Abstract
ATP synthase catalyzes ATP synthesis at the expense of an electrochemical ion gradient across a membrane that can be generated by different exergonic reactions. Sulfur reduction is the main energy-yielding reaction in the hyperthermophilic strictly anaerobic Crenarchaeon Ignicoccus hospitalis. This organism is unusual in having an inner and an outer membrane that are separated by a huge intermembrane compartment. Here we show, on the basis of immuno-EM analyses of ultrathin sections and immunofluorescence experiments with whole I. hospitalis cells, that the ATP synthase and H2:sulfur oxidoreductase complexes of this organism are located in the outer membrane. These two enzyme complexes are mandatory for the generation of an electrochemical gradient and for ATP synthesis. Thus, among all prokaryotes possessing two membranes in their cell envelope (including Planctomycetes, Gram-negative bacteria), I. hospitalis is a unique organism, with an energized outer membrane and ATP synthesis within the periplasmic space. In addition, DAPI staining and EM analyses showed that DNA and ribosomes are localized in the cytoplasm, leading to the conclusion that in I. hospitalis energy conservation is separated from information processing and protein biosynthesis. This raises questions regarding the function of the two membranes, the interaction between these compartments, and the general definition of a cytoplasmic membrane.
Keywords: Archaea, ATP synthase, ATPase, immunolabeling, sulfur reductase
The hyperthermophilic Crenarchaeon Ignicoccus hospitalis KIN4/IT is a strictly anaerobic chemolithoautotrophic sulfur reducer that grows optimally at 90 °C. It conserves energy by the reduction of elemental sulfur with molecular hydrogen and uses CO2 as sole carbon source (1). Together with Nanoarchaeum equitans, it forms an “intimate association,” the only known stable coculture of two Archaea (2, 3). Like all Ignicoccus species (4), I. hospitalis cells possess an unusual architecture, with two compartments that can clearly be distinguished in composition and morphologic appearance. As shown in a number of EM studies (5–7), the densely packed cytoplasm is surrounded by two membranes, an “inner membrane” and an “outer membrane.” These two membranes enclose an intermembrane compartment with a variable width from 20 to 500 nm, resulting in a volume exceeding that of the cytoplasm (5). Its low electron density suggests that it is devoid of cellular material like ribosomes or DNA, and it was therefore named “periplasm” (7). The inner membrane, called the cytoplasmic membrane, releases numerous vesicles into the periplasmic space and also engulfs vesicles into the cytoplasm (7). Both membranes exhibit similar lipid composition, with the exception that the outer membrane lacks caldarchaeol cores (8). In addition, the latter contains multiple copies of a pore-forming complex (9), whereas a surface layer (S-layer), typical for most Crenarchaeota, is lacking (10). Therefore, the architecture of the I. hospitalis cell envelope is unique among Archaea. Moreover, owing to its huge intermembrane compartment and an outer membrane without LPS and porins (11–14), it is fundamentally different from other prokaryotic cell envelopes with two membranes (e.g., Gram-negative bacteria).
To date, in prokaryotes no outer membranes but only cytoplasmic membranes have been described as harboring ATP synthase complexes, the key components in cellular bioenergetics (15). These complexes (bacteria, mitochondria, and chloroplasts: F1FO ATP synthases; Archaea: A1AO ATP synthases) consist of a hydrophilic (F1, A1) and a membrane-bound domain (FO, AO) (16). Driven by an electrochemical ion gradient (17), the membrane-bound domain translocates ions (H+; Na+) across the membrane, resulting in ATP synthesis by the hydrophilic, catalytic domain. The enzyme is also able to reverse this process by hydrolyzing ATP. In contrast to other ATP hydrolyzing enzymes, this complex is sensitive to specific inhibitors. According to the genome annotation of I. hospitalis, several ATP hydrolyzing enzymes are present (18); only one set of subunits, however, was predicted (A, B, C, D, E, F, a, and c) to build a functional ATP synthase (19) and thought to be located in the inner membrane of I. hospitalis (18). The latter assumption was also based on the fact that primary H+ or Na+ pumps are absent in outer membranes of mitochondria, chloroplasts, and Gram-negative bacteria (12, 20), so that a gradient sufficient to drive ATP synthesis cannot be generated. Therefore, outer membranes are generally believed to be “non–energy-conserving” (13). To date, neither a proton motive force across an outer membrane nor ATP synthesis within a periplasmic space has been described.
In this article we show that in I. hospitalis the outer membrane is energized and that ATP synthesis is spatially separated from DNA replication, transcription, and protein biosynthesis. These results raise questions regarding the function of the two membranes in I. hospitalis, the interaction between the two cell compartments, the general definition of a cytoplasmic membrane, and a possible energy transfer from I. hospitalis to N. equitans.
Results
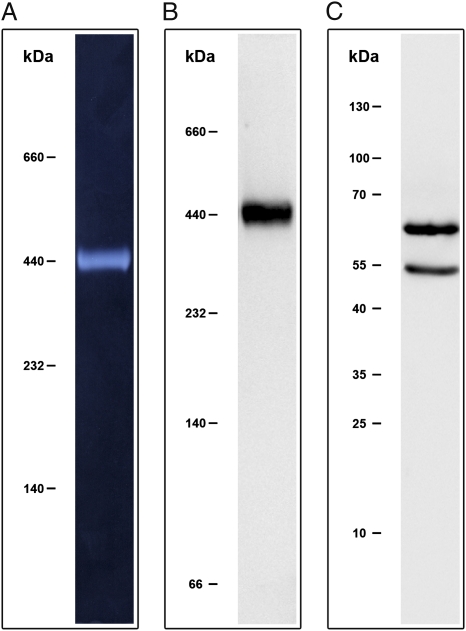
Purification and Identification of the 440-kDa Subcomplex of ATP Synthase.
To clarify how I. hospitalis conserves energy, we started to purify and characterize its A1AO ATP synthase. We solubilized membrane proteins of I. hospitalis by addition of n-dodecyl-β-D-maltopyranoside (DDM). The solubilisate exhibited a specific ATP hydrolysis activity of 1.7 U/mg protein. This activity was completely inhibited by the addition of diethylstilbestrol (DES, 1.5 mM) and to approximately 40% by N′, N′-dicyclohexylcarboiimide (DCCD, 1.5 mM), a property characteristic for a coupled A1AO ATP synthase complex. After separating the solubilisate by high-resolution clear native electrophoresis (hrCNE), the protein complexes were checked for their ability to hydrolyze ATP in an in-gel enzymatic assay (Fig. 1A). One prominent active complex with an apparent molecular mass of ≈440 kDa was obtained. This complex was purified by ion exchange chromatography, ultrafiltration, and size exclusion chromatography. After tryptic digestion, MALDI-TOF analysis revealed two subunits, A and B, to be part of the I. hospitalis ATPase (scores 577 and 316, respectively). Using specific antibodies generated against the complex, Western blot analyses after hrCNE gave a single signal at an apparent mass of 440 kDa for the native complex (Fig. 1B). After its extraction and separation of the subunits under denaturing conditions (SDS/PAGE), Western blot analysis showed that the masses of the major subunits A and B (approximately 65 and 55 kDa; Fig. 1C), correspond to the predicted masses of the annotated A and B subunits of the ATPase (67 and 52 kDa).
Fig. 1.
Detection of ATP synthase by ATP hydrolysis in-gel assay and Western blotting. (A) ATP hydrolysis activity test (90 min at 80 °C) after native protein complex separation (hrCNE) of the I. hospitalis solubilisate. (B) Immunoblot of an hrCNE gel using the 440 kDa complex antibody of I. hospitalis (dilution 1:5,000). (C) Immunoblot of an SDS/PAGE gel using both antibodies raised against the A and B subunits of M. jannaschii (dilution 1:10,000). Immunolabeling was visualized by HRP-conjugated secondary antibodies (Sigma-Aldrich; dilution, 1:5,000).
Localization of ATP synthase in I. hospitalis.
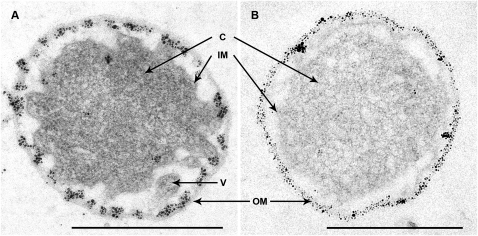
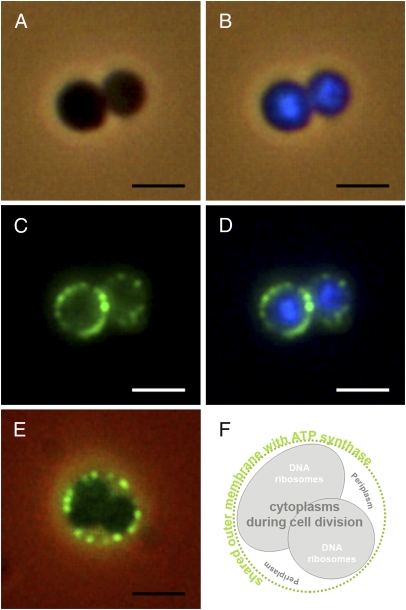
By EM of ultrathin sections of I. hospitalis cells, we investigated the subcellular localization of ATP synthase. Surprisingly, immunolabeling with the antibody raised against the purified 440-kDa ATPase complex showed a highly predominant labeling of the outer membrane of I. hospitalis cells (Fig. 2A). The same result was obtained with antibodies raised against the heterologous membrane-bound subunit a (Fig. 2B) and against the catalytic subunit A of Methanocaldococcus jannaschii (Fig. S1). In all cases, less than 10% of the signals could be detected within the cytoplasm, the inner membrane, and the periplasmic space (including the membrane-coated vesicles). This clearly indicates that the by far major part of ATP synthase molecules of I. hospitalis is located in the outer membrane. To confirm this result, we carried out fluorescence light microscopy experiments (Fig. 3 A–E) using DNA staining with DAPI (Fig. 3B) and concomitantly the antibody against the 440-kDa ATPase complex (Fig. 3C). According to these images the DNA is, as expected, exclusively located inside the inner membrane (i.e., in the cytoplasm), whereas we observed the fluorescence of the antibodies bound to the A1AO ATP synthase to a great extent in the outer membrane, as illustrated in the merge of the two fluorescence images (Fig. 3D). A similar observation was made for dividing I. hospitalis cells, with two daughter cytoplasms existing within the outer membrane (7) (Fig. 3 E and F).
Fig. 2.
Localization of A1AO ATP synthase on I. hospitalis cells by EM of ultrathin sections. (A) Labeling with antibodies specifically raised against the purified 440-kDa ATPase complex, (B) Labeling with antibodies raised against the membrane-bound subunit a. For both images the secondary antibody with ultrasmall gold particles is visualized by silver enhancement (dilution, 1:200). Dilution of the primary antibodies, 1:200. C, cytoplasm; IM, inner membrane; V, vesicles in the periplasm; OM, outer membrane. (Scale bars, 1 μm.)
Fig. 3.
Epifluorescence microphotographs of I. hospitalis cells. (A) Phase contrast image. (B) Merged image of phase contrast and DAPI-stained cytoplasm (blue). (C) Localization of ATP synthase immunolabeled with the specific 440-kDa ATPase complex antibody; secondary antibody coupled to Alexa Fluor 488 (green). (D) Merge of Fig. 2 B and C. (E) Merge of a phase contrast image and an immunofluorescence image of a dividing I. hospitalis cell. (F) Scheme of the dividing cell in Fig. 2E. (Scale bars, 2 μm.)
On the basis of these results, we conclude that the outer membrane of I. hospitalis is energized and that energy conservation takes place across this membrane. As a consequence, it is necessary to assume that the ion potential-generating enzyme is also located here. As an obligate chemolithoautotroph, I. hospitalis conserves energy exclusively by reduction of elemental sulfur using molecular hydrogen as electron donor (1). Therefore, we aimed at localizing H2:sulfur oxidoreductase complex, which generates a proton motive force used for ATP generation by A1AO ATP synthase.
Localization of H2:Sulfur Oxidoreductase in I. hospitalis.
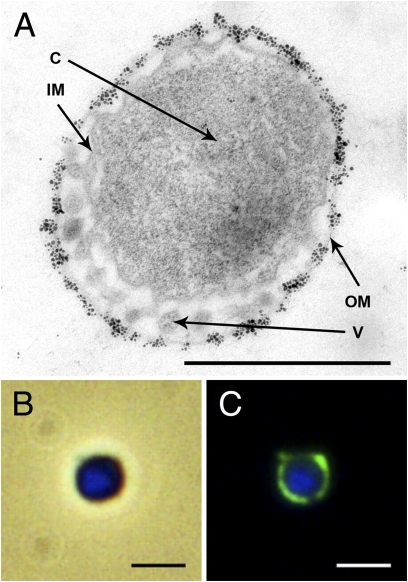
Labeling of ultrathin sections with specific antibodies against H2:sulfur oxidoreductase complex from Pyrodictium abyssi strain TAG11 (21) (Fig. 4A) demonstrated that this complex was also located in the outer membrane of I. hospitalis. A similar result was obtained by immunofluorescence light microscopy (Fig. 4 B and C). In a parallel experiment, the solubilisate was analyzed by Western blotting after separation under native conditions (hrCNE). A strong signal in an in-gel hydrogenase assay and in Western blot analyses with specific antibodies gave direct evidence for the presence of an active H2:sulfur oxidoreductase complex (Fig. S2 A and B). In contrast, no signals were detected when both experiments were carried out with purified ATP synthase complex. These data prove that H2:sulfur oxidoreductase complex (primary proton pump) as well as ATP synthase are located in the outer membrane of I. hospitalis.
Fig. 4.
Localization of H2:sulfur oxidoreductase complex on I. hospitalis cells with antibodies specifically raised against the purified complex of P. abyssi strain TAG11. (A) EM of ultrathin sections: the immunolabeling was performed similar to Fig. 2; C, cytoplasm; IM, inner membrane; V, vesicles in the periplasm; OM, outer membrane. (Scale bar, 1 μm.) (B) Merged image of phase contrast and DAPI-stained cytoplasm (blue). (C) Colocalization of H2:sulfur oxidoreductase complex and cytoplasm visualized by immunofluorescence (secondary antibody coupled to Alexa Fluor 488, green) and DAPI staining. (Scale bars in B and C, 2 μm.)
Discussion
To date, the existence of an energized outer membrane in prokaryotes harboring more than one membrane system has not been previously described. For Gram-negative bacteria, it is known that the cytoplasmic membrane is energized but that energy can be transmitted to the outer membrane via, for example, the TonB system (12, 20, 22). A reverse transport has never been observed, with the exception of the uptake of “host derived-ATP” by members of the genera Rickettsia or Chlamydia (23). Other bacteria that have, at a first glance, some structural similarity to Ignicoccus, are the Planctomycetes: they lack murein but possess two membranes, named intracytoplasmic and cytoplasmic (24). Hence, two compartments are found: the inner “riboplasm,” containing DNA and ribosomes, and the outer “paryphoplasm” (24). The latter was postulated on the basis of histochemical experiments to contain RNA, but its nature was not further investigated. In fact, the distribution of key metabolic enzymes in Planctomycetes cells is not yet known, and therefore no data are available regarding which of the two membranes is energized.
Among the Archaea, the cells of all Ignicoccus species are distinct and unique in possessing a two-membrane system (1, 4, 5, 7), defining two structurally distinguishable compartments: the cytoplasm and the huge periplasm. The outer membrane of I. hospitalis was suggested to only contain lipids and one major protein (9), thus surrounding a more or less empty intermembrane compartment like an inert blanket. This model for the outer membrane was consistent with the observation that the inner membrane compartment divides independently from the outer membrane (7). Other Archaea, like members of Sulfolobales, Desulfurococcales (with the exception of all Ignicoccus species), and Methanococcales, possess a single membrane only and a “quasi-periplasmic space” between this cytoplasmic membrane and the S-layer (10). In these organisms, immunolabeling data show that A1AO ATPase is, as expected, located in the cytoplasmic membrane (e.g., Sulfolobus solfataricus; Fig. S3). Therefore, it was unexpected and surprising to find ATP synthesis within the periplasm of I. hospitalis. It demonstrates that several assumptions (18) about the function and localization of the different cell envelope components of I. hospitalis were wrong. First, it is not the inner but rather the outer membrane that is energized (H2:sulfur oxidoreductase) and the site of energy conservation (A1AO ATP synthase). Second, at least a certain amount of energy-consuming cellular processes must take place in the periplasmic space. Because a specific biophysical separation of the inner and outer compartment is not yet possible, we can only speculate about such metabolic reactions: although DNA (Fig. 3 B and D) and ribosomes are found in the cytoplasm, they are absent in the periplasm, and therefore it can be excluded that transcription, translation, and DNA replication take place here. In contrast, reactions like the first steps of the CO2 fixation pathway (25), or uptake and activation of external molecules like acetate or succinate (26), have to be considered to be located in the periplasm, because high amounts of energy are needed for these reactions (25).
According to our results, I. hospitalis possesses a very complex and unique ultrastructure, with two independent functional partitions inside a prokaryotic cell: the inner membrane separates energy conservation in the periplasm from information processing and protein biosynthesis in the cytoplasm. The latter processes are highly dependent on a constant energy supply, but there is hardly any energy-conserving ATP synthase in the surrounding inner membrane. Therefore, ATP has to be channeled from the periplasm to the cytoplasm, for example, by diffusion as in the case of the outer membrane of mitochondria, or by a potential-driven antiport of ADP and ATP, as known for the inner membrane of mitochondria (27). The resurvey of the I. hospitalis genome (18) and analysis of major membrane proteins (28) did not give any hint for known ATP transport systems. However, such transporters are most likely hard to detect on the gene level because the similarity among the known ADP/ATP carriers (mainly found in endosymbionts and mitochondria) (23, 27, 29) is quite low, and to our knowledge no such system has been identified to date for any Archaeon. Besides an import of ATP, transport of molecules across the inner membrane requires an electrochemical gradient sufficient to drive these processes. Because the amount of H2:sulfur oxidoreductase and ATP synthase is negligible, other (as yet unknown) processes generating an electrochemical gradient must be present in the inner membrane of I. hospitalis. From the genome sequence, however, no obvious candidates for ATP-dependent ion pumps or other means to energize the membrane are apparent (18).
Consequently, neither the inner nor the outer membrane of I. hospitalis satisfies all criteria of a cytoplasmic membrane. Although the outer membrane is energized by a primary proton pump and contains ATP synthase, the inner membrane surrounds the compartment containing the machinery of information processing and biosynthesis reactions. This extraordinary cellular architecture, whereby features of a classical cytoplasmic membrane are distributed to two membranes, raises the fundamental question of how to define a cytoplasmic membrane in general and in particular in I. hospitalis.
Our results may also help shed light on the interaction of I. hospitalis with N. equitans (3) and a central question encountered with N. equitans: how is energy supply ensured in an organism that possesses no primary proton pumps? N. equitans only contains (according to the genome data and annotation) a rudimentary, incomplete ATP synthase (30) which was supposed to be unable to synthesize ATP on its own (15), although this has to be proven experimentally. The ATP formation in the periplasm of I. hospitalis facilitates the ATP uptake for N. equitans: it avoids the otherwise complex import from the cytoplasm of I. hospitalis across three membranes (7), into the cytoplasm of N. equitans.
Finally, our results can even lead to an extreme hypothesis: it is assumed that the eukaryotic cell may have originated from an archaeal ancestor, which incorporated bacterial organisms (31). If this is correct (which is controversially discussed), would not an organism like I. hospitalis, with its huge and energized periplasmic space, be an ideal candidate for such an ancestor, which provides ATP and further metabolites but without an interaction between the cytoplasms of symbiont and host?
Materials and Methods
Cultivation Conditions.
I. hospitalis KIN4/IT cells were cultivated in 1/2 SME (synthetic sea water) Ignicoccus medium at 90 °C as previously described (1, 2), with elemental sulfur as electron donor and a gas phase consisting of H2/CO2 (250 kPa; 80:20 vol/vol). Mass cultivation was carried out in a 300-L enamel-protected fermentor with the following minor modifications: stirring was decreased from 150 to 100 rpm, and the gassing rate was increased from 15 L/min to 60 L/min N2/H2/CO2 (65:15:20, vol/vol/vol) after reaching a cell density of ≈5 × 106 cells/mL.
Cells of M. jannaschii JAL-1T and S. solfataricus DSM1616T were grown as previously reported (16, 32).
Electron Microscopy.
For EM analysis, fresh cells were cultivated in cellulose capillaries and cryo-immobilized by high-pressure freezing as previously described (33). After freeze-substitution fixation in 95% ethanol/0.5% glutaraldehyde/1% formaldehyde/0.5% uranyl acetate and 5% water, samples were embedded in Epon; for immunolabeling on ultrathin sections, the primary antibody was detected using secondary antibodies with ultrasmall gold and silver enhancement (7). Electron micrographs were digitally recorded using a slow-scan CCD camera (TVIPS) attached to a CM12 transmission electron microscope (FEI).
Light and Fluorescence Microscopy.
Phase contrast and fluorescence microscopy were carried out with an Olympus BX 60 fluorescence microscope (BV and NB filter) as previously described (2). Micrographs were recorded digitally using a PixelFly CCD camera (1,024 × 1,024 pixels; PCO). Digital images were edited with Photoshop CS3 (Adobe). Phase contrast and fluorescence images were merged with ImageJ (open source).
DNA of I. hospitalis cells was stained with freshly prepared DAPI staining solution, containing (for 100 μL): 30 μL 2 M Na-acetate (pH 4.7), 50 μL 100 mM Na2-EDTA, 10 μL DAPI (0.2 mg/mL), and 10 μL 1% SDS solution. For optimal staining, 1 μL of this solution was added to 8 μL of an I. hospitalis culture.
Labeling of Whole I. hospitalis Cells for Immunofluorescence Analyses.
For immunofluorescence, fresh log-phase cells were fixed by addition of formaldehyde (30% in PBS), washed with PBSG (PBS + 50 mM glycine) and PBS. Subsequently, PBS was replaced by PBSBT (PBS + 2% BSA + 0.02% Tween 20), and the primary antibody was added (1:250). After incubation for 2 h, cells were washed with PBS, incubated with the secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG; Invitrogen, 1:500 in PBS) for 2 h, and finally washed twice in PBS. All procedures were carried out at room temperature. The primary antibodies for ATP synthase subunits A, B, and a of M. jannaschii and for H2:sulfur oxidoreductase complex from P. abyssi were generated as previously described (16, 21, 34). Antibodies against the 440-kDa subcomplex of I. hospitalis were raised in rabbits (Davids Biotechnology) from the purified native complex.
Membrane Preparation.
Frozen or freshly cultivated and centrifuged I. hospitalis cells were resuspended in sterile lysis buffer (25 mM Tris/HCl, 10 mM MgCl2, and 0.1 mM PMSF, pH 7.5) at 4 °C. Cells were disrupted by two French press passages (3.5 MPa; Aminco), and the cell debris were removed by centrifugation (Beckman Avanti J-25, JA25.50 rotor, 3,000 × g, 15 min, 4 °C). Membranes were sedimented from the extract by ultracentrifugation, washed, and resuspended in 25 mM 2-(N-morpholine)-ethane sulphonic acid (Mes)/NaOH, 10 mM MgCl2, 0.1 mM PMSF, and 10% glycerol (vol/vol), pH 6.0, as previously described (17). All protein concentrations were determined with a BCA Protein Assay Kit (Pierce Biotechnology).
Solubilization and Purification of the 440-kDa Protein Complex.
Proteins from the resuspended membranes were solubilized with 1.5 mg DDM (Anatrace) per milligram protein and a final concentration of 2.5% (wt/vol) DDM. After the addition of DDM, the membranes were incubated for 2 h at 40 °C and stored overnight at room temperature. Solubilized membrane proteins were separated from residual membrane particles by ultracentrifugation (Beckman Optima LE-80K, 70.1 Ti rotor, 120,000 × g, 90 min, 4 °C). The 440-kDa complex was further enriched by negative purification via cation exchange chromatography [HiTrap SP HP column, GE Healthcare; CXC buffer A: 25 mM Mes/NaOH, 10 mM MgCl2, 0.1 mM PMSF, 10% glycerol (vol/vol), and 0.05% DDM (wt/vol), pH 6.0; CXC buffer B: CXC buffer A + 1 M NaCl]. The flow-through, containing the active 440-kDa complex, was diluted 1:10 with AXC buffer A [25 mM Tris/HCl, 10 mM MgCl2, 0.1 mM PMSF, 10% glycerol (vol/vol), and 0.05% DDM (wt/vol), pH 8.0; AXC buffer B: AXC buffer A + 1 M NaCl] and then applied onto an anion exchange chromatography column (MonoQ 5/50 G; GE Healthcare). Proteins were eluted by a linear gradient from 0 to 40% AXC buffer B (20 column volumes). The fractions showing ATP hydrolysis activity were pooled and concentrated in ultrafiltration spin column devices (Vivaspin, 100,000 molecular weight cut-off; Sartorius). A final purification of the 440-kDa complex was carried out by size exclusion chromatography (HiLoad 16/60 Superdex 200 pg; SEC buffer: AXC buffer A + 150 mM NaCl) of the concentrated AXC eluate.
ATPase Activity and Inhibition Assays.
ATPase activity tests and inhibitor studies, using DES and DCCD, were performed as previously described (17).
Electrophoresis, In-Gel Enzyme Activity Assays, and Immunodetection.
Protein complexes were separated by hrCNE and tested for ATP hydrolysis by an ATPase in-gel assay (35). The assay is based on the precipitation of inorganic phosphate, released by hydrolysis of ATP, with reactive lead ions from the test buffer (lead nitrate). This results in opaque precipitations at the corresponding positions of ATP hydrolyzing enzymes in the gel. The test was carried out with the following modifications: hrCNE gels (20 × 20 cm; 5–13% acrylamide) were run with 5 mA/gel for 16 h at 4 °C, and the assays were performed at 80 °C for 1 h in the dark. The hydrogenase activity was detected after hrCNE by incubation of the gel with methylene blue staining solution (200 mM Tris, pH 8.5; 2.5 mM methylene blue) for 40 min under anoxic conditions. The gel was washed twice with H2-saturated Tris/HCl buffer (100 mM, pH 8.5) and incubated for 20 min at 80 °C. Active enzymes were detected by a bleaching of the blue color at the corresponding gel positions. Tricine SDS/PAGE and Western blot analyses were carried out as previously described (36, 37).
Supplementary Material
Acknowledgments
We thank K. Y. Pisa, U. Friedrich, and R. Wirth for stimulating discussions; M. Thomm for ongoing support; R. Dirmeier and K. Y. Pisa for providing antibodies; and T. Heimerl, A. Röhl, N. Wasserburger, C. Neuner, G. Leichtl, T. Hader, and K. Eichinger for technical support. This work was supported by Deutsche Forschungsgemeinschaft Grants HU 703/2-1 (to H.H. and R.R.), SFB807 (to V.M.), and SFB699 (to R.R.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911711107/DCSupplemental.
References
- 1.Paper W, et al. Ignicoccus hospitalis sp. nov., the host of ’Nanoarchaeum equitans’. Int J Syst Evol Microbiol. 2007;57:803–808. doi: 10.1099/ijs.0.64721-0. [DOI] [PubMed] [Google Scholar]
- 2.Huber H, et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 3.Jahn U, et al. Nanoarchaeum equitans and Ignicoccus hospitalis: New insights into a unique, intimate association of two Archaea. J Bacteriol. 2008;190:1743–1750. doi: 10.1128/JB.01731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber H, et al. Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov. and Ignicoccus pacificus sp. nov. Int J Syst Evol Microbiol. 2000;50:2093–2100. doi: 10.1099/00207713-50-6-2093. [DOI] [PubMed] [Google Scholar]
- 5.Rachel R, Wyschkony I, Riehl S, Huber H. The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea. 2002;1:9–18. doi: 10.1155/2002/307480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Näther DJ, Rachel R. The outer membrane of the hyperthermophilic Archaeon Ignicoccus: Dynamics, ultrastructure and composition. Biochem Soc Trans. 2004;32:199–203. doi: 10.1042/bst0320199. [DOI] [PubMed] [Google Scholar]
- 7.Junglas B, et al. Ignicoccus hospitalis and Nanoarchaeum equitans: Ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch Microbiol. 2008;190:395–408. doi: 10.1007/s00203-008-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jahn U, Summons R, Sturt H, Grosjean E, Huber H. Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I. Arch Microbiol. 2004;182:404–413. doi: 10.1007/s00203-004-0725-x. [DOI] [PubMed] [Google Scholar]
- 9.Burghardt T, Näther DJ, Junglas B, Huber H, Rachel R. The dominating outer membrane protein of the hyperthermophilic Archaeum Ignicoccus hospitalis: A novel pore-forming complex. Mol Microbiol. 2007;63:166–176. doi: 10.1111/j.1365-2958.2006.05509.x. [DOI] [PubMed] [Google Scholar]
- 10.König H, Rachel R, Claus H. In: Archaea: Molecular and Cellular Biology. Cavicchioli R, editor. Washington, DC: American Society of Microbiology Press; 2007. pp. 315–340. [Google Scholar]
- 11.Beveridge TJ. Structures of Gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun V. Energy transfer between biological membranes. ACS Chem Biol. 2006;1:352–354. doi: 10.1021/cb600256k. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls DG, Ferguson SJ. Bioengergetics 3. London, UK: Academic Press; 2002. [Google Scholar]
- 14.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewalter KY, Müller V. Bioenergetics of archaea: Ancient energy conserving mechanisms developed in the early history of life. Biochim Biophys Acta. 2006;1757:437–445. doi: 10.1016/j.bbabio.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 16.Lingl A, et al. Isolation of a complete A1AO ATP synthase comprising nine subunits from the hyperthermophile Methanococcus jannaschii. Extremophiles. 2003;7:249–257. doi: 10.1007/s00792-003-0318-7. [DOI] [PubMed] [Google Scholar]
- 17.Pisa KY, Huber H, Thomm M, Müller V. A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 2007;274:3928–3938. doi: 10.1111/j.1742-4658.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- 18.Podar M, et al. A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 2008;9:R158. doi: 10.1186/gb-2008-9-11-r158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vonck J, Pisa KY, Morgner N, Brutschy B, Müller V. Three-dimensional structure of A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus by electron microscopy. J Biol Chem. 2009;284:10110–10119. doi: 10.1074/jbc.M808498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun V, Endriss F. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals. 2007;20:219–231. doi: 10.1007/s10534-006-9072-5. [DOI] [PubMed] [Google Scholar]
- 21.Dirmeier R, Keller M, Frey G, Huber H, Stetter KO. Purification and properties of an extremely thermostable membrane-bound sulfur-reducing complex from the hyperthermophilic Pyrodictium abyssi. Eur J Biochem. 1998;252:486–491. doi: 10.1046/j.1432-1327.1998.2520486.x. [DOI] [PubMed] [Google Scholar]
- 22.Brinkman KK, Larsen RA. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. J Bacteriol. 2008;190:421–427. doi: 10.1128/JB.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz-Esser S, et al. ATP/ADP translocases: A common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 2004;186:683–691. doi: 10.1128/JB.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuerst JA. Intracellular compartmentation in Planctomycetes. Annu Rev Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 25.Huber H, et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2008;105:7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahn U, Huber H, Eisenreich W, Hügler M, Fuchs G. Insights into the autotrophic CO2 fixation pathway of the Archaeon Ignicoccus hospitalis: comprehensive analysis of the central carbon metabolism. J Bacteriol. 2007;189:4108–4119. doi: 10.1128/JB.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008;1778:1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Burghardt T, et al. Insight into the proteome of the hyperthermophilic Crenarchaeon Ignicoccus hospitalis: The major cytosolic and membrane proteins. Arch Microbiol. 2008;190:379–394. doi: 10.1007/s00203-008-0399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler HH, Neuhaus HE. Non-mitochondrial ATP transport. Trends Biochem Sci. 1999;24:64–68. doi: 10.1016/s0968-0004(98)01334-6. [DOI] [PubMed] [Google Scholar]
- 30.Waters E, et al. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- 32.Huber H, Prangishvili D. In: The Prokaryotes. Dworkin M, Falcow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. Vol 3. New York: Springer; 2006. pp. 23–51. [Google Scholar]
- 33.Rieger G, Müller K, Hermann R, Stetter KO, Rachel R. Cultivation of hyperthermophilic archaea in capillary tubes resulting in improved preservation of fine structures. Arch Microbiol. 1997;168:373–379. [Google Scholar]
- 34.Lemker T, Ruppert C, Stöger H, Wimmers S, Müller V. Overproduction of a functional A1 ATPase from the archaeon Methanosarcina mazei Gö1 in Escherichia coli. Eur J Biochem. 2001;268:3744–3750. doi: 10.1046/j.1432-1327.2001.02284.x. [DOI] [PubMed] [Google Scholar]
- 35.Wittig I, Carrozzo R, Santorelli FM, Schägger H. Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis. 2007;28:3811–3820. doi: 10.1002/elps.200700367. [DOI] [PubMed] [Google Scholar]
- 36.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 37.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.