Abstract
A novel concept in eukaryotic signal transduction is the use of nutrient transporters and closely related proteins as nutrient sensors. The action mechanism of these “transceptors” is unclear. The Pho84 phosphate transceptor in yeast transports phosphate and mediates rapid phosphate activation of the protein kinase A (PKA) pathway during growth induction. We have now identified several phosphate-containing compounds that act as nontransported signaling agonists of Pho84. This indicates that signaling does not require complete transport of the substrate. For the nontransported agonist glycerol-3-phosphate (Gly3P), we show that it is transported by two other carriers, Git1 and Pho91, without triggering signaling. Gly3P is a competitive inhibitor of transport through Pho84, indicating direct interaction with its phosphate-binding site. We also identified phosphonoacetic acid as a competitive inhibitor of transport without agonist function for signaling. This indicates that binding of a compound into the phosphate-binding site of Pho84 is not enough to trigger signaling. Apparently, signaling requires a specific conformational change that may be part of, but does not require, the complete transport cycle. Using Substituted Cysteine Accessibility Method (SCAM) we identified Phe160 in TMD IV and Val392 in TMD VIII as residues exposed with their side chain into the phosphate-binding site of Pho84. Inhibition of both transport and signaling by covalent modification of Pho84F160C or Pho84V392C showed that the same binding site is used for transport of phosphate and for signaling with both phosphate and Gly3P. Our results provide to the best of our knowledge the first insight into the molecular mechanism of a phosphate transceptor.
Keywords: nutrient sensing, protein kinase A, growth induction, Saccharomyces cerevisiae
In yeast and subsequently in other eukaryotic species, several nutrient-transporter-related nutrient receptors or “transceptors” have been discovered (1). They can be nontransporting homologues of nutrient transporters as in the case of the yeast glucose sensors Snf3 and Rgt2 (2) and amino acid sensor Ssy1 (3–5), the glucose-6-phosphate sensor UhpC in E. coli (6), or the SGLT3 glucose sensor in mammalian cells (7). Alternatively, several actively transporting nutrient carriers have also been found to act as nutrient sensors, as in the case of the yeast Mep2 ammonium carrier (8–10), Gap1 amino acid carrier (11) and Pho84 phosphate carrier (12), the Arabidopsis nitrate transporter NRT1.1 (13), and the Drosophila PATH (14) and mammalian SNAT2 amino acid transporters (15). The precise functioning of both the transporting and nontransporting transceptors is not well understood.
Phosphate transport in S. cerevisiae is carried out by five phosphate transporters, Pho84, Pho87, Pho89, Pho90, and Pho91 (16). A quintuple deletion strain lacking all carriers is deficient in phosphate transport and inviable. Pho84 provides a major contribution to phosphate transport and is strongly induced under phosphate starvation conditions (16). Whereas a specific role of Pho84 in transcriptional repression of PHO pathway target genes by phosphate has been contradicted (16), Pho84 was shown to mediate rapid activation of PKA pathway targets during growth induction by phosphate in phosphate-deprived fermenting yeast cells (12). Addition of phosphate also triggers phosphorylation and ubiquitination of Pho84 followed by endocytic internalization and degradation in the vacuole. This process may be triggered by Pho84-mediated activation of the PKA pathway (17).
The plasma membrane bound Git1 carrier imports glycerophosphoinositol (GlyPIno) and upon overexpression also phosphate into yeast cells (16, 18). GlyPIno is generated in phosphatidylinositol catabolism by surface phospholipases and released into the medium. Extracellular hydrolysis of GlyPIno is insignificant (19).
In the present paper we show that phosphate-containing compounds that are not transported by Pho84 can trigger phosphate signaling. Transport by another carrier, like Git1, does not trigger signaling. We have identified amino acid residues that are exposed with their side chain into the phosphate-binding site of Pho84 and show that the same site is used for transport and signaling. We also provide evidence that binding of the phosphate substrate into the phosphate-binding site of Pho84 is not enough to trigger signaling, indicating that agonist action requires induction of a specific conformational change. It may be part of, but clearly does not require the complete transport cycle. To the best of our knowledge, these results provide previously undescribed insight into the action mechanism of a phosphate transceptor.
Results
Screening for Organic Phosphate Compounds That Can Activate PKA Signaling Through the Pho84 Transceptor.
Addition of phosphate in the presence of glucose to phosphate-starved yeast cells triggers rapid activation of the protein kinase A pathway, which can be measured conveniently by activation of trehalase, a well-established protein kinase A target (12). We have screened phosphate-containing compounds unlikely to be transported by Pho84 for possible agonist action on the signaling function of Pho84. We identified multiple organic phosphate compounds that could trigger PKA signaling and were unlikely to be transported into yeast cells (Table 1). Phosphate esters were the most effective compounds. Other phosphorus-containing compounds, like phosphonoacetic acid (PAA), did not produce a significant activation (Table 1). Unexpectedly, we found that glycerol-3-phosphate (Gly3P) was rapidly taken up by yeast cells, and we have explored its uptake and signaling mechanism further in detail. We tested the other compounds of Table 1 also for possible use as a source of phosphate in the phoΔnull strain, which completely lacks phosphate transport. Only Gly3P and GlyPIno were able to support growth indicating that the other compounds can either not be transported or cannot be metabolized intracellularly to serve as a source of phosphate. These results are provided in SI Text.
Table 1.
Maximal increase in trehalase activity after addition of inorganic phosphate or different organic phosphate esters and phosphorus compounds to phosphate-starved cells.
| Strain | |||
| Compound |
Wild type |
pho3Δ pho5Δ |
pho84Δ pho3Δ pho5Δ |
| Agonists | |||
| Phosphate | 110.8 ± 27.2 | 110.7 ± 19.8 | 15.4 ± 10.5 |
| Fru1,6P | 80.4 ± 16.3 | 72.5 ± 6.0 | 7.0 ± 2.5 |
| Fru6P | 81.9 ± 17.5 | 71.0 ± 17.6 | 5.3 ± 1.6 |
| Gly3P | 78.8 ± 11.7 | 77.5 ± 10.5 | 5.1 ± 4.5 |
| Glu6P | 76.6 ± 16.5 | 68.5 ± 15.4 | 12.2 ± 3.7 |
| Glu1P | 75.6 ± 15.0 | 72.4 ± 18.0 | 6.1 ± 3.7 |
| ATP | 56.4 ± 11.5 | 54.2 ± 11.2 | ≤1.0 |
| GlyPIno | 46.1 ± 12.5 | 45.8 ± 10.2 | ≤1.0 |
| Methylphosphonate | 33.4 ± 11.5 | 34.5 ± 11.6 | ≤1.0 |
| Non- or very poor agonists | |||
| Dimethylphosphonate | 15.6 ± 7.6 | 16.8 ± 7.3 | ≤1.0 |
| Diethylphosphate | 14.1 ± 11.5 | 14.5 ± 10.7 | ≤1.0 |
| Phosphonoformic acid | 12.3 ± 10.7 | 13.4 ± 11.8 | ≤1.0 |
| Phosphonoacetic acid (PAA) | 17.2 ± 13.3 | 16.5 ± 11.0 | ≤1.0 |
| Phosphonopropionic acid | 13.7 ± 11.9 | 13.8 ± 11.2 | ≤1.0 |
| Trimethylphosphate | 8.9 ± 14.3 | 8.5 ± 11.1 | ≤1.0 |
| Phosphocholine | 4.9 ± 10.3 | 4.5 ± 10.5 | ≤1.0 |
| Trimethyl phosphonoacetate | ≤1.0 | ≤1.0 | ≤1.0 |
| Trimethyl phosphonoformate | ≤1.0 | ≤1.0 | ≤1.0 |
Basal trehalase activity before addition of phosphate or phosphate analogue was 59.5 ± 5.4 for the wild type strain, 56.1 ± 4.5 for the pho3Δ pho5Δ strain, and 23.5 ± 4.7 for the pho84Δ pho3Δ pho5Δ. This basal level of activity has been subtracted from the peak value after addition of the compound to reveal specifically its agonist signaling capacity. Standard deviations are indicated.
Addition of 10 mM Gly3P to phosphate-starved wild type cells triggered rapid PKA signaling (Fig. 1A) similar to the effect of phosphate addition (12). The yeast Saccharomyces cerevisiae expresses the secreted phosphatases Pho3 and Pho5, but a pho3Δ pho5Δ strain showed only a slight reduction in activation of PKA signaling by Gly3P (Fig. 1A) or by the other organic phosphate esters (Table 1). This shows that the activation is not due to phosphate released extracellularly by the secreted phosphatases from the organic phosphate esters. We have performed extensive control experiments to rule out that the activation observed with the organic phosphate compounds is due to contaminating phosphate in the commercial preparations or to residual, secreted phosphatase activity. These experiments are described in SI Text. The observation that glycerophosphoinositol (GlyPIno) can trigger PKA signaling is consistent with the conclusion that external hydrolysis is not involved because GlyPIno is unable to serve as a source of phosphate or inositol in a strain deficient in GlyPIno uptake (19).
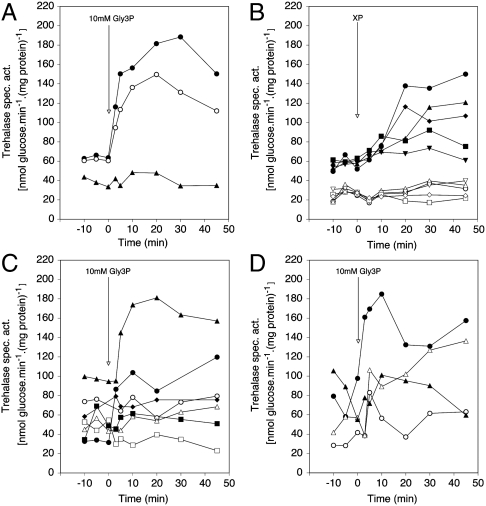
Fig. 1.
The PKA target trehalase is activated after addition of Gly3P or other organic phosphate esters to phosphate-starved cells. (A) Activation of the PKA target trehalase after addition of 10 mM Gly3P to phosphate-starved cells of a wild type and a pho3Δ pho5Δ strain with or without deletion of PHO84. Strains: wild type (•), pho3Δ pho5Δ (○), pho84Δ pho3Δ pho5Δ (▴). (B) Activation after addition of 10 mM of other organic phosphate esters to phosphate-starved cells of a wild type (closed symbols) and a pho84Δ pho3Δ pho5Δ (open symbols) strain. Compounds: Gly3P (•,○), Glu6P (▴,▵), Methylphosphonate (▪,□), ATP (⧫,◊), Phosphonoacetic acid (PAA) (▾,▿). (C) Activation after addition of 10 mM Gly3P to phosphate-starved cells of a wild type strain and strains expressing either none or only a single phosphate carrier from the ADH1 promoter. GAL-PHO84 is repressed in the glucose-containing phosphate starvation medium. Strains: wild type (•), phoΔnull git1Δ +pGAL-PHO84 (○), phoΔnull git1Δ +pPHO84 (▴), +pPHO87 (▵), +pPHO90 (▪), +pPHO91 (□) or +pGIT1 (⧫). (D) Activation of PKA signaling with 10 mM Gly3P in mutants with reduced PKA activity. Wild type (TPK1-3 BCY1) (•), tpk1Δ tpk2Δ TPK3 (○), tpk1Δ tpk2w1 tpk3Δ (▴), tpk1Δ tpk2w1 tpk3Δ bcy1Δ (Δ).
Activation of PKA Signaling by Gly3P Requires Pho84 and is Only Supported by Pho84.
Deletion of PHO84 completely abolished PKA signaling by Gly3P (Fig. 1A). This was also observed for the other organic phosphate esters tested (Fig. 1B and Table 1). The residual activation observed with phosphate (Table 1) is due to Pho87 that also acts to some extent as phosphate transceptor (12). Also the basal level of trehalase activity was substantially reduced by deletion of PHO84. These results indicate that Pho84 is required for activation of PKA signaling by Gly3P and other organic phosphate esters, and in particular that apparently neither Git1 nor Pho91 (which transport Gly3P, see further) can trigger activation of PKA signaling. To gain further evidence for the capacity of Pho84 to support Gly3P-induced PKA signaling, we overexpressed PHO84, PHO87, PHO89, PHO90, PHO91, or GIT1 in a pho84Δ pho87Δ pho89Δ pho90Δ pho91Δ (phoΔnull) git1Δ strain. (Inviability of the control sextuple deletion strain was rescued with a plasmid expressing GAL-PHO84). Assessment of Gly3P-induced PKA signaling in these strains showed that 1) only overexpression of PHO84 and not that of any of the other carriers supported signaling and 2) that overexpression of PHO84 apparently caused a strong increase in the basal level of PKA activity, resulting in an upshift of the whole trehalase activation profile compared to that in the wild type strain (Fig. 1C). These results support a specific role of Pho84 as sensor for Gly3P in PKA signaling. Consistent results were obtained for trehalose mobilization in the same strains. They are provided in SI Text.
As for Phosphate, Activation of PKA Signaling by Gly3P Depends on the Catalytic But Not the Regulatory Subunits of PKA.
In S. cerevisiae, the catalytic subunits of PKA are encoded by the genes TPK1, TPK2, and TPK3, while the regulatory subunits are encoded by the BCY1 gene (20, 21). Strains expressing only TPK3 or only one, partially weakened allele of a TPK gene (tpkw1) display strongly reduced activity of the PKA pathway (22). When Gly3P was added to phosphate-starved cells of a tpk2w1 strain (Fig. 1D) or a strain expressing only TPK3 (Fig. 1D) trehalase activation was strongly reduced. Addition of Gly3P to phosphate-starved cells of the tpk2w1 bcy1Δ strain resulted in a similar activation of PKA signaling compared with the tpk2w1 BCY1 strain (Fig. 1D). These results are strikingly similar to those previously obtained with the same strains for phosphate activation (12), supporting that Gly3P and phosphate activate PKA signaling through the same signaling pathway. We have also measured another target of the PKA pathway, SSA3 expression, by Q-PCR, and we show that it is affected by phosphate and Gly3P in a similar, PKA-dependent manner as trehalase activity. We also performed micro-array analysis of genome-wide gene expression in phosphate-starved cells after addition of phosphate or Gly3P, which confirmed that the two compounds trigger a very similar response, which is dependent on PKA activity. These results are provided in SI Text.
Gly3P is Transported by the Git1 and Pho91 Carriers but Not by Pho84.
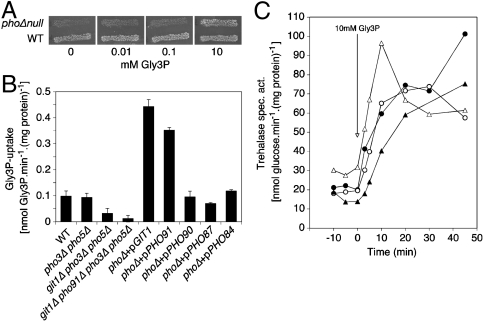
A yeast strain deficient in the five phosphate carriers, Pho84, Pho87, Pho89, Pho90, and Pho91, is unable to grow in a medium with phosphate because the phosphate cannot be transported into the cells. Such a strain can be rescued by inducible expression of the Pho84 carrier from the GAL promoter (16). We found that this phosphate carrier null strain can also be rescued by addition of Gly3P in the medium (Fig. 2A).
Fig. 2.
Uptake of Gly3P occurs through Git1 and Pho91 and is not required for activation of the PKA target trehalase. (A) Gly3P can serve as source of phosphate for a strain lacking all phosphate carriers. Growth of the wild type strain and the phoΔnull strain on SD medium (contains 7 mM phosphate) supplemented with different concentrations of Gly3P. (B) Uptake rate of 14C-Gly3P in phosphate-starved cells of the wild type, pho3Δ pho5Δ, git1Δ pho3Δ pho5Δ and git1Δ pho91Δ pho3Δ pho5Δ strains and of the phoΔnull strain overexpressing a single phosphate carrier. (C) Activation of the PKA target trehalase after addition of Gly3P to phosphate-starved cells does not require the Gly3P transporters Git1 and Pho91. Strains: wild type (•), git1Δ pho3Δ pho5Δ (○), pho91Δ pho3Δ pho5Δ (▴), git1Δ pho91Δ pho3Δ pho5Δ (Δ).
Direct measurement of Gly3P transport using radioactive Gly3P in short-term uptake experiments (5 min) confirmed that phosphate-starved cells of a wild type yeast strain take up Gly3P (Fig. 2B). It was shown previously that transport of GlyPIno by Git1 is competitively inhibited by Gly3P (18), suggesting that this carrier might be responsible for Gly3P transport. We have deleted the GIT1 gene in the pho3Δ pho5Δ strain to avoid interference with possible hydrolysis of Gly3P by the secreted phosphatases. Deletion of PHO3 and PHO5 had no effect on Gly3P short-term uptake in a wild type strain (Fig. 2B). Additional deletion of Git1 caused a drop in Gly3P uptake (Fig. 2B) suggesting that Git1 is responsible for part of the Gly3P uptake. Subsequently, we have constructed strains with deletions of the five phosphate carrier genes PHO84, PHO87, PHO89, PHO90, and PHO91, as well as GIT1, and expressing either PHO84, PHO87, PHO90, PHO91, or GIT1 from the ADH1 promoter on a plasmid. Determination of Gly3P uptake showed that only Git1 and Pho91 supported strong Gly3P uptake (Fig. 2B). Although the other carriers apparently have some residual Gly3P uptake activity when overexpressed, their contribution to Gly3P uptake in a wild type strain appears to be very low. This was confirmed by deletion of PHO91 in the git1Δ pho3Δ pho5Δ strain, which resulted in near-complete elimination of Gly3P transport activity (Fig. 2B). Consistent results were obtained for growth recovery of phosphate-starved cells with Gly3P as only source of phosphate and for the increase in intracellular phosphate in phosphate-starved cells after supply of Gly3P (see SI Text).
Gly3P-Induced PKA Signaling Does Not Depend on Gly3P Uptake.
Examination of Gly3P-induced PKA signaling in the git1Δ pho3Δ pho5Δ, pho91Δ pho3Δ pho5Δ, and git1Δ pho91Δ pho3Δ pho5Δ strains revealed a very similar increase compared with a wild type strain (Fig. 2C). This indicates that neither Git1 and Pho91 nor Gly3P transport into the cell is required for Gly3P-induced activation of PKA signaling. Consistent results were obtained with GlyPIno, which is transported by Git1 (18). They are provided in SI Text.
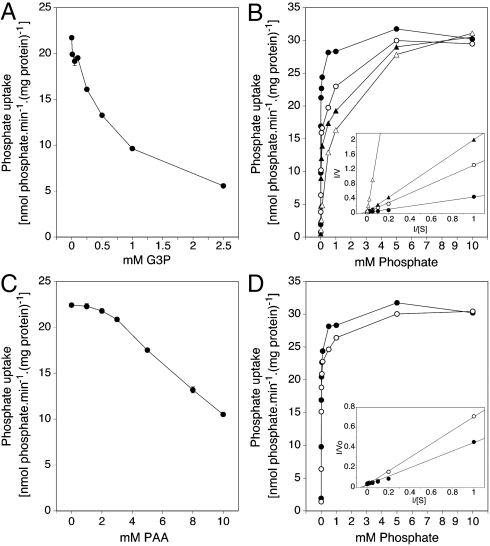
The Agonist Gly3P and the Nonagonist Phosphonoacetic Acid (PAA) are Both Competitive Inhibitors of Phosphate Transport by Pho84.
We have determined whether the signaling agonist Gly3P is able to inhibit phosphate transport in a strain only expressing Pho84. The results show that increasing concentrations of Gly3P inhibit transport of a fixed concentration of phosphate (0.1 mM) with increasing efficiency (Fig. 3A) and that increasing phosphate concentrations counteract inhibition by several, fixed Gly3P concentrations (Fig 3B). This shows that Gly3P acts as a competitive inhibitor of phosphate transport by Pho84. Therefore, both compounds must directly interact with the phosphate-binding site of Pho84. We performed the same experiments with the nonagonist PAA that showed that this compound also acts as competitive inhibitor of phosphate transport by Pho84 and thus must also bind into the phosphate-binding site of Pho84 in a similar way as phosphate (Fig 3C and D). Since PAA is unable to trigger PKA signaling, the mere binding of a compound into the phosphate-binding site of Pho84 is apparently not enough to trigger signaling.
Fig. 3.
Gly3P and PAA are competitive inhibitors of phosphate transport by Pho84. Strain: phoΔnull git1Δ +pPHO84. (A) Uptake rate of 32Pi (0.1 mM) in the presence of different Gly3P concentrations. (B) Uptake rate of 32Pi as a function of the initial phosphate concentration in the absence (•) and presence of 0.5 mM (○), 1 mM (▴), and 2 mM (▵) Gly3P. These values have also been summarized in a Lineweaver-Burk plot (inset); no inhibitor (•), 0.5 mM (○), 1 mM (▴) and 2 mM (▵) Gly3P. The Km value of Pho84 was 14.8 μM. The correlation coefficients for the linear regression plots in the absence and presence of 0.5 mM, 1 mM and 2 mM Gly3P were 0.996, 0.999, 0.999 and 0.999, respectively. (C) Uptake rate of 32Pi in the presence of different PAA concentrations. (D) Uptake rate of 32Pi as a function of the initial phosphate concentration in the absence (•) and presence (○) of 5 mM PAA. These values have also been summarized in a Lineweaver-Burk plot (inset); no inhibitor (•), 5 mM PAA (○). The correlation coefficients for the linear regression plots in the absence and presence of PAA were 0.996 and 0.998, respectively.
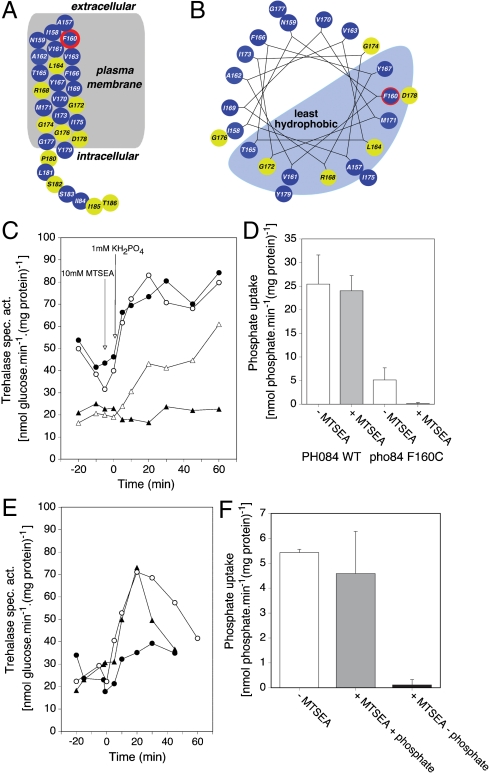
Substituted Cysteine Accessibility Method (SCAM) Identifies Phe160 (TMD IV) and Val392 (TMD VIII) as Residues Exposed into the Phosphate-Binding Site.
SCAM analysis is a well-established methodology to identify amino acid residues of which the side chain is exposed into the ligand-binding site of receptors or the substrate-binding site of transporters (23, 24). We have replaced different residues of TMD IV and TMD VIII in Pho84 individually by cysteine using site-directed mutagenesis. We first selected TMD IV because of several reasons as explained in SI Text. We tested transport and signaling with the different cysteine-substituted mutant proteins. An overview of the results for the whole TMD IV is shown in Fig. 4A and in helical wheel representation in Fig. 4B. Mutagenesis to cysteine of conserved residues in the glycine-rich sequence (Gly172, Gly174, and Gly176) and of the conserved R168 residue resulted in inactive proteins. Pho84 also shows significant sequence similarity with the Snf3 and Rgt2 glucose sensors and with Hxt glucose transporters. An alignment of TMD IV, with the conserved domains indicated and the results of the mutagenesis, is shown in SI Text.
Fig. 4.
SCAM analysis of TMD IV reveals Phe160 as being exposed with its side chain into the phosphate-binding site of Pho84. (A) Location of the cysteine substitution mutations in TMD IV of Pho84. (B) Representation in helical wheel analysis. Amino acid residues that were mutagenized to cysteine are indicated in color according to the transport and signaling capacity of the mutant protein with 1 mM phosphate. Normal transport and signaling with or without preaddition of 10 mM MTSEA (Blue), strongly reduced or completely abolished transport and signaling (Yellow), and partially reduced transport and signaling in the cysteine substitution protein and deficient transport and signaling after preaddition of 10 mM MTSEA (Blue and Red). (C) Activity of trehalase after addition of 1 mM phosphate to phosphate-starved cells of the wild type strain (•,○) and the Phe160 cysteine substitution mutant (▴,▵) without (○,▵) and with (•,▴) preaddition of 10 mM MTSEA. (D) Transport of 1 mM phosphate in phosphate-starved cells of the wild type and Phe160 cysteine substitution mutant with (Gray) and without (White) preaddition of 10 mM MTSEA. (E) Activity of trehalase after addition of 1 mM phosphate to phosphate-starved cells of the Phe160 cysteine substitution mutant without (○) and with preaddition of 10 mM MTSEA in the absence (•) or presence of 50 mM phosphate (▴). (F) Transport of 1 mM phosphate in phosphate-starved cells of the Phe160 cysteine substitution mutant without (White) and with preaddition of 10 mM MTSEA in the presence (Gray), or absence (Black) of 50 mM phosphate.
For the cysteine-substituted proteins where transport and signaling were not or only partially affected, we tested transport and signaling with preaddition of MTSEA. This is a sulfhydryl-binding compound that only reacts efficiently with ionized thiolates in an aqueous environment, such as the binding site for water-soluble ligands or substrates, and not with unionized thiolates, such as those residing within the hydrophobic bilayer of the membrane. Only for Pho84F160C, preaddition of MTSEA blocked the partial subsequent activation of trehalase with phosphate (Fig. 4C) as well as the partial transport of phosphate observed with this mutant protein (Fig. 4D). Phe160 is located at the outside border of the TMD. This result is consistent with the side chain of Phe160 being exposed into the phosphate-binding site of Pho84 and binding of MTSEA to the cysteine substitution protein responsible for blocking its phosphate-binding site. We also performed the classical control protection experiment (23) in which we added increasing concentrations of phosphate together with MTSEA to cells expressing only Pho84F160C. After washing of the cells, we measured signaling and transport with phosphate. Addition of phosphate effectively counteracted the binding of MTSEA, resulting in recovery of both signaling (Fig. 4E) and transport (Fig. 4F). This confirms that phosphate and MTSEA compete for the same binding site, into which Phe160 is exposed with its side chain. Since the Pho84F160C protein showed a partial reduction of trehalase activation and phosphate transport, we performed additional experiments to exclude that the mutation affected a binding site elsewhere in the protein through distortion of the general conformation of the protein. We mutagenized Phe160 to other residues similar in size as cysteine, i.e., alanine, serine, and methionine, but this did not result in an MTSEA-sensitive allele of Pho84 (see SI Text).
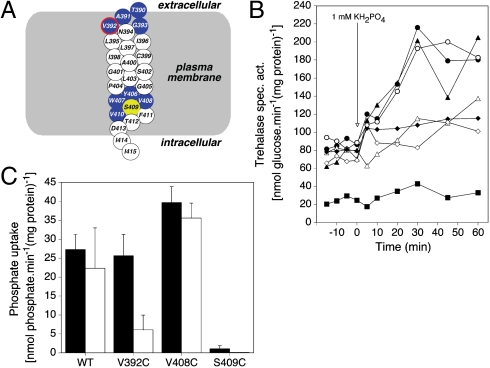
Because recent work has identified Ser388 and Val389 in TMD VIII of the Gap1 amino acid transceptor as residues of which the side chain is exposed into the substrate-binding site (25), we also tested selected amino acid residues in TMD VIII of Pho84 (Fig. 5A and B). We identified Pho84V392C as MTSEA-sensitive, both for PKA signaling (Fig. 5B) and for transport (Fig. 5C). These results indicate that also in Pho84 TMD VIII is apparently located adjacent to the substrate-binding site, as in Gap1. Since both transport and signaling are abolished in the Pho84F160C and Pho84V392C mutant proteins after addition of MTSEA, Pho84 appears to use the same phosphate-binding site for transport and signaling. Interestingly, we also identified a mutant protein, Pho84V408C, for which PKA signaling with phosphate was reduced (Fig. 5B), whereas phosphate transport was enhanced (Fig. 5C). Hence, the mutagenesis results also indicate that the functions of transport and signaling in the Pho84 transceptor can be separated.
Fig. 5.
SCAM analysis of selected residues in TMD VIII reveals Val392 as being exposed with its side chain into the phosphate-binding site of Pho84. (A) Location of the cysteine substitution mutations in TMD VIII of Pho84. Color-coding as in Fig. 4A and B. (B) Activity of trehalase after addition of 1 mM phosphate to phosphate-starved cells of the wild type strain (•,○) and the Val392C (▴,▵), Val408C (⧫,◊), and Ser409C (▪) mutants without (•,▴,▪) and with (○,▵) preaddition of 10 mM MTSEA. (C) Transport of 1 mM phosphate in phosphate-starved cells of the wild type and the Val392C, Val408C, and Ser409C mutants without (Black) and with (White) preaddition of 10 mM MTSEA.
Discussion
Signaling by Pho84 Does Not Require the Complete Transport Cycle.
We have shown that organic phosphate esters like Gly3P can activate the signaling function of Pho84 without being transported by Pho84. To the best of our knowledge, no previous plasma membrane sensor has been discovered in eukaryotic cells that responds to organic phosphate agonists. Unsubstituted phosphate esters were the best activators. Presence of another bond than P-O or substitution of the phosphate hydroxyl groups greatly reduced or abolished activation. We demonstrated experimentally that Gly3P is not transported by Pho84, and the same seems to be true also for the other phosphate esters, because they could not be used as source of phosphate in a strain lacking secreted phosphatases. The presence of signaling without transport is a strong further argument for the receptor function of Pho84. Actually, in this case Pho84 functions in the same way as the nontransporting transceptors Snf3, Rgt2 and Ssy1. It clearly shows that complete transport of the substrate is not required for signaling by the Pho84 transceptor. Gly3P acts as a competitive inhibitor of phosphate transport by Pho84 and must therefore bind into the same binding site as the regular substrate phosphate. Recent work also identified nontransported agonists for PKA signaling by the amino acid transceptor Gap1 (25). Hence, it appears that the nutrient transceptors controlling the PKA pathway in yeast function in a similar way. We also identified the Git1 and Pho91 carriers as responsible for Gly3P uptake, but in spite of their capacity to transport Gly3P into the cells they are unable to support rapid PKA signaling. This not only shows that these carriers are unable to function as transceptors for PKA signaling but also shows that intracellular Gly3P and phosphate derived from its intracellular hydrolysis are unable to trigger rapid PKA signaling. These results therefore further reinforce the unique role of Pho84 in the sensing of phosphate at the level of the plasma membrane during recovery from phosphate starvation.
Signaling by Pho84 Requires a Specific Conformational Change.
We have identified PAA as a competitive inhibitor of phosphate transport by Pho84 implying that it binds into the same substrate-binding site as phosphate. However, PAA did not act as an agonist for PKA signaling. This indicates that mere binding of a compound into the substrate-binding site of Pho84 is not enough to trigger its signaling function. Apparently, the substrate ligand has to induce a specific conformation to trigger the signaling pathway just like in regular receptors. The signaling conformation may be part of the regular transport cycle, but the agonist action of Gly3P indicates that the complete transport cycle is clearly not required for signaling. A similar conclusion was drawn for the amino acid transceptor Gap1 for which a competitive inhibitor of transport also was identified without agonist function for signaling (25). This conclusion does not agree with the model proposed by Wu et al. (26) for the action mechanism of the nontransporting amino acid sensor Ssy1. These authors proposed that the outward-facing conformation, not the inward-facing conformation, was the signaling conformation and that binding of a nutrient ligand to the outward-facing conformation stabilized this conformation and therefore enhanced signaling. It remains to be investigated, however, whether the nontransporting transceptors like Ssy1 act in the same way as the transporting transceptors Pho84 and Gap1.
Pho84 Uses the Same Phosphate-Binding Site for Transport and Signaling.
It is unclear how transporter-related receptors bind their ligands in order to trigger signaling. They can use the same substrate-binding site as the one used for transport in regular nutrient transporters or they could have developed a new ligand-binding site. The transporting transceptors, like Pho84 and Gap1, offer the possibility to directly test whether the same binding site is used for transport and signaling. With SCAM analysis we have now identified residues in Pho84 and previously in Gap1 (25) of which the side chain is exposed into the substrate-binding site of the transceptor. This allows permanent modification of these residues by covalent linkage of the cysteine thiol group with MTSEA. Both in Pho84 and in Gap1 this abolished the transport and the signaling by the transceptor, strongly suggesting that both transceptors use the same binding site for transport and signaling. Multiple control experiments, including the classical MTSEA/ligand competition experiment (23), support this interpretation of the results.
We can conclude that the Pho84 phosphate transceptor functions like a regular receptor for its signaling function. The agonist triggers a specific conformational change that may be part of the conformational changes occurring during transport but clearly does not require the complete transport cycle. The same phosphate-binding site appears to be involved in the initiation of transport and signaling, but specific phosphate analogues and mutations in the protein allow separation of the two functions.
Materials and Methods
Strains and Plasmids, Strain and Plasmid Constructions, Growth and Phosphate Starvation Conditions.
These are provided in SI Text.
Transport Assays and Biochemical Determinations.
Gly3P uptake was assayed by means of radioactively labeled 14C-glycerol-3-phosphate (PerkinElmer). Details on the transport assays and biochemical determinations are provided in SI Text.
SCAM Analysis.
The plasmid EB1368 with PHO84 was used as a template for site-directed mutagenesis. Residues in TMD IV of Pho84 were mutagenized individually to cysteine using the Quick Change site-directed mutagenesis kit (Stratagene). For SCAM analysis, 10 mM (final concentration) 2-aminoethyl methanethiosulfonate, hydrobromide (MTSEA) (Toronto Research Chemicals) was added to the cells 3 min before the addition of phosphate for both transport and trehalase assay.
Reproducibility of the Results.
All experiments were repeated at least twice. Standard deviations are shown for comparisons between independent data points (transport measurements). Representative results are shown for comparisons between collections of interdependent data points (time course measurements). The extent of trehalase activation is variable between different experiments, but the differences observed between specific conditions or between strains are highly reproducible.
Supplementary Material
Acknowledgments.
We thank Erin O’Shea (Cambridge, MA) for strains and plasmids, Jana Patton-Vogt (Pittsburgh) for the gift of GlyPIno and discussion of unpublished results, Bengt Persson (Kalmar, Sweden) for stimulating discussions, and Nico Van Goethem for help with preparation of the figures. This work was supported by a Marie Curie fellowship from the European Commission (Y.P.), by a Government of India, Department of Biotechnology Overseas Associateship Award (BT/IN/BTOA/19/2006) (P.T.), and by grants from the Fund for Scientific Research—Flanders, Interuniversity Attraction Poles Network P6/14 and the Research Fund of the Katholieke Universiteit Leuven (Concerted Research Actions) (J.M.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906546107/DCSupplemental.
References
- 1.Holsbeeks I, Lagatie O, Van Nuland A, Van de Velde S, Thevelein JM. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem Sci. 2004;29:556–564. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didion T, Regenberg B, Jorgensen MU, Kielland-Brandt MC, Andersen HA. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- 4.Klasson H, Fink GR, Ljungdahl PO. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol. 1999;19:5405–5416. doi: 10.1128/mcb.19.8.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iraqui I, et al. Amino acid signaling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwöppe C, Winkler HH, Neuhaus HE. Connection of transport and sensing by UhpC, the sensor for external glucose-6-phosphate in Escherichia coli. Eur J Biochem. 2003;270:1450–1457. doi: 10.1046/j.1432-1033.2003.03507.x. [DOI] [PubMed] [Google Scholar]
- 7.Diez-Sampedro A, et al. A glucose sensor hiding in a family of transporters. Proc Natl Acad Sci USA. 2003;100(20):11753–11758. doi: 10.1073/pnas.1733027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz MC, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas K, Morschhauser J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol. 2005;56:649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- 10.Van Nuland A, et al. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol Microbiol. 2006;59:1485–1505. doi: 10.1111/j.1365-2958.2005.05043.x. [DOI] [PubMed] [Google Scholar]
- 11.Donaton MC, et al. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2003;50:911–929. doi: 10.1046/j.1365-2958.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- 12.Giots F, Donaton MC, Thevelein JM. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2003;47:1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x. [DOI] [PubMed] [Google Scholar]
- 13.Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008;54(5):820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- 14.Goberdhan DC, Meredith D, Boyd CA, Wilson C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132(10):2365–2375. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 15.Hyde R, Cwiklinski EL, MacAulay K, Taylor PM, Hundal HS. Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. J Biol Chem. 2007;282(27):19788–19798. doi: 10.1074/jbc.M611520200. [DOI] [PubMed] [Google Scholar]
- 16.Wykoff DD, O’Shea EK. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundh F, et al. Molecular mechanisms controlling phosphate-induced downregulation of the yeast Pho84 phosphate transporter. Biochemistry. 2009;48:4497–4505. doi: 10.1021/bi9001198. [DOI] [PubMed] [Google Scholar]
- 18.Patton-Vogt JL, Henry SA. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics. 1998;149:1707–1715. doi: 10.1093/genetics/149.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almaguer C, Cheng W, Nolder C, Patton-Vogt J. Glycerophosphoinositol, a novel phosphate source whose transport is regulated by multiple factors in Saccharomyces cerevisiae. J Biol Chem. 2004;279:31937–31942. doi: 10.1074/jbc.M403648200. [DOI] [PubMed] [Google Scholar]
- 20.Toda T, et al. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 22.Cameron S, Levin L, Zoller M, Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988;53:555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- 23.Javitch JA. Probing structure of neurotransmitter transporters by substituted-cysteine accessibility method. Methods Enzymol. 1998;296:331–346. doi: 10.1016/s0076-6879(98)96025-6. [DOI] [PubMed] [Google Scholar]
- 24.Mueckler M, Makepeace C. Cysteine-scanning mutagenesis and substituted cysteine accessibility analysis of transmembrane segment 4 of the Glut1 glucose transporter. J Biol Chem. 2005;280:39562–39568. doi: 10.1074/jbc.M509050200. [DOI] [PubMed] [Google Scholar]
- 25.Van Zeebroeck G, Bonini BM, Versele M, Thevelein JM. Transport and signaling via the amino acid binding site of the yeast Gap1 amino acid transceptor. Nat Chem Biol. 2009;5(1):45–52. doi: 10.1038/nchembio.132. [DOI] [PubMed] [Google Scholar]
- 26.Wu B, et al. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J Cell Biol. 2006;173:327–331. doi: 10.1083/jcb.200602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.