Abstract
Lens epithelium-derived growth factor (LEDGF) fusion proteins can direct HIV-1 DNA integration to novel sites in the host genome. The C terminus of LEDGF contains an integrase binding domain (IBD), and the N terminus binds chromatin. LEDGF normally directs integrations to the bodies of expressed genes. Replacing the N terminus of LEDGF with chromatin binding domains (CBDs) from other proteins changes the specificity of HIV-1 DNA integration. We chose two well-characterized CBDs: the plant homeodomain (PHD) finger from ING2 and the chromodomain from heterochromatin binding protein 1α (HP1α). The ING2 PHD finger binds H3K4me3, a histone mark that is associated with the transcriptional start sites of expressed genes. The HP1α chromodomain binds H3K9me2,3, histone marks that are widely distributed throughout the genome. A fusion protein in which the ING2 PHD finger was linked to the LEDGF IBD directed integrations near the start sites of expressed genes. A similar fusion protein in which the HP1α chromodomain was linked to the LEDGF IBD directed integrations to sites that differed from both the PHD finger fusion–directed and LEDGF-directed integration sites. The ability to redirect HIV-1 DNA integration may help solve the problems associated with the activation of oncogenes when retroviruses are used in gene therapy.
Keywords: chromatin, histone marks, lentiviral vectors
Retroviruses can integrate their DNAs at many sites in host DNA (1), but different retroviruses have different integration site preferences (2–9). HIV-1 and simian immunodeficiency virus DNAs preferentially integrate into expressed genes, murine leukemia virus (MLV) DNA preferentially integrates near transcriptional start sites (TSSs), and avian sarcoma leukosis virus (ASLV) and human T cell leukemia virus (HTLV) DNAs integrate nearly randomly, showing a slight preference for genes. This suggests that the preintegration complexes (PICs) of the different retroviruses interact with different factors in host cell chromatin and that the integration site preferences are determined by the distribution of the different host cell factors. Studies of the specificity of integration of yeast retrotransposons, which are distantly related to retroviruses, provide strong support for this interpretation (10–12). The host factors that MLV, ASLV, and HLTV PICs interact with are not known.
HIV-1 integrase (IN) binds specifically to lens epithelium-derived growth factor (LEDGF). A crystal structure shows that the integrase binding domain (IBD), located in the C-terminal portion of LEDGF, makes important contacts with both the catalytic core and N-terminal domains of IN (13). The N-terminal portion of LEDGF contains a PWWP domain and AT hooks that bind chromatin and DNA, respectively (14) (Fig. S1). The PWWP domain is the primary chromatin binding determinant, although the protein or modification to which it binds is currently unknown (15, 16). LEDGF promotes the association of the PIC with the host genome, enhancing the integration of HIV-1 DNA (16–20).
In assays that depend on the integration of viral DNA, viral titer is reduced 5–100-fold in the absence of LEDGF (18, 19, 21). In some of these experiments, the level of LEDGF was reduced by treatment with siRNA. Relatively small amounts of residual LEDGF can support HIV-1 DNA integration (18, 22). If the PWWP domain of LEDGF is removed, the truncated protein cannot support HIV-1 replication. However, fusion proteins in which the PWWP domain of LEDGF is replaced with the linker histone H1 or with a peptide derived from the chromatin binding Kaposi’s sarcoma herpes virus LANA protein do support efficient HIV-1 replication (23). This result means that these fusion proteins are able to bind both chromatin and the PIC in a way that supports viral integration. We show that replacing the PWWP domain and AT hooks of LEDGF with other chromatin binding domains (CBDs) supports viral integration and redirects the viral DNA integration to sites in the genome that reflect the chromatin binding patterns of the CBDs. The ability to redirect HIV-1 DNA integration may help solve one of the problems associated with using retroviral vectors in gene therapy: the unintentional activation of host cell genes by the nearby insertion of vector DNA (13, 24).
Results
CBD–IBD Fusions Can Functionally Replace LEDGF to Support Efficient HIV-1 DNA Integration.
We chose two CBDs that are well defined both structurally and functionally, selecting one that binds primarily to euchromatin and one that can bind to more tightly packed heterochromatin. The first CBD was the plant homeodomain (PHD) finger from ING2, which is known to bind histone 3 lysine 4 trimethylations (H3K4me3) (25, 26). The other CBD was the chromodomain from heterochromatin binding protein 1α (HP1α, also known as Cbx5), which has been shown to bind to histone 3 that is either di- or trimethylated on lysine 9 (H3K9me2,3). There is evidence that H3K9me2,3 marks are widely distributed throughout the genome and are found both in poorly expressed and actively expressed genes (27).
We generated fusion proteins in which the two CBDs were joined to the LEDGF IBD. To avoid problems from the expression of small amounts of LEDGF that can occur in siRNA knockdowns, we expressed the CBD–IBD fusions in a mouse embryo fibroblast cell line that was derived from a LEDGF knockout mouse (MEF-KO) (19). As a positive control, we expressed human LEDGF (huLEDGF). The expression plasmid contains a CMV promoter, a DNA segment encoding huLEDGF or the CBD–IBD fusion, and an internal ribosome entry site (IRES) followed by the coding region for GFP (19). The expression plasmid was introduced into MEF-KO cells by nucleofection, and cells that expressed similar levels of GFP were selected by FACS (Fig. S2). The huLEDGF and the CBD–IBD fusions were HA tagged. Expression of the fusion proteins was confirmed by Western blot. Judged by the Western blots, the sorted cells contained similar amounts of huLEDGF and the CBD–IBD proteins. GFP-expressing cells were infected with an HIV-1 vector that expresses luciferase. Although there is some variation in the assay, the titer in MEF-KO cells that expressed high levels of huLEDGF was ≈100-fold higher than the titer in unmodified parental MEF-KO cells, which agrees with the results reported by Shun et al. (19).
Titer, measured by luciferase activity, depends both on the efficiency of integration (the number of integrated proviruses) and on their level of expression. Expression of the ING2 PHD finger IBD fusion (ING2–IBD) in MEF-KO cells increased the titer of the HIV-1 vector by ≈60-fold (three separate experiments showed increases of 67-, 58-, and 59-fold), and the HP1α chromodomain IBD fusion (HP1α–IBD) by ≈80-fold (two separate experiments showed increases of 77- and 80-fold), demonstrating that integration was efficient and that a significant fraction of the proviruses were expressed (Table S1).
huLEDGF Directs HIV-1 DNA Integration to Expressed Genes in MEF-KO Cells.
Genomic DNAs were prepared from MEF-KO cells 48–72 h after infection. Viral–host junctions were selectively amplified from these DNAs by restriction enzyme digestion, followed by linker-mediated PCR. To reduce bias caused by the distribution of restriction enzyme recognition sites in the mouse genome, three separate digestions were done on each DNA sample: (i) Mse I, (ii) Tsp509I, and (iii) a mixture of three restriction enzymes (AvrII, SpeI, and NheI). Because PCR amplification can create multiple copies of single integration sites, all copies of specific host–virus DNA junctions were computationally removed, leaving a set of unique integration sites that were used in the data analysis.
We used 454 pyrosequencing to obtain more than 17,000 independent huLEDGF directed integrations in single-copy DNA. A smaller number of integrations were identified in known repeat sequences. The ratio of integrations in repeat DNA to single-copy DNA was approximately 1:40. This ratio significantly underestimates the number of integrations in repeat sequences because the currently available draft of the sequence of the mouse genome does not include all repeat sequences and because identical integration sites were computationally removed in the analysis. There is also the possibility that the distribution of restriction sites will create bias against amplifying integrations in repeat sequences. In terms of the pattern of integrations into genes, and near landmarks like CpG islands and TSSs, the distribution of the huLEDGF-directed integrations in the mouse genome was similar to that previously reported for integration in human and mouse cells (Table 1) (5, 6, 8, 9, 19).
Table 1.
Overall integration patterns and experimental reproducibility
| Integration site percentages |
|||
| CBD–IBD fusion (integration sites) | In RefGene | CpG island ±2.5 KB | TSS ±2.5 KB |
| huLEDGF (17,009) | 67.6%* | 2.2%* | 3.8% |
| ING2–IBD Sanger (66) | 80.0%* | 40.0%*† | 63.1%*† |
| ING2–IBD runA (7,433) | 68.3%* | 23.7%*† | 50.5%*† |
| ING2–IBD runB (8,136) | 67.1%* | 24.5%*† | 50.3%*† |
| ING2–IBD A+B (15,491) | 67.6%* | 24.1%*† | 50.3%*† |
| HP1α–IBD (14,803) | 34.2%† | 1.2%† | 2.7%*† |
| Random (10,000) | 33.6%† | 1.3%† | 4.1% |
P < 0.001, χ2 test, vs. 10,000 random sites.
P < 0.001, χ2 test, vs. huLEDGF sites.
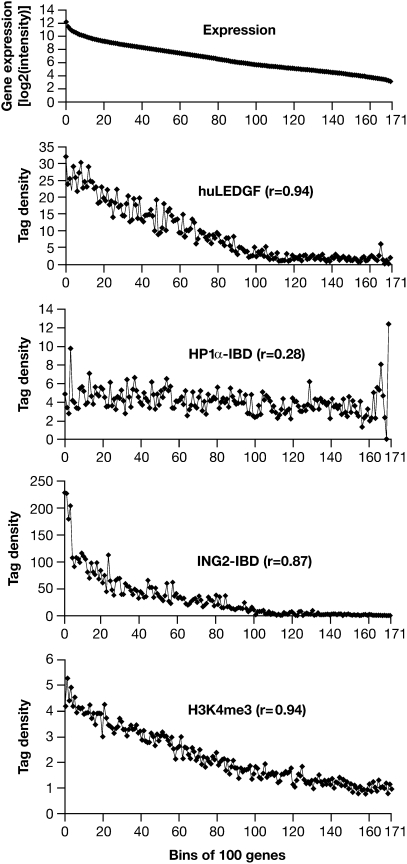
To determine whether there is a relationship between the integration sites and gene expression, arrays were used to measure gene expression in the MEF-KO cells. A total of 17,306 well-characterized RefSeq Genes were sorted into groups of 100 according to their RNA levels, and the integration data were sorted on the basis of these data. As expected, huLEDGF directed integrations to expressed genes. The most highly expressed genes have been reported to be less efficient targets than genes that are moderately well expressed. Because huLEDGF-directed integrations can occur anywhere in a gene, simply counting the integrations in each gene favors larger genes. When we normalized the integration data according to the size of the genes (discussed below), there was a simple positive correlation (r = 0.94) between the level of expression and the number of huLEDGF-directed integrations (Fig. 1).
Fig. 1.
Gene expression and integration site preferences. Top: The uppermost panel (Expression) represents >17,000 well-characterized mouse genes placed into bins of 100 according to the level of expression measured in the MEF-KO cells. The average level of expression for the genes in each bin is given on the y axis (log2 scale). Bottom: Distribution of H3K4me3 marks from the data of Barski et al. (30) plotted against bins of 100 genes according to the level of expression in human CD4+ cells. Upper Middle, Middle, Lower Middle: These three panels show, on the y axis, the number of integrations in each bin of 100 genes for LEDGF and the two CBD–IBD fusions. The genes were placed in bins according to their level of expression in MEF-KO cells, shown in the uppermost panel. The integration data have been normalized to a total of 10,000 integrations. The data for the ING2 PHD finger fusion were based on the number of integrations within 2.5 kb of a TSS for the 100 genes in each bin, so each point represents the number of total integrations in 0.5 Mb. For huLEDGF and the HP1α chromodomain fusions, which direct integrations throughout genes, we first enumerated the total number of integrations in all of the genes in each bin. Because the size of the genes varies (highly expressed genes tend to be smaller), the total target size in different bins also varies. To solve this problem, the data were renormalized on the basis of a total target size of 1 Mb in each bin. This means that the ING2 fusion data cannot be directly compared with the HP1α fusion and huLEDGF data (see text). Note that the various panels have different scales on the y axis.
ING2 PHD Finger Fusion Directs HIV-1 DNA to Integrate Near TSSs.
A total of ≈15,000 independent ING2–IBD-directed integrations in single-copy DNA were obtained from a combination of Sanger sequencing and two completely independent experiments analyzed by 454 pyrosequencing. Approximately half (50.3%) of the ING2–IBD-directed integration sites were within 2.5 kb of a TSS. As expected (5, 8, 19), huLEDGF directed only a small fraction of integrations (3.8%) to within 2.5 kb of TSSs, a frequency that did not differ significantly from random (Table 1). These data show that replacing the PWWP domain and AT hooks of LEDGF with a PHD finger redirects HIV-1 DNA integration.
Retroviral DNAs preferentially integrate at relatively short, weakly conserved palindromes, and different retroviruses preferentially integrate their DNAs in different palindromes. The palindrome preferences of the various retroviruses seem to reflect preferences of their respective INs (6, 28, 29). The palindrome preference of HIV-1 does not change in cells that lack LEDGF (8, 19). The HIV-1 DNA integrations that were directed by the CBD–IBD fusions had the same palindromic preference as those directed by LEDGF.
Integration Site Data Are Reproducible.
The two independent 454 datasets obtained with the ING2–IBD were compared. Both the overall pattern of integration (Table 1) and the preferred sites for integration (i.e., hotspots) were highly reproducible (Fig. S3). We initially defined hotspots as three or more integrations within 10 kb and highly favored hotspots as five or more integrations. Because most of the PHD finger fusion–directed integrations were near TSSs, the PHD finger hotspots were recalculated using three or more integrations within 2.5 kb of a TSS as a hotspot, and five or more integrations as a highly favored hotspot. For comparative analyses, each of the datasets was normalized to 10,000 integrations. Each of the 19 hotspots with the greatest number of integrations in the combined ING2–IBD fusion dataset was a highly favored hotspot in both of the independent 454 experiments (Fig. S3A), although there was some variation in the rank order of hotspots between assays. Of the 177 and 178 highly favored hotspots in the two 454 datasets, 169 are present in both datasets (Fig. S3B).
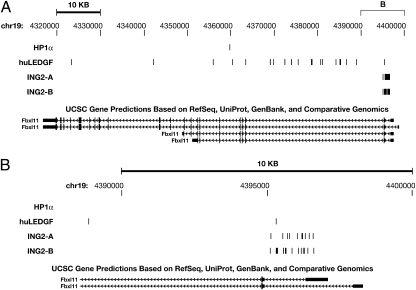
Some of the ING2–IBD-directed integrations were very tightly clustered. Fig. 2 shows clusters in the Malat1 gene (Malat1 is the strongest ING2–IBD hotspot, discussed below). Because identical integration sites were seen in the two independent ING2–IBD experiments (Fig. 2C), it is likely that at least some of the integrations occur close to where the CBD–IBD binds. The data also show that CBD–IBD-directed integrations are highly reproducible with respect to the overall pattern of integration, suggesting that the hotspots are the preferred CBD binding sites.
Fig. 2.
ING2 PHD finger fusion directs tightly clustered integrations. The figure was prepared by marking integration sites on the University of California, Santa Cruz database of the mouse genome (mm9). The nucleotide positions are given at the top of each panel, and the transcript(s) from the gene are shown at the bottom. A scale bar for the DNA segment is shown at the top of the figure. The data from the two ING2–IBD 454 experiments (run A and run B) are shown separately in all panels. Integrations are shown as vertical bars. (A) Region of mouse chromosome 19 that contains the Malat1 gene, which contains both ING2–IBD-directed integrations. The approximate positions of the segment shown in B and C are marked at the top. (B) A small segment of Malat1 in which there were clustered ING2–IBD-directed integrations; (C) a second cluster in a different segment of the Malat1 gene where there were ING2–IBD-directed integrations at exactly the same sites in the two 454 experiments.
ING2 PHD Finger Fusion Directs Integrations to Expressed Genes.
There was a strong positive correlation between the integration sites targeted by the ING2 PHD finger fusion and the level of gene expression (the correlation coefficient was 0.87). There are H3K4me3 marks near the TSSs of essentially all highly expressed genes, and H3K4me3 marks correlate with level of expression (r = 0.94) (Fig. 1). To facilitate data comparison, the total number of integrations directed by huLEDGF and the CBD–IDB fusions was normalized to 10,000. Fig. 1 shows the number of ING2–IBD-directed integrations within 2.5 kb of a TSS (for 100 genes, the total target size is 0.5 Mb). huLEDGF and the HP1α–IBD directed integration throughout genes (discussed below). The total number of integrations in each group of 100 genes was determined. However, because many of the most highly expressed genes are smaller than average, the total size of the target DNA in the groups varied. For the first group of 100 (the 100 most highly expressed genes), the total target size was ≈1.2 Mb. For groups near the middle of the expression distribution, the total target size was 5–8 Mb. To compensate for this difference, the huLEDGF and HP1α–IBD data were renormalized so that the total target size in each group was 1 Mb.
There were ING2–IBD-directed integrations in ≈80% of the 200 most highly expressed genes, and in ≈75% of the next 200. A number of the 400 most highly expressed genes had only a single ING2–IBD-directed integration, and it is likely that additional experiments would show integrations in additional highly expressed genes. Only approximately one quarter of the 200 most highly expressed genes had five or more ING2–IBD-directed integrations. The fact that the ING2–IBD fusion preferentially directed integrations to a specific subset of the 400 most highly expressed genes explains the relatively large number of integrations in the first four points in the graphs of integration vs. expression (Fig. 1).
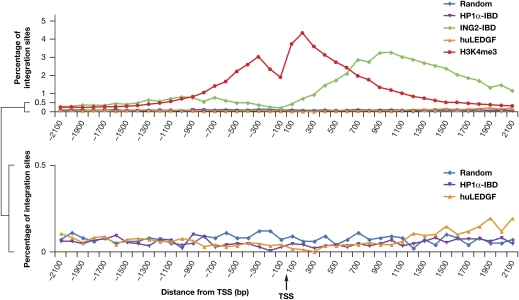
The ING2–IBD fusions directed more integrations downstream of TSSs than upstream, with almost no integrations at TSSs (Fig. 3). However, the distribution of the integration sites for the ING2–IBD in MEF-KO cells was significantly broader than the distribution of H3K4me3 marks in human CD4+ cells. The relatively broad distribution of ING2–IBD integration sites around TSSs can be explained if there is preferential integration into the genes that have the largest number of H3K4me3 marks. There can be no more than two H3K4me3 marks per nucleosome, which means that the regions immediately adjacent to TSSs can have only a limited number of marks. Genes that have many H3K4me3 marks will therefore have marks (and hence integrations) relatively far from the TSS. This is what is seen in the Malat1 gene, which expresses a noncoding RNA and is the most favored hotspot in both of the ING2–IBD fusion 454 datasets. Approximately 0.6% of all ING2–IBD directed integrations were in Malat1 (Fig. 2 and Fig. S3). The ING2–IBD-directed integrations in Malat1 were spread over the gene, rather than being localized near the TSS. If Malat1 has a very large number of H3K4me3 marks, some of these marks must be on nucleosomes within the body of the gene. In fact, the data of Barski et al. (30) show that H3K4me3 marks are distributed throughout the Malat1 gene in human CD4+ cells (Fig. S4). The distribution of integration sites in other genes that are strong ING2–IBD hotspots supports this idea. For example, the hotspot in the Fbxl11 gene was near the TSS, but not all of the integrations cluster within a few hundred base pairs of the TSS (Fig. 4).
Fig. 3.
Distribution of integration sites near TSSs. (Upper) The y axis shows the percentage of the total integrations for the individual CBD–IBD fusions that occur within 100 bp intervals from a TSS. The TSS is in the middle, integrations upstream of a TSS are on the left (negative numbers), and those downstream are on the right (positive numbers). The ING2–IBD causes preferential integration near TSSs. The distribution of H3K4me3 marks in the genome of human CD4+ cells (30) is shown for comparison. (Lower) Data for the HP1α–IBD- and huLEDGF–IBD-directed integrations, which were indistinguishable from the baseline in the upper graph, shown on an expanded scale.
Fig. 4.
Genes that contain hotspots for CBD–IBD fusions. The format is the same as in Fig. 2. (A) The entire Fbxl11 gene. At this resolution, the multiple integration sites near the promoter appear as thick black lines. (B) A closer view of the region near the Fbxl11 TSS, showing the individual integration sites.
Integrations Directed by the HP1α Chromodomain Fusion Differ from Those Directed by the PHD Finger Fusion and huLEDGF.
As discussed earlier, we chose the ING2 PHD finger because we expected it to preferentially bind near the TSSs of expressed genes, and the HP1α chromodomain because it should not. We obtained ≈14,800 independent integrations into single-copy DNA for the HP1α–IBD fusion. The HP1α–IBD directed a larger fraction of the integrations into repeat DNA (1:5) than the PHD finger fusions (1:20). Given that HP1α is known to bind heterochromatin, and that many of the repeated sequences are in heterochromatin, an increase was expected.
When we analyzed the pattern of integrations directed by the HP1α–IBD in single-copy DNA, the pattern was similar to a computer generated random distribution. There was no preference for integrations into genes or near CpG islands or TSSs (Table 1). Although there were hotspots, the most favored hotspots have far fewer integrations than the most favored PHD finger hotspots, even after correcting for the overall number of integrations.
To determine whether the HP1α–IBD-directed integrations differed from random, we reset the hotspot window to 30 kb. In the random dataset of the same size (14,803 integrations) there should be relatively few 30-kb segments with four integrations and none with more than four (we found seven instances of four integrations in a simulation). In contrast, in the HP1α–IBD-directed integration dataset, there were 18, 14, 11, 10 (n = 2), 9 (n = 3), and 8 (n = 2) integrations in the 10 most favored 30-kb windows, showing that HP1α–IBD-directed integrations were not random.
Although some of the most favored HP1α−IBD-directed hotspots were in or near genes, others occurred in regions that do not contain identified genes. This is in contrast to the results obtained with the PHD finger fusion and huLEDGF, in which the most favored hotspots were in genes. For integrations directed into genes by the HP1α–IBD, there was little correlation with gene expression (r = 0.28) (Fig. 1).
Discussion
The integration data for huLEDGF and the two CBD–IBD fusions show that HIV-1 DNA integration can occur in most of the chromatin, including genes that are poorly expressed and regions of the genome that lie outside of genes. On the basis of our limited ability to analyze integrations into repeated sequences, it seems that some CBD–IBD fusions can direct HIV-1 DNA to integrate in repeat DNA and heterochromatin with reasonable efficiency. It should not be surprising that HIV-1 DNA can integrate into heterochromatin. Some yeast retrotransposons, for example TY5, normally integrate their DNA into heterochromatin (10).
Cellular LEDGF directs efficient integration of HIV-1 DNA, which leads to good expression of the integrated proviruses and high-level viral replication. It has been estimated that more than 90% of the complete linear viral DNAs made in cultured cells are successfully integrated (31, 32). However, the fact that both of the CBD–IBD fusions we tested strongly enhanced titer in MEF-KO cells shows that both support efficient integration and high-level expression of the resulting proviruses. This result suggests that the location of a provirus in the genome has a fairly modest effect on expression. The simple interpretation is that the structure/sequence of the HIV-1 provirus has been strongly selected for efficient expression in a variety of different locations. Although the specific integration sites may have some effect on expression, it is most likely of secondary importance.
Although we do not know precisely how far HIV-1 DNA integrates from the site where a CBD–IBD binds, the data support the interpretation that most of the directed integrations occur relatively near the binding site. This interpretation is based on the identification of tightly clustered (<100 bp) integration sites with the ING2 PHD finger fusion. This suggests that the distribution of the directed integrations could be used to determine where the CBD binds to the chromatin. Such an approach has been applied in yeast. The “calling card” system uses modified yeast proteins to direct the integration of the DNA of a yeast retrotransposon to places in the genome where particular fusion proteins bind (11).
The substantial increase in titer seen in the MEF-KO cells that expressed the CBD–IBD fusions, taken together with the differences in the integration patterns seen with huLEDGF and the two CBD–IBD fusions, suggests that there is a relatively low backround of “untethered” integration events in cells that express functional CBD–IBD fusions. The reproducibility of the ING2–IBD integration site data shows that the CBD–IBD fusions can exert considerable control over the integration sites of an HIV-1–based vector. The fact that CBD–IBD fusions efficiently direct integration to different sites in the host genome suggests that this type of approach could be used to direct the DNA genomes of lentiviral vectors to “safe” places in the genome during gene therapy. This should reduce the risks of insertional mutagenesis, a serious concern with all retroviral vectors. Controlling integration site selection in cells from patients will take additional work, but recent progress developing a modified LEDGF–IBD/HIV-1 IN pair whose interactions differ significantly from wild-type seems to be a promising step. This type of approach has two obvious advantages: (i) the vectors will not efficiently infect cells that express only normal LEDGF, and (ii) it will be possible to redirect HIV-1 DNA integration without eliminating endogenous LEDGF (13).
Materials and Methods
Cell Culture and Virus Production.
The E2(−/−) MEF-KO cell line, which was isolated from a Psip1 knockout mouse embryo, has been described previously (19). The cells were maintained in DMEM supplemented with 5% FBS, 5% newborn calf serum, and penicillin (50 U/mL) plus streptomycin (50 μg/mL). Replication defective HIV-1 virus was produced in 293T cells grown on the same media. Briefly, an HIV-1 vector containing a 50-bp deletion in the env coding region with a luciferase reporter gene inserted in place of the nef gene, pNLNgoMIVR-Emod Luc, was transfected, together with a VSV-G envelope expression plasmid, using the calcium phosphate method (31, 33). VSV-G pseudotyped HIV-1 viral supernatants were collected 48–72 h after transfection, cleared of debris by centrifugation, and frozen. Virus was quantified using the HIV-1 p24 ELISA kit (Perkin-Elmer).
Expression Plasmids, Nucleofection, FACS, and Infection.
LEDGF expression vectors were constructed in the pIRES2-eGFP plasmid (19). The parental plasmid, pIRES2-eGFP LEDGFWT HA, contains wild-type huLEDGF with a C-terminal HA tag. pIRES2-eGFPΔPWWPΔAT HA, which encodes a deleted form of LEDGF that lacks both the N-terminal PWWP domain and the AT hooks, was digested with XhoI and EcoRI. The DNA segments encoding the experimental CBDs were prepared by PCR amplification from cDNAs with primers encoding an upstream XhoI site and ATG initiation codon and a downstream EcoRI site. DNA segments encoding the experimental CBDs were digested with XhoI and EcoRI. The resulting DNA fragments were inserted in place of the DNA that encoded the N terminus of LEDGF. To express wild-type huLEDGF or the CBD–IBD fusions, the DNAs were introduced into MEF-KO cells by nucleofection (Amaxa). The cells were allowed to recover for 24–36 h and were sorted for GFP expression using a Becton Dickinson FACS Aria 1 cell sorter. Approximately 1–2 × 106 GFP-positive cells were plated on a 100-mm dish and infected with 500 ng VSV-G pseudotyped pNLNgoMIVR-Emod Luc and 8 μg/mL polybrene. After 48–72 h, cells were harvested for titer and integration site analyses.
Luciferase Assays.
For each CBD–IBD fusion, an aliquot of ≈106 cells was assayed for luciferase using the Bright-Glo Luciferase Assay System (Promega). Titer was calculated as the fraction of the activity (measured as luminescence) in cells that expressed huLEDGF.
Cloning and Sequencing of Integration Sites.
Genomic DNA was isolated from transduced MEF-KO cells using a QIAamp DNA Blood Mini Kit (Qiagen) or by extracting HIRT pellets (34). Linker-mediated PCR (LM-PCR) was used to amplify the junctions of integration sites and genomic DNA, as described previously (4, 29). Genomic DNA samples (2 μg) were digested with restriction enzymes (i) MseI, (ii) Tsp509I, or (iii) a combination of AvrII/NheI/SpeI and then ligated to linkers. The linkers are designed to allow amplification of the host–viral DNA junctions from the downstream LTR into the adjacent host DNA (the linker and primer sequences are given in Table S2). BglII digestion was used to prevent the amplification of an internal PCR product from the upstream LTR into the HIV-1 genome. LM-PCR was performed with one primer specific for U5 and R and the other primer specific for the ligated linker using the following conditions: preincubation at 94 °C for 2 min, then 25 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. The PCR products were diluted 1:50, and nested PCR was performed under the same conditions using a second set of primers, one specific for U5 and the other for the ligated linker. DNA barcodes (multiplex identifiers, MIDs) were included in the nested LTR primer to allow different samples to be sequenced at the same time on the 454 machine. PCR was performed in multiple aliquots to reduce founder effects (15 μL × 12), increasing the diversity of the library. Nested PCR products were subjected to 454 pyrosequencing or TOPO cloning followed by conventional Sanger sequencing. The 454 pyrosequencing was performed at the Laboratory of Molecular Technology of the National Cancer Institute at Frederick, using a standard GS FLX amplicon emPCR kit II (Roche) according to the manufacturer’s suggested protocol.
Bioinformatics Analysis and Expression Profiling
Descriptions of the bioinformatics analysis and expression profiling are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank John Coffin and Alex Ruthenburg for helpful discussions; Kathleen Noer and Roberta Matthai of the Center for Cancer Research-Frederick Flow Cytometry Core for cell sorting; Alan Kane and Jiro Wada for help with the figures; and Jeff Skaar and Teresa Burdette for help with the manuscript. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH) (National Cancer Institute). A portion of this funding was under Contract no. HHSN261200800001E. Research in the laboratory of A.E. was supported by NIH Grant AI039394, and M.-C.S. was supported in part by NIH Training Grant T32 AI007386. Research in the laboratory of C.D.A. was supported by NIH Method to Extend Research in Time Award GM53122 and The Rockefeller University. C.M.H. was supported by the Emerald Foundation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914142107/DCSupplemental.
See Commentary on page 2735.
References
- 1.Hughes SH, et al. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978;15:1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- 2.Derse D, et al. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewinski MK, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell RS, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narezkina A, et al. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall HM, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schröder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 10.Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Heinz ME, Crosby SD, Johnston M, Mitra RD. ‘Calling Cards’ method for high-throughput identification of targets of yeast DNA-binding proteins. Nat Protoc. 2008;3:1569–1577. doi: 10.1038/nprot.2008.148. [DOI] [PubMed] [Google Scholar]
- 12.Leem YE, et al. Retrotransposon Tf1 is targeted to Pol II promoters by transcription activators. Mol Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hare S, et al. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1665. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llano M, et al. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 16.Shun MC, et al. Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J Virol. 2008;82:11555–11567. doi: 10.1128/JVI.01561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci. 2008;65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llano M, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 19.Shun MC, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandekerckhove L, et al. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346:415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Meehan AM, et al. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009;5:e1000522. doi: 10.1371/journal.ppat.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 25.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 26.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Julias JG, Ferris AL, Boyer PL, Hughes SH. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J Virol. 2001;75:6537–6546. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas JA, Ott DE, Gorelick RJ. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J Virol. 2007;81:4367–4370. doi: 10.1128/JVI.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He J, Landau NR. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative viral receptor. J Virol. 1995;69:4587–4592. doi: 10.1128/jvi.69.7.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.