Abstract
DNA ligase IV (LIG4) is an essential component of the nonhomologous end-joining (NHEJ) repair pathway and plays a key role in V(D)J recombination. Hypomorphic LIG4 mutations in humans are associated with increased cellular radiosensitivity, microcephaly, facial dysmorphisms, growth retardation, developmental delay, and a variable degree of immunodeficiency. We have generated a knock-in mouse model with a homozygous Lig4 R278H mutation that corresponds to the first LIG4 mutation reported in humans. The phenotype of homozygous mutant mice Lig4R278H/R278H (Lig4R/R) includes growth retardation, a decreased life span, a severe cellular sensitivity to ionizing radiation, and a very severe, but incomplete block in T and B cell development. Peripheral T lymphocytes show an activated and anergic phenotype, reduced viability, and a restricted repertoire, reminiscent of human leaky SCID. Genomic instability is associated with a high rate of thymic tumor development. Finally, Lig4R/R mice spontaneously produce low-affinity antibodies that include autoreactive specificities, but are unable to mount high-affinity antibody responses. These findings highlight the importance of LIG4 in lymphocyte development and function, and in genomic stability maintenance, and provide a model for the complex phenotype of LIG4 syndrome in humans.
Keywords: genomic instability, immunodeficiency, lymphocytes, nonhomologous end joining, immune dysregulation
Nonhomologous end joining (NHEJ) is one of the two major DNA repair pathways in mammalian cells that protect the genome against DNA double-stranded breaks (DSBs) generated by genomic insults such as ionizing radiation (IR) or reactive oxygen species or that arise during V(D)J recombination (1) and Ig heavy chain (IGH) class switch recombination (CSR) (2). V(D)J recombination is the process by which developing T and B lymphocytes assemble their antigen receptor variable region exons. In this process, the recombinase activating gene (RAG)1 and RAG2 proteins introduce DSBs at recombination signal sequences (RSS) that flank coding variable (V), diversity (D), and joining (J) elements. This process generates hairpin-sealed coding ends and blunt phosphorylated signal ends. These DNA ends are recognized and resolved by proteins of the NHEJ pathway, with formation of coding joins and recombination signal (RS) joins, respectively. In particular, the KU70/KU80 heterodimer binds directly the DNA ends, allowing activation of the DNA-PK catalytic subunit (DNA-PKcs) (3) and phosphorylation of Artemis, which mediates opening of hairpin-sealed DNA coding ends (4, 5). In the last phase of NHEJ, the DNA ligase IV (LIG4)/XRCC4 complex mediates ligation of the DNA ends (6). In addition, the XRCC4-like factor (XLF), also known as Cernunnos, participates in this process (7, 8).
In humans, mutations in Artemis, DNA-PKcs, and Cernunnos/XLF have been associated with combined immunodeficiency (4, 7, 9, 10). Mutations in the DNA ligase IV (LIG4) gene underlie the LIG4 syndrome, a rare autosomal disorder characterized by cellular radiosensitivity, microcephaly, neurological abnormalities, bone marrow failure, and increased susceptibility to malignancies (11). A variable degree of immunodeficiency has been reported in patients with LIG4 syndrome, ranging from apparent lack of immunological defects (12) to severe defects in T and B cell development, resulting in SCID (13–15) or in Omenn syndrome (16). The molecular basis for this phenotypic heterogeneity is still unclear.
In mice, Lig4 deficiency is embryonically lethal and is associated with cellular radiosensitivity, massive neuronal apoptosis, and arrest in T and B cell development (17, 18). Both the embryonic lethality and neuronal apoptosis, but not the lymphoid development defect and the cellular radiosensitivity, can be rescued by simultaneous p53 deficiency (19). Lig4 haploinsufficiency contributes to translocations and cancer in certain cell cycle checkpoint deficient backgrounds (20).
We have generated a knock-in mouse model with a homozygous Lig4 arginine to histidine (R278H) mutation that corresponds to the mutation identified in the first LIG4-deficient patient, who developed T cell leukemia associated with increased cellular radiosensitivity (12). Here, we demonstrate that mice homozygous for this mutation represent a model of the complex cellular and clinical phenotype observed in patients with LIG4 syndrome.
Results
Generation and Characterization of Lig4R/R Cells and Mice.
A targeting construct carrying the CGC to CAT mutation at codon 278 of the Lig4 gene (resulting in the R278H amino acid substitution) and a neomycin-resistance gene (NeoR) flanked by LoxP sites was used for homologous recombination in TC1 (129/Svev) embryonic stem (ES) cells (Fig. S1A). Southern blot analysis revealed ES clones carrying the Lig4 R278H mutant allele (Fig. 1A). Upon deletion of the NeoR cassette, subclones were injected for germline transmission to generate mice heterozygous for the R278H mutation (Lig4+/R mice).
Fig. 1.
Lig4 R278H/R278H (Lig4R/R) targeting of ES cells, R278H protein expression, and ionizing radiation sensitivity of the homozygous targeted ES cells. (A) Southern blot of EcoRI-digested DNA from the tails of germline mice. Probe B (Fig. S1A) was used to detect wild-type (+/+), heterozygous (+/R), and R278H/R278H (R/R) bands. (B) Western blot analysis of Lig4 protein expression in thymocytes from Ku70−/− Lig4−/− (indicated as −/−), Lig4+/+(+/+), Lig4+/R (+/R), and Lig4R/R (R/R) mice. Expression of tubulin is shown as a loading control. (C) Ionizing radiation (IR) sensitivity in +/+, R/R, and Xrcc4−/− MEFs. R/R MEFs are as IR sensitive as Xrcc4−/− MEFs. (D) Weight and size (mean ± SE) of 4-week-old +/+ (n = 10), +/R (n = 10), and R/R (n = 10) littermates. P values were determined by unpaired Student's t test.
Lig4+/R mice were intercrossed to produce homozygous mutant Lig4R/R mice. Lig4R/R mice, in contrast to Lig4−/− mice, were viable and generated according to Mendelian inheritance. Western-blot analysis, performed on total lysate of thymocytes from Lig4R/R mice, showed that the Lig4 R278H mutant is expressed, albeit at slightly reduced levels as compared to wild-type Lig4 (Fig. 1B). Lig4R/R mouse embryonic fibroblasts (MEFs) displayed increased radiation sensitivity (Fig. 1C), similar to XRCC4-deficient MEFs and to human cell lines carrying the Lig4 R278H mutation (11, 21). As an initial assay for potential NHEJ defects, we generated Lig4R/R ES cells by sequential gene targeting of the second allele of a Lig4+/R ES line (Fig. S1A) and then assayed ability to support V(D)J recombination and RS joins formation via a transient assay. These studies revealed that RS joining was severely impaired in Lig4R/R ES cells relative to that of wild-type TC1 ES cells (Fig. S2); although, in contrast to prior findings in Lig4−/− mice, we did find normal joins among the few recovered. In preliminary assays, we also found that coding joining appeared markedly impaired but that relatively normal joins could be recovered. Together, these studies suggest a severe but incomplete V(D)J recombination/NHEJ defect in Lig4R/R cells. At birth and at later ages, Lig4R/R mice are strikingly smaller than their wild-type littermate controls (Fig. 1D and Fig. S1 B and C), similar to Ku70- and Ku80-deficient mice (22). Fertility of Lig4R/R mice is severely compromised; therefore, generation of Lig4R/R mice was based on interbreeding of Lig4+/R mice.
T Cell Development and Function in Lig4R/R Mice.
Both the thymus and the spleen of Lig4R/R mice were small (Fig. S3A). Overall preservation of the thymic architecture and of the corticomedullary demarcation was observed in Lig4R/R mice (Fig. S3B). Total thymic cellularity and the absolute number of CD4− CD8− double-negative (DN), CD4+ CD8+ double-positive (DP), and single-positive (SP) CD4+ or CD8+ thymocytes were drastically reduced in Lig4R/R mice (Fig. 2A). Staining with anti-CD4 and anti-CD8 antibodies showed accumulation of DN thymocytes at the CD44− CD25+ DN3 stage of development (Fig. 2A), as expected for the severe defect in NHEJ and V(D)J recombination indicated by the transient assays of Lig4R/R ES cells (Fig. S2). However, consistent with the leakiness of the functional defect detected in the ES cell assays, we observed generation of very low numbers of DP and SP thymocytes (Fig. 2A).
Fig. 2.
Severe lymphopenia and T cell phenotype in Lig4R/R mice. (A) (Upper Left) Absolute number of total, double-negative (DN), double-positive (DP) and CD4 and CD8 single-positive thymocytes in wild-type (+/+) and Lig4R/R (R/R) mice. (Upper Right) Representative FACS analysis of thymocytes from an R/R mouse and its littermate control (+/+). (Lower Left) Representative analysis of CD44 and CD25 expression within CD4− CD8− DN thymocytes from an R/R mouse and its +/+ littermate control. (Lower Right) Distribution of DN cells at various stages of differentiation (DN1 to DN4) in R/R and in +/+ mice. (B) (Upper Panel) Absolute number of total splenocytes (Left) and of CD4+ and CD8+ splenic T cells (Right) in R/R mice and +/+ littermate controls (Lower Left) Expression of CD44 and CD62L within CD4+ (top) and CD8+ (bottom) splenic T cells from an R/R mouse and its +/+ littermate control. (Lower Right) Distribution of naive (CD44lo CD62Lhi) and effector memory (CD44hi CD62Llo) cells within CD4+ and CD8+ splenic T cells. Results are shown as the mean ± SE. Gating was on live lymphocytes and the percentages of the indicated populations are relative to the lymphocyte gate. P values were determined by unpaired Student's t test.
Analysis of endogenous T cell receptor (TCR) β coding joins and RS joins involving TCRVβ14 that were isolated from genomic DNA of Lig4R/R thymocytes revealed extensive deletions within RS joins (Fig. S4). In contrast to normal thymocytes where substantial numbers of nonproductive joins are recovered, nearly all coding joins recovered from Lig4R/R thymocytes were productive (Fig. S4). Although there could be several explanations for this finding, one possibility is that most joins in Lig4R/R encompass large deletions (23–25) that go beyond the PCR primers so that only highly selected normal joins are detected by the assay.
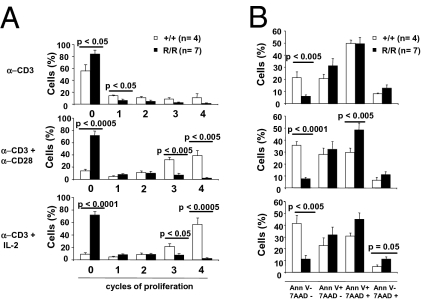
Total cellularity and the absolute number of SP CD4+ and CD8+ cells were also markedly reduced in the spleen of Lig4R/R mice (Fig. 2B). Preliminary results indicate that CD4+ and CD8+ peripheral T cells from Lig4R/R mice have a CD3hi CD24lo phenotype, similar to mature T lymphocytes from littermate controls (Fig. S5A). However, in contrast to what was observed in control mice, the vast majority of both SP CD4+ and SP CD8+ splenic T cells of Lig4R/R mice displayed a CD44+ CD62L− phenotype (Fig. 2B), suggesting in vivo activation. A dramatic defect of T cell proliferation, as assessed by 5,6-carboxyfluorescein succinimidyl ester (CFSE) dilution, was observed when Lig4R/R splenic T cells were cultured in vitro for 72 h in the presence of anti-CD3 mAb in combination with either anti-CD28 mAb or IL-2 (Fig. 3A and Fig. S5B). Furthermore, when simultaneously stained for annexin-V (AnnV) and 7-AAD, in vitro activated Lig4R/R splenic T cells showed markedly reduced viability (Fig. 3B).
Fig. 3.
Reduced proliferation and decreased viability of splenic T cells from Lig4R/R mice on in vitro activation. (A) Percentage of Lig4R/R (R/R) and control (+/+) splenic T cells at different cycles of proliferation, upon culture with anti-CD3, anti-CD3 plus anti-CD28, or anti-CD3 plus IL-2. Proliferation was assessed by CFSE dilution, gating on live cells. (B) Distribution of the percentages of live (AnnV− 7AAD−), early apoptotic (AnnV+ 7AAD−), late apoptotic (AnnV+ 7AAD+), and dead (AnnV− 7AAD+) cells upon culture with the indicated stimuli. Results are shown as mean percentage ± SE. P values were determined by unpaired Student's t test.
To characterize the ability of Lig4 R278H mutation to support generation and maintenance of a polyclonal T cell repertoire, we performed spectratyping analysis in thymic and splenic T cells of Lig4R/R mice and their Lig4+/+ littermates. Thymocytes from Lig4R/R mice showed a largely polyclonal distribution of the TCR third complementarity-determining region (CDR3) length for each TCRβ variable (TCRVB) gene family analyzed. However, oligoclonal patterns, indicative of a restriction in T cell repertoire, were detected in most of the TCRVB families analyzed in Lig4R/R splenic T cells (Fig. S6 and SI Text). These results, along with the activated/memory phenotype of peripheral T cells, are suggestive of peripheral expansion, leading to oligoclonality. Similar abnormalities have been reported in murine models of Omenn syndrome (26, 27). However, at variance with these models, Lig4R/R mice revealed only modest inflammatory infiltrates (mainly composed of T lymphocytes and neutrophils) in the gut and the liver (Fig. S7A). CD4+ CD25hi Foxp3+ regulatory T cells (Tregs) were detected in mesenteric lymph nodes from Lig4R/R mice (Fig. S7B and SI Text), although their function could not be tested due to low absolute number. In addition, splenic T cells from Lig4R/R mice showed no obvious skewing in their cytokine expression profile both under resting conditions and when cultured in vitro with anti-CD3 plus anti-CD28 mAbs, with the exception of reduced levels of IL-13 and IFN-γ secretion (Fig. S7C and SI Text).
B Cell Development and Function in Lig4R/R Mice.
The proportion of B220+ cells in the bone marrow of Lig4R/R mice was significantly reduced, when compared to littermate controls (Fig. 4A). The majority of B220+ IgM− cells from Lig4R/R mice expressed CD43, indicating an incomplete block at the pro-B cell stage (Fig. 4A). The proportion of B220+ IgM+ B cells in the spleen of Lig4R/R mice was markedly reduced; however, a residual number of these cells were consistently detected (Fig. S8A). These results are in contrast to the inability of Lig4−/− B cell progenitors (even on a p53-deficient background) to progress to the B220+ IgM+ stage both in vivo and in vitro (19). The splenic B cells of Lig4R/R mice showed a restricted repertoire (Fig. S9 and SI Text). IgG2b, IgG3, and IgA serum levels were significantly reduced in 8-week-old Lig4R/R mice; however, residual levels of serum IgM and relatively normal levels of IgG1, IgG2a, and IgE were detected (Fig. 4B). Although Lig4R/R mice had a lower number of transitional, follicular, and marginal zone splenic B cells, they had a normal number of B220high B cells that expressed intracytoplasmic IgG1 (Fig. 4C). Naive Lig4R/R mice spontaneously produced higher titers of 2,4,6-trinitrophenol (TNP)-specific IgM and IgG as compared to Lig4+/+ mice (P < 0.0005 and P < 0.005, respectively) (Fig. 4D). However, following immunization with the T-independent type II antigen TNP-Ficoll or with T-dependent antigen TNP–keyhole limpet hemocyanin (KLH), Lig4R/R mice produced lower amounts of antigen-specific IgM and IgG antibodies than Lig4+/+ mice (Fig. 4D), and no high-affinity TNP-specific IgG antibodies were detected in Lig4R/R mice after secondary immunization with TNP-KLH (Fig. S8B). Spontaneous production of low-affinity, polyreactive antibodies is often associated with autoimmunity. Indeed, increased levels of anti-ssDNA and anti-chromatin antibodies were detected in Lig4R/R mice (Fig. 4E). Overall, these data indicate that Lig4R/R mice have profound abnormalities in B cell development and antigen-specific antibody production; however, they spontaneously produce low-affinity IgM and IgG antibodies that contain self-reactive specificities.
Fig. 4.
Severe block in B cell development, Ig serum levels, antigen-specific antibody responses, and autoantibody production in Lig4R/R mice. (A) (Upper) Dot plot analysis of bone marrow cells labeled with B220 and IgM antibodies and CD43 expression on an electronically gated B220+ IgM− subset. (Lower) Percentage (mean ± SE) of bone marrow B cells of Lig4R/R (R/R) and Lig4+/+ (+/+) mice. P values were determined by unpaired Student’s t test. (B) Ig serum levels in 8-week-old R/R and +/+ mice. Statistical significance was assessed using two-way Student’s t test. (C) Absolute number (± SE) of follicular (Fo), marginal zone (MZ), and transitional (Tr) cells and intracellular (ic) IgG1+ cells in the spleens of 8-week-old R/R and +/+ mice (n = 3 per group). Markers used to define B cell subpopulations are reported in SI Text. (D) IgM and IgG antibody responses to the T-independent antigen TNP-Ficoll (Left) and the T-dependent antigen TNP-KLH (Right) in +/+ and R/R mice. (E) IgG autoantibodies to single-stranded DNA (ssDNA) (Left) and to chromatin (Right) in 8-week-old R/R and +/+ mice. Statistical significance was assessed using a Mann–Whitney U test.
Genomic Instability and Tumor Susceptibility.
Lig4R/R mice showed increased morbidity when compared to Lig4+/R and Lig4+/+ littermates (Fig. 5A). When moribund Lig4R/R mice were killed, a significant proportion (13/44) of them showed evidence of thymic tumors (Fig. 5A) with either a DP or a SP phenotype; no cases of DN thymic lymphomas were observed. Southern-blot analysis of 8 of 13 T cell tumors revealed clonal rearrangements or deletion of the Jβ1 cluster in the vast majority of the tumors and clonal rearrangements involving the Jβ2 cluster in several of them (T3, T5, T6, and T7 in Fig. 5B). No cases of B cell lymphomas were documented; however, colon adenocarcinoma and medulloblastoma were observed in 4 and 1 mice, respectively. To test whether the reduced life span and the increased occurrence of tumors were associated with genomic instability, positively selected CD43+ splenic T cells from Lig4R/R mice and from heterozygous Lig4+/R mice were cultured in vitro with Con A, and the presence of chromosomal abnormalities was evaluated by telomere-specific fluorescent in situ hybridization (T-FISH). Approximately 20% of the metaphases in Lig4R/R T cells contained aberrancies with the majority being chromosomal breaks and translocations, as compared to <2% chromosomal aberrancies in Lig4+/R cells (Fig. 5C).
Fig. 5.
Reduced survival, genomic instability of peripheral T cells, and spontaneous tumor development in Lig4R/R mice. (A) Kaplan–Meier survival curve of wild-type (+/+), Lig4+/R (+/R), and Lig4R/R (R/R) mice. Circles identify the age at which individual mice were found to have T cell tumors when killed. (B) Southern-blot analysis of T cell tumor clonality. Genomic DNA from eight tumor samples (T1–T8) from R/R mice was digested with EcoRI and probed with a 0.9-kb NcoI probe (probe B) downstream of Jβ1.6 and a 0.7-kb ClaI-BamHI probe (probe A) 3′ of Jβ2, respectively (47), and a control probe LR8 (48). Kidney genomic DNA was used as an internal control for germline configuration. (C) Telomere-FISH (T-FISH) analysis of genomic instability of Lig4R/R peripheral T cells upon in vitro activation. CD43+ splenic T cells from +/R and R/R mice were stimulated with Con A, hybridized with a telomere probe, counterstained with DAPI, and analyzed for chromosomal aberrations. Percentages ( ± SD) of metaphases containing chromosomal breaks and of the total aberrations observed (%) are shown. CB, chromosomal break; cb, chromatid break; T, translocation.
Discussion
Mutations of the LIG4 gene have been reported in 14 patients (11–16, 28–30) who shared growth retardation and microcephaly, but showed significant heterogeneity in the degree of immunological impairment and in the occurrence of tumors. Variability of the clinical phenotype has been attributed to different degrees of impairment of LIG4 protein expression and function associated with the various mutations. In the attempt to better define the pathophysiology of the phenotypic manifestations of LIG4 syndrome, we have generated and characterized a knock-in mouse model carrying a homozygous R278H mutation that corresponds to the first LIG4 mutation identified in humans (12).
Using a plasmid-rejoining assay in fibroblast cell lines derived from patients homozygous for the LIG4 R278H mutation, NHEJ activity was significantly impaired, but not abrogated, and the fidelity of RS joins was markedly reduced (11, 21, 31). In keeping with these observations, we have found that Lig4 R278H protein expression is only modestly reduced in the thymus of Lig4R/R mice and that RS and coding joins formation is not abrogated, consistent with greatly, but not completely, impaired NHEJ activity. Profound abnormalities of thymic architecture, with inability to support generation of a broadly polyclonal repertoire of T cells (26, 32), have been associated with severe immunopathology both in patients and in mice with hypomorphic RAG mutations. In contrast, the diversity of thymic T cell repertoire and thymic architecture are largely preserved in Lig4R/R mice, probably because the NHEJ defect is not complete. Furthermore, generation of CD4+ CD25hi Foxp3+ cells (i.e., bona fide Tregs) was preserved. Overall, these findings may explain the lack of significant immunopathology in Lig4R/R mice.
Total thymic cellularity was drastically reduced, and survival of in vitro activated T lymphocytes was impaired in Lig4R/R mice, thus contributing to the severe peripheral T cell lymphopenia that was particularly prominent among CD8+ lymphocytes. A SCID phenotype has been observed in patients with LIG4 mutations in whom no qualitative V(D)J recombination defects were present in the few circulating T lymphocytes (14), and a leaky SCID phenotype has been recently reported in another mouse model of Lig4 deficiency (Lig4Y288C/Y288C mice) characterized by poor lymphocyte survival despite residual Lig4 activity (33). Overall, these data suggest that hypomorphic LIG4 mutations may cause profound immunodeficiency by both affecting cell survival and impairing V(D)J recombination.
Lig4R/R mice show a dramatic defect in B-cell development and a severe, but incomplete in vivo CSR defect, as demonstrated by residual levels of some Ig isotypes. Normal or increased levels of IgM, IgG1, and IgA were previously demonstrated also in Lig4Y288C/Y288C mice (33) and might reflect accelerated plasma cell differentiation associated with B cell lymphopenia (34). Stimulation of B lymphocytes through toll-like receptors and cytokine secretion by T lymphocytes in a lymphopenic and immunodeficient environment may be involved in inducing CSR in vivo in Lig4R/R mice. Normal or increased IgM and IgG levels have been reported in some patients with severe immunodeficiency due to LIG4 mutations (13, 14). Among other animal models of defective NHEJ, B cell development and CSR recombination are largely preserved in Cernunnos-deficient mice (35), and switching to IgG1 is also maintained in DNA-PKcs-deficient mice harboring IgH and IgL knock-in alleles (36). Lig4R/R mice spontaneously produce higher amounts of low-affinity antibodies that include self-reactive specificities, but are unable to mount robust antibody responses following antigenic challenge. Production of autoantibodies has been also reported in Lig4Y288C/Y288C mice (33) and in patients with leaky SCID due to LIG4 mutations (13). Several mechanisms may account for this B-cell-mediated immune dysregulation. Peripheral CD4+ CD25hi Foxp3+ cells were detected in Lig4R/R mice (Fig. S7B), arguing against impaired Treg-mediated control of autoimmunity, although functional activity of these cells could not be tested due to the low number. On the other hand, Lig4 mutations should affect receptor editing and hence impinge on a key mechanism of B cell tolerance. Finally, severe B cell lymphopenia has been shown to result in increased serum levels of B-cell activating factor (BAFF) (37), and this could facilitate rescue and expansion of low-affinity self-reactive B cells in Lig4R/R mice as shown in other models (37, 38). Additional experiments are needed to address these hypotheses.
Lig4R/R cells show radiation sensitivity and genomic instability at similar levels as observed in Lig4−/−, Xrcc4−/−, and Ku70−/− or Ku80−/− cells (22, 24). In Lig4R/R mice, these features are associated with development of thymic tumors. Similar findings have been reported in Lig4Y288C/Y288C mice (33). Peripheral lymphoid malignancies have been reported in 4 of 14 patients with LIG4 mutations, including the original patient with a homozygous LIG4 R278H mutation (12, 13, 28, 30). Development of DP or SP thymic tumors has been reported also in other murine models of impaired NHEJ, in particular in Ku70−/− (22, 39) and in DNA-PKcs-deficient mice (40–42), although with significant variability that may reflect differences in genetic background and/or environmental factors. Importantly, these models as well as the Lig4R/R and the Lig4Y288C/Y288C mice share “leaky” defects in T cell development. These findings are consistent with the need for productive TCR gene rearrangement to induce proliferation, which may then make the cells susceptible to secondary hits in the NHEJ-deficient background, and also may lead to generation of subsequent developmental stages more susceptible to transformation.
In summary, Lig4R/R mice represent a model for a naturally occurring mutation in the LIG4 gene in humans. They recapitulate most of the phenotypic features of LIG4 syndrome and may thus serve as a model to explore more in detail the pathophysiology of human LIG4 syndrome.
Materials and Methods
Generation of Lig4R/R Mice.
Lig4+/R mice were generated by gene targeting (Fig. S1 and SI Text) and intercrossed to generate homozygous Lig4R/R mice. Timed Lig4+/R matings were performed to generate day 12–13 Lig4R/R MEFs. Expression of Lig4 protein was assessed as described (2).
Radiation Sensitivity.
To assess the radiation sensitivity of Lig4+/+, Lig4R/R, and Xrcc4−/− MEFs, colony survival assays were performed as described (35). Three independent experiments were performed, and results were expressed as average (± SD) of colonies detected.
V(D)J Recombination Assays.
Analysis of endogenous Vβ14DJβ1 junctions was performed on DNA from Lig4+/+ and Lig4R/R thymocytes using primers and conditions previously described (43). Transient V(D)J recombination assays were performed as described (44).
Immunophenotypic Analysis and Histopathology.
Single cell suspensions from thymi, spleens, and bone marrow were prepared and stained with specific anti-mouse antibodies (SI Text). Data were acquired on a FACSCalibur or LSR flow cytometer (BD Biosciences) and analyzed using FlowJo software for Mac version 8.3 (Treestar). Histopathology of the thymus, liver, and gut was performed as reported in SI Text.
In Vitro Lymphocyte Proliferation.
Splenic T cells were purified from Lig4+/+ and Lig4R/R mice, using the Negative Selection Mouse T Cell Enrichment Kit (Stem Cell Technologies) according to the manufacturer’s protocol. Proliferation in response to anti-CD3 mAb (5 μg/mL) ± anti-CD28 mAb (2 μg/mL) or IL-2 (100 IU/mL) assayed by CFSE (Molecular Probes) dilution was performed as described (45). Apoptosis was measured by staining with the Annexin V-PE Apoptosis detection kit (BD Biosciences).
Ig Levels, Antibody Responses, and Autoantibodies.
Ig serum levels and antibody responses to TNP-Ficoll and to TNP-KLH were examined in 8-week-old Lig4+/+ and Lig4R/R mice as described (46). Low- and high-affinity anti-TNP IgG antibodies, anti-ssDNA, and anti-chromatin antibodies were measured by ELISA as described in SI Text.
Analysis of Genomic Instability and Telomere-FISH Analysis.
Positively selected CD43+ splenic T cells from Lig4R/R mice and from heterozygous Lig4+/R mice were plated at 1 × 106/mL and activated in vitro with 2.5 μg/mL Con A in RPMI medium 1640 supplemented with 10% FCS, penicillin/streptomycin (100 units/mL), 10 mM Hepes, 2 mM glutamine, and 10 μM 2-mercaptoethanol for 48 h. Metaphase spreads were prepared and analyzed by telomere staining (T-FISH) as described (2).
Southern Blot Analysis of Thymic Tumors.
Genomic DNA from the tumor was isolated and digested with EcoRI. Southern blotting was performed as described using TCRβ genomic probes A (0.7 kb ClaI-BamHI) and B (0.9 kb NcoI-NcoI) (47).
Supplementary Material
Acknowledgments
We thank Drs. Michel Nussenzweig, Harvey Cantor, and Jianzhu Chen for critical reading of the manuscript. This work was partially supported by the National Institutes of Health Grant P01 AI076210-01A1 (to L.D.N. and F.W.A.) and by the Manton Foundation (L.D.N.). F.W.A. is a Howard Hughes Investigator. C.T.Y. is a recipient of a V Foundation Scholar Award and a grant from the Emerald Foundation. J.H.W. and S.Z. are recipients of a special fellowship from the Leukemia & Lymphoma Society of America. J.H.W. is also the recipient of National Institutes of Health Training Grant CA 09382-26.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914865107/DCSupplemental.
References
- 1.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: Similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 2.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 3.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 4.Moshous D, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 6.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 7.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Riballo E, et al. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 10.van der Burg M, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 12.Riballo E, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 13.Enders A, et al. A severe form of human combined immunodeficiency due to mutations in DNA ligase IV. J Immunol. 2006;176:5060–5068. doi: 10.4049/jimmunol.176.8.5060. [DOI] [PubMed] [Google Scholar]
- 14.Buck D, et al. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur J Immunol. 2006;36:224–235. doi: 10.1002/eji.200535401. [DOI] [PubMed] [Google Scholar]
- 15.van der Burg M, et al. A new type of radiosensitive T-B-NK+ severe combined immunodeficiency caused by a LIG4 mutation. J Clin Invest. 2006;116:137–145. doi: 10.1172/JCI26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunebaum E, Bates A, Roifman CM. Omenn syndrome is associated with mutations in DNA ligase IV. J Allergy Clin Immunol. 2008;122:1219–1220. doi: 10.1016/j.jaci.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 17.Frank KM, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 18.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 19.Frank KM, et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 20.Sharpless NE, et al. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
- 21.Riballo E, et al. Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J Biol Chem. 2001;276:31124–31132. doi: 10.1074/jbc.M103866200. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 23.Taccioli GE, et al. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 24.Rooney S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, et al. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 26.Khiong K, et al. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J Clin Invest. 2007;117:1270–1281. doi: 10.1172/JCI30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marrella V, et al. A hypomorphic R229Q Rag2 mouse mutant recapitulates human Omenn syndrome. J Clin Invest. 2007;117:1260–1269. doi: 10.1172/JCI30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Omran TI, Cerosaletti K, Concannon P, Weitzman S, Nezarati MM. A patient with mutations in DNA ligase IV: Clinical features and overlap with Nijmegen breakage syndrome. Am J Med Genet A. 2005;137A:283–287. doi: 10.1002/ajmg.a.30869. [DOI] [PubMed] [Google Scholar]
- 29.Gruhn B, et al. Successful bone marrow transplantation in a patient with DNA ligase IV deficiency and bone marrow failure. Orphanet J Rare Dis. 2007;2:5. doi: 10.1186/1750-1172-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toita N, et al. Epstein-Barr virus-associated B-cell lymphoma in a patient with DNA ligase IV (LIG4) syndrome. Am J Med Genet A. 2007;143:742–745. doi: 10.1002/ajmg.a.31644. [DOI] [PubMed] [Google Scholar]
- 31.Smith J, et al. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poliani PL, et al. Early defects in human T-cell development severely affect distribution and maturation of thymic stromal cells: Possible implications for the pathophysiology of Omenn syndrome. Blood. 2009;114:105–108. doi: 10.1182/blood-2009-03-211029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nijnik A, et al. Impaired lymphocyte development and antibody class switching and increased malignancy in a murine model of DNA ligase IV syndrome. J Clin Invest. 2009;119:1696–1705. doi: 10.1172/JCI32743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agenès F. B lymphocyte life span, rate of division and differentiation are regulated by total cell number. Eur J Immunol. 2003;33:1063–1069. doi: 10.1002/eji.200323550. [DOI] [PubMed] [Google Scholar]
- 35.Li G, et al. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manis JP, Dudley D, Kaylor L, Alt FW. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 2002;16:607–617. doi: 10.1016/s1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 37.Lesley R, et al. Reduced competitiveness of autoantigen-engaged B cells due to increased dependence on BAFF. Immunity. 2004;20:441–453. doi: 10.1016/s1074-7613(04)00079-2. [DOI] [PubMed] [Google Scholar]
- 38.Thien M, et al. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity. 2004;20:785–798. doi: 10.1016/j.immuni.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Li GC, et al. Ku70: A candidate tumor suppressor gene for murine T cell lymphoma. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 40.Custer RP, Bosma MS, Bosma MJ. Severe combined immunodefiency (SCID) in the mouse. Pathology, reconstitution, neoplasms. Am J Pathol. 1985;120:464–477. [PMC free article] [PubMed] [Google Scholar]
- 41.Jhappan C, Morse HC, 3rd, Fleischmann RD, Gottesman MM, Merlino G. DNA-PKcs: A T-cell tumour suppressor encoded at the mouse scid locus. Nat Genet. 1997;17:483–486. doi: 10.1038/ng1297-483. [DOI] [PubMed] [Google Scholar]
- 42.Kurimasa A, et al. Catalytic subunit of DNA-dependent protein kinase: Impact on lymphocyte development and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bassing CH, et al. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 44.Rooney S, et al. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J Exp Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parish CR, Warren HS. Use of the intracellular fluorescent dye CFSE to monitor lymphocyte migration and proliferation. Curr Protoc Immunol. 2002;4 doi: 10.1002/0471142735.im0409s49. Unit 4.9. [DOI] [PubMed] [Google Scholar]
- 46.Tsitsikov EN, Gutierrez-Ramos JC, Geha RS. Impaired CD19 expression and signaling, enhanced antibody response to type II T independent antigen and reduction of B-1 cells in CD81-deficient mice. Proc Natl Acad Sci USA. 1997;94:10844–10849. doi: 10.1073/pnas.94.20.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khor B, Sleckman BP. Intra- and inter-allelic ordering of T cell receptor beta chain gene assembly. Eur J Immunol. 2005;35:964–970. doi: 10.1002/eji.200425806. [DOI] [PubMed] [Google Scholar]
- 48.Rooney S, et al. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci USA. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.