 significantly enhances excitatory synapse number on dendritic shafts, even though changes in spine morphology are observed. This, in turn, leads to increases in mEPSC frequency in neurons lacking Pum2 and decreases in neurons overexpressing Pum2-EYFP. On investigation with antibodies specific for postsynaptic components of inhibitory synapses, we did not find any changes in inhibitory synapse number.
Fig. 3.
Pum2 regulates excitatory synapse number. Neurons were transfected with control shRNA (A), shPum2 (B), or Pum2-EYFP (C) or were cotransfected with shPum2 and Pum2R (D) at 15 DIV, fixed, and either GFP fluorescence (A–D, Top) -detected or -stained for the indicated proteins: postsynaptic homer (A–D, Top Middle) and presynaptic synapsin (A–D, Bottom Middle) at 18 DIV. (A–D, Bottom) Merged images. Representative dendrites are shown in A–D; representative protrusions for each group are boxed in red and enlarged. The length of each box is 50 μm. (E) Quantification of synapsin and homer puncta. Neurons from three independent experiments were transfected at 15 DIV, fixed, and stained at 18 DIV. In each experiment, at least four neurons from each group were quantified. Neurons lacking Pum2 (red bars) showed significant increases (***P < 0.001) in both synapsin and homer puncta as compared with control shRNA-treated neurons (black bars). In contrast, Pum2-EYFP-transfected neurons (green bars) demonstrated a significant reduction in both homer puncta (***P < 0.01) and synapsin puncta (**P < 0.01) as compared with control neurons (black bars). However, neurons cotransfected with shPum2 and Pum2R (blue bars) displayed no significant changes in either homer or synapsin puncta when compared with control. n.s., not significant. When colocalizing homer and synapsin puncta were quantified, shPum2-treated neurons had significantly more than control (P < 0.001), whereas neurons expressing Pum2-EYFP had significantly fewer (P < 0.05) than control. Neurons cotransfected with shPum2 and Pum2R had comparable colocalizing puncta as control.
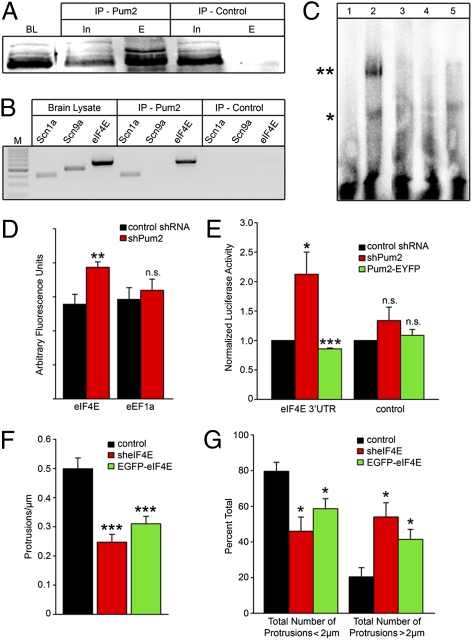
Because Pum2 is an RBP that represses the translation of target mRNAs (17), we hypothesized that these developmental changes observed on Pum2 misexpression were attributable to alterations in the translational state of putative Pum2 target mRNAs. We immunoprecipitated (IP) Pum2-containing RNPs using an affinity-purified Pum2 antibody (Fig. 4A) and subjected the eluted RNA to RT-PCR to test for candidate Pum2-associated mRNAs (2, 4). Pum2 associated with both the eIF4E and scn1a mRNAs, encoding the eIF4E translation factor and a voltage-gated sodium channel, respectively (Fig. 4B). Although scn9a contains a 3′-UTR similar to that of scn1a, including the Pum2 consensus binding site (18), we could not detect this message in our Pum2 IPs (Fig. 3B). To confirm that Pum2 was specifically interacting with the eIF4E mRNA, we performed an EMSA using the 3′-UTR of eIF4E and recombinant Pum2 protein. When combined, Pum2 led to a shift in the migration of the radiolabeled probe (Fig. 4C, lane 2, asterisk). Incubation with the anti-Pum2 antibody caused a supershifted band, indicating direct interaction of the two molecules (Fig. 4C, lane 2, double asterisk). In contrast, the interaction did not occur when the mRNA was incubated with the N terminus of the Pum2 protein lacking the RNA-binding domain (Fig. 4C, lane 3). The interaction was also successfully abolished by competition with an unlabeled probe, again verifying the specificity of the binding of Pum2 and eIF4E but not with an unrelated RNA (Fig. 4C, lanes 4 and 5, respectively). We next set out to determine whether eIF4E protein levels were altered in neurons lacking Pum2. Neurons were transfected at 15 DIV and stained with a mouse anti-eIF4E antibody 3 days later. Fluorescence intensity was measured in the soma and expressed as arbitrary units. In neurons lacking Pum2, eIF4E levels were significantly increased from 55.60 (±5.86 SE) arbitrary units to 77.32 (±3.21 SE; P < 0.01; Fig. 4D). Levels of eEF1A protein, whose mRNA has been predicted to be a Pum2 target (19), were not changed in response to loss of Pum2 (Fig. 4D). We also could not detect eEF1A mRNA in our Pum2 IPs. We finally set out to determine if Pum2 acts as a translational repressor of eIF4E. The 3′-UTR of the eIF4E mRNA was fused to the Renilla luciferase gene and coexpressed, together with firefly luciferase, for internal normalization in cortical neurons. Luciferase activity was significantly higher (2.12 ± 0.38 SE normalized luciferase activity units; P < 0.05; Fig. 4E, red bars) in Pum2-deficient neurons compared with control (Fig. 4E, black bars). Neurons overexpressing Pum2-EYFP demonstrated reduced levels of Renilla luciferase activity (0.86 ± 0.01 SE normalized luciferase activity units; P < 0.001; Fig. 4E, green bars). Control experiments lacking the eIF4E 3′-UTR did not show significant changes in Renilla luciferase activity (Fig. 4E).
Fig. 4.
Pum2 binds and regulates specific transcripts. (A) Immunopurified Pum2-specific antibodies immunoprecipitate Pum2 protein. BL, forebrain lysate; IP-Pum2, immunoprecipitation using Pum2 antibodies; IP-control, control immunoprecipitation using preimmune sera; In, input; E, elution. (B) Specific mRNAs are enriched in the IP-Pum2 eluates. Pum2-specific antibodies were used to IP Pum2-containing RNPs, and RT-PCR was performed to identify candidate targets. Scn1a and eIF4E mRNAs were detected in brain lysates as well as in the IP-Pum2 but not in the IP-control. Scn9a mRNA was detected in the brain lysate but not in the IP-Pum2 or IP-control. (C) EMSA showing specific binding of Pum2 to eIF4E. The eIF4E probe in the absence of Pum2 is depicted in lane 1. Lane 2 shows the Pum2-eIF4E RNA complex (*) as well as a supershift of the complex on incubation with anti-Pum2 antibody (**). Pum2 lacking the RNA-binding domain (Pum Nt) does not bind to eIF4E (lane 3; see Fig. S2). Binding is confirmed by competition with unlabeled eIF4E probe (lane 4) and unrelated RNA (lane 5). (D) Pum2 down-regulation leads to increased eIF4E protein levels. Neurons were treated with shPum2 (red boxes) or control shRNA (black boxes) and stained for either eIF4E or eEF1A. Significantly more (P < 0.001) eIF4E protein is detected as compared with control. eEF1a protein levels remained unchanged. n.s., not significant. (E) Pum2 binds the 3′-UTR of the eIF4E mRNA and represses translation. The eIF4E 3′-UTR was fused to the Renilla luciferase gene and transfected via electroporation. In neurons lacking Pum2 (shPum2, red bars), significantly more (*P < 0.05) luciferase activity was detected (black bars). In neurons expressing Pum2-EYFP (green bars), significantly less (***P < 0.001) luciferase activity was found eIF4E regulates dendritic spine morphology. (F) sheIF4E-treated neurons (red bars) and EGFP-eIF4E-expressing neurons (green bars) had a significant (***P < 0.001) reduction in GFP-positive protrusion density compared with control (black bars). (G) Quantification of GFP-filled dendritic protrusion length. Control neurons (black bars) had significantly more protrusions less than 2 μm in length compared with neurons lacking eIF4E (red bars; *P < 0.05) and neurons overexpressing EGFP-eIF4E (green bars; *P < 0.05).
The influence of Pum2 on eIF4E translation prompted us to investigate whether misexpression of eIF4E led to neuronal phenotypes similar to those produced by misexpression of Pum2. Neurons were transfected with plasmids expressing either eIF4E (EGFP-eIF4E) or shRNAs specific for eIF4E transcript (sheIF4E). We first determined the levels of the up- and down-regulations in neurons compared with the endogenous eIF4E by quantitative Western blot (Fig. S2 D and E). We then analyzed dendritic branching in neurons on either overexpression or down-regulation of eIF4E. To our surprise, a significant increase in the number of line crossings (P < 0.05) for all radial distances in sheIF4E-transfected neurons was observed (Fig. S2F). Overexpression of EGFP-eIF4E led to a small reduction of dendritic branching. Similar results were obtained using a second vector expressing shRNA targeting a different region of eIF4E, suggesting that the observed phenotype was related to eIF4E down-regulation. To determine the effects on mature neurons, 15 DIV neurons were transfected with control shRNA, sheIF4E, or EGFP-eIF4E and fixed 3 days later (Fig. 4 F and G). Neurons misexpressing eIF4E demonstrated a similar phenotype when compared with neurons misexpressing Pum2. We observed a significant reduction in protrusion density in neurons lacking eIF4E or overexpressing EGFP–eIF4E (Fig. 4F): Control neurons had 0.50 (±0.04 SE) protrusions per micrometer, whereas neurons lacking eIF4E had 0.25 (±0.03 SE; P < 0.001) and EGFP–eIF4E-expressing neurons had 0.31 (±0.03 SE; P < 0.001) protrusions per micrometer, respectively. Both treatment groups also displayed altered protrusion morphology (Fig. 4G). These effects are comparable to those observed with misexpression of Pum2, in which we found longer filopodia-like structures. When measured, 79.57% of all protrusions were under 2 μm in length in control neurons. In neurons lacking eIF4E, 45.99% of all protrusions were less than 2 μm in length. Similar results were obtained on EGFP-eIF4E overexpression, in which 58.6% of all protrusions were less than 2 μm in length. When expressed as a percentage of the total number of quantified protrusions, the shift toward longer filopodia-like structures greater than 2 μm in length was significant for both shPum2 and Pum2–EYFP (P < 0.05; Fig. 4G).
In summary, our work demonstrates a critical contribution of Pum2 to dendrite development and synaptic function in cultured hippocampal neurons. In mature neurons, Pum2 regulates the number of excitatory synapses along the dendritic shaft. Furthermore, Pum2 significantly affects mEPSC frequency. The fact that Pum2-deficient neurons show an increase in mEPSC frequency suggests that these neurons have increased synapse numbers. This is indeed what we found. However, we cannot exclude the possibility that there might be an increased presynaptic transmitter vesicle release probability. Further work is needed to resolve this interesting aspect. Our work also suggests an interesting mechanism of translational control within transport particles (2, 20). In Drosophila, Pum binds a series of transcripts, with one example being eIF4E (2, 4). eIF4E has been shown to be part of translational particles near synapses (2). Knock-down of Pum led to an increase in eIF4E, and it was concluded that Pum was negatively regulating the translation of eIF4E. Our work now presents convincing evidence that vertebrate Pum2 negatively regulates the translation of eIF4E and other transcripts, such as scn1a, in hippocampal neurons. The newly synthesized eIF4E would then, in turn, critically regulate local protein synthesis at the synapse, and thereby dendritic spine morphogenesis and synaptic function.
A study was published during the time this paper was undergoing revision showing that Pum2 can bind the 5′-cap, and thereby competes with eIF4E to control RNA translation (21). Interestingly, this capacity seems to be linked to critical amino acids located upstream of the RNA binding motifs in the Pum2 protein. Taken together, these results (21), together with our data, suggest that Pum2 and eIF4E are linked in a complex manner. We show that Pum2 binds and regulates the translation of the eIF4E mRNA. Independent of this role, the two proteins compete for interaction with the 5′-cap of Xenopus RINGO/SPY RNA to repress (in the case of Pum2) or enhance (in the case of eIF4E) translation of the target mRNAs (21). Disrupting Pum2 or eIF4E expression appears to disrupt this complex relation that leads to our observed phenotypes in neurons.
It will be interesting to determine whether the 5′-cap-binding activity of Pum2 is restricted to specific mRNAs. Potential RNA targets for both mammalian Pum1 and Pum2 have been described (22). Although the molecular interactions between Pum1 and 2 are still elusive, Pum proteins show similar substrate specificity and the sets of associated mRNAs strongly overlap. In addition, Pum proteins may regulate ≈15% of the cells’ transcriptome, suggesting a more general functional role in the control of translation by recruiting previously undescribed or uncharacterized protein components. In fact, the protein composition of Pum2 complexes remains elusive (23). For this reason, future investigators hope to identify the molecular composition of Pum2-containing RNPs in neurons and to unravel unique regulatory mechanisms governing translational control. Because long-term memory in adult Drosophila requires Pum (6), our work provides important insight into the underlying mechanism of how Pum regulates synaptic translation, thereby affecting dendritic spine morphogenesis and synaptic function in hippocampal neurons. Future experiments will have to provide experimental data verifying this exciting hypothesis.
Materials and Methods
Constructs, Hippocampal Cell Culture and Transient Transfection.
For details of all constructs, cloning, and antibodies, see SI Methods. Rat hippocampal neurons were cultured and transiently transfected as described (7, 24, 25).
EMSAs.
EMSA was performed as previously described (26) with modifications (see SI Methods).
Dual Luciferase Assay.
The 3′-UTR of mouse eIF4E transcript was amplified by PCR and cloned downstream of the Renilla luciferase gene into the psiCHECK-2 vector (Promega) expressing both Renilla and firefly luciferase. As control, empty luciferase reporter plasmid without the 3′-UTR was used (For furhter details, see SI Methods).
IP and RT-PCR.
Adult rat brain was homogenized in ice-cold extraction buffer [EB; 25 mM Hepes (pH 7.4), 150 mM KCl, 20 mM MgCl2, 8% (v/v) glycerol, 0.1% Nonidet P-40, 1 mM DTT, RNase inhibitor, and standard protease inhibitor mixture]. One hundred micrograms of affinity-purified anti-Pum2 antibody or an adequate amount of the respective preimmune serum was incubated with 100 μg of Protein A Sepharose beads (Amersham Bioscience) for 2 h at 4 °C. The latter were preincubated/blocked with 60 μg of tRNA for 45 min rotating at 4 °C. After four washes with 1× EB, half of the immunoprecipitate (50 μg of the beads) was analyzed via Western blot and the other half was treated with TRIzol (Invitrogen) to isolate total RNA following the manufacturer’s instructions. After RT, the cDNA was used as a template for PCR. For primer sequences, see Table S1.
Sholl and Data Analysis.
Sholl analysis was performed as previously described (13, 27). For further details, see SI Methods.
Electrophysiology.
For details of the whole-cell patch clamp recording setup, and experimental design, see SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. Eric Klann, Reinhard Lührmann, Werner Sieghart, and Nahum Sonenberg for kindly providing reagents and for helpful discussions. We thank Kristina Kosenburger, Julia Sandholzer, and Martina Schwarz for their excellent technical assistance. We are grateful to Drs. Jürgen Sandkühler, Ralf Dahm, Alejandra Gardiol, Sigismund Huck, and Alessandro Quattrone for comments and critical discussions. This work was supported by Austrian Science Funds (to P.M. and M.A.K), by a Human Frontier Science Program Organization network grant and the European Science Foundation program RNAQuality (to M.A.K.), and by a Cassa di Risparmio di Trento e Rovereto grant Foundation (to P.M.) The financial support of the University of Trento (Progetto Biologia) through Prof. Marco Andreatta is gratefully acknowledged (to P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See companion paper: 10.1073/pnas.0911451107.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907128107/DCSupplemental.
References
- 1.Ye B, et al. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 2.Menon KP, et al. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 3.Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci. 2009;29:5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 7.Vessey JP, et al. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goetze B, et al. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J Cell Biol. 2006;172:221–231. doi: 10.1083/jcb.200509035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeitelhofer M, et al. Improved protocol for high-efficiency transfection of shRNA-encoding plasmids into primary hippocampal neurons. J Neurosci Res. 2009;87:289–300. doi: 10.1002/jnr.21840. [DOI] [PubMed] [Google Scholar]
- 13.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- 14.Comery TA, et al. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hering H, Sheng M. Dendritic spines: Structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 16.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 17.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 18.White EK, Moore-Jarrett T, Ruley HE. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA. 2001;7:1855–1866. [PMC free article] [PubMed] [Google Scholar]
- 19.Fox M, Urano J, Reijo Pera RA. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Sigrist SJ, et al. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature. 2000;405:1062–1065. doi: 10.1038/35016598. [DOI] [PubMed] [Google Scholar]
- 21.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galgano A, et al. Comparative analysis of mRNA targets for human PUF-family pro-teins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Zeitelhofer M, et al. High-efficiency transfection of mammalian neurons via nucleofection. Nat Protoc. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- 25.Macchi P, et al. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J Neurosci. 2003;23:5778–5788. doi: 10.1523/JNEUROSCI.23-13-05778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luzi E, Eckstein F, Barsacchi G. The newt ribozyme is part of a riboprotein complex. Proc Natl Acad Sci USA. 1997;94:9711–9716. doi: 10.1073/pnas.94.18.9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y, et al. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr Biol. 2007;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.