Abstract
Myeloid antigen-presenting cells (APC) express CD1d molecules that present exogenous and endogenous lipid antigens that activate CD1d-restricted T cells, natural killer T (NKT) cells. NKT cell activation has been shown to mediate the potent downstream activation of other immune cells through cell–cell interactions and rapid, prolific cytokine production. Foreign antigens are not required for NKT cell activation. The endogenous lipids bound to CD1d are sufficient for activation of NKT cells in the setting of Toll-like receptor-induced cytokines. The most potent NKT cell antigens identified are glycosphingolipids (GSL). The GSL repertoire of endogenous ligands bound to CD1d molecules that are expressed in myeloid APC at steady state and in the setting of activation has not been delineated. This report identifies the range of GSL bound to soluble murine CD1d (mCD1d) molecules that sample the endoplasmic reticulum/secretory routes and cell surface-cleaved mCD1d that also samples the endocytic system. Specific GSL species are preferentially bound by mCD1d and do not solely reflect cellular GSL. GM1a and GD1a are prominent CD1d ligands for molecules following both the ER/secretory and lysosomal trafficking routes, whereas GM2 was eluted from soluble CD1d but not lysosomal trafficking CD1d. Further, after LPS activation, the quantities of soluble CD1d-bound GM3 and GM1a markedly increased. A unique α-galactose-terminating GSL was also found to be preferentially bound to mCD1d at steady state, and it increased with APC activation. Together, these studies identify the range of GSL presented by CD1d and how presentation varies based on CD1d intracellular trafficking and microbial activation.
Keywords: antigen presentation, cluster of differentiation 1, lipids
The cluster of differentiation (CD1) family of antigen-presenting molecules is a unique class of proteins that, in contrast to major histocompatibility (MHC) class I and II molecules, presents lipids, glycolipids, and lipopeptide antigens to T cells. CD1d-restricted T cells are often referred to as natural killer T (NKT) cells and have been implicated in host defense during infection (1–3), allergic responses (4), immune tolerance (5), and tumor immunosurveillance (6), and they are thought to play a role in autoimmune disease (7, 8). Their protean effects result in large part from their ability to rapidly recruit, stimulate, or influence the function of innate leukocytes, dendritic cells, and adaptive T and B cells (3).
One of the most potent NKT cell antigens, α-galactosylceramide (α-GalCer), has been an indispensable tool in the study of CD1d molecule and NKT cell biology, but it is not found in microbial pathogens. Related microbial and plant-derived α-glycosyl ceramides and glycerol-based lipids that are stimulatory are found in microbial organisms and pollen but are significantly less potent than α-GalCer (9–12). It has been well-documented that foreign antigens are not required for the activation of NKT cells. Self-reactivity or autoreactivity has been shown in murine and human NKT cells and was found to be necessary for NKT cell thymic selection (13, 14). Endogenous CD1d antigens have been shown to play an important role in peripheral NKT cell immunity. Several studies have shown that, in the context of microbial infection, CD1d endogenous antigens are sufficient to mediate NKT activation. IL-12 produced by dendritic cells (DC) on stimulation was sufficient to activate NKT cells (1). This mechanism of NKT cell activation suggests that activation of DC by many Toll-like receptor (TLR) agonists can mediate NKT cell activation in a foreign-antigen independent manner (10, 15–18). In addition to the inflammatory cytokine production generated in response to TLR activation, APC undergo changes in their lipid biosynthetic pathways that alter their endogenous lipid repertoire and more potently activate NKT cells (17–19). These findings highlight the importance of endogenous antigen repertoire in NKT cell development and in the peripheral immune system under inflammatory conditions.
Similar to microbial lipid CD1d antigens, the majority of the endogenous antigens identified belong to two lipid classes, glycerol-based phospholipids and glycosphingolipids (GSL). Phosphatidylinositol (PI), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and lyso-PC have been shown to stimulate a small subset of NKT hybridomas in vitro (20–22). A number of mammalian GSL have been identified as NKT cell antigens. Several gangliosides, including GM1 and GD3, have been shown to be presented by CD1 (7, 23, 24). More recently, sulfatide has been shown to be presented by all CD1 isoforms (25, 26). Sulfatide has been shown to be produced in activated THP-1 cells, suggesting a possible increased role in the setting of inflammation (19). The GSL isoglobotrihexosylceramide (iGb3) can stimulate both murine and human NKT cells (27), but the physiologic role of iGb3 remains unclear (28, 29).
A variety of lipids have been eluted from human CD1d molecules including phospholipids, sphingomyelin, and GSL, suggesting that CD1 molecules can accommodate diverse lipids (22, 30, 31). The majority of studies to date that examined the repertoire of lipids bound by CD1d have used transfected B cell lines and identified PI, PC, lysophospholipids, GM3, and sphingomyelin (20, 22, 30, 31). These studies have been informative and have drawn attention to these lipid classes. However, the spectrum of CD1d ligands in myeloid lineage APC has not been examined. The range of GSL and phospholipids expressed by a specific cell type may have a significant impact on the lipid ligands bound by CD1 molecules. For example, the GSL repertoire in B cells has been shown to undergo serial changes with B cell maturation (32, 33). These changes in GSL expression are likely caused by serial changes in the activation of the corresponding glycosyl transfereases as has been previously shown (33). The GSL expression pattern of human lymphocytes, monocytes, and granulocytes has also been shown to be cell specific (34). The cellular microenvironment and lysosomal proteins specific to myeloid lineage APC may also play an important role in the GSL bound and presented by mCD1d, as has been well-demonstrated for MHC class II-presented peptides (35). APC activation has previously been suggested to result in more stimulatory CD1d-bound lipids (17, 18).
We sought to define the pool of GSL bound by CD1d on myeloid APC at steady state and determine if these endogenous GSL ligands are edited or altered by APC activation or CD1 trafficking. Here, we present the development and use of a sensitive and specific method to isolate and identify the array of GSLs bound to murine CD1d expressed in APCs at steady state, with distinct CD1 trafficking routes, and under inflammatory conditions. These studies reveal the GSL repertoire that is bound by mCD1d, how that repertoire compared with the total pool of cellular GSL, and how it is modified by CD1 trafficking and APC activation.
Results
Murine Monocyte/Macrophage RAW 264.7 Cells Contain a Broad Range of GSL.
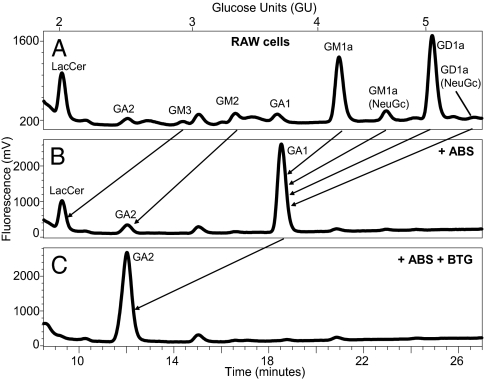
The choice of the murine monocyte/macrophage RAW cell line was based on similarities to APC that express CD1d in vivo and their ability to respond to inflammatory stimuli. First, the GSL profile for RAW cells at steady state was examined. RAW cells were directly extracted with increasingly polar solvent conditions. The isolated lipids were then digested with ceramide glycanase, and the released glycans were fluorescently labeled, allowing for the identification of the GSL head groups based on their HPLC retention times (Table 1) as well as for confirmatory studies with exoglycosidase digests. A schematic illustrating the glycan headgroup structures for each GSL is provided in Fig. S1. RAW cells express a broad array of GSL (Fig. 1A). The relative abundance of each GSL was calculated from the eluted peak areas. The most prominent GSL glycans identified were, in order of relative abundance, GD1a (44.2%), GM1a (36.3%), GM2 (5%), and GM1a (NeuGc; 4.5%) (Fig. 1A; Table 2). Confirmatory exoglycosidase experiments confirmed the identity of these peaks. The eluted GSL glycans were digested with Athrobacter ureafaciens sialidase (ABS) that cleaves terminal sialic acid residues. As expected, the peaks that corresponded to sialic acid containing glycan headgroups such as GM1a, GM2, and GM3 (one sialic acid residue) and GD1a (two sialic acid residues) were lost with a concomitant increase in the size of the peaks that correspond to the expected glycans that have lost these residues such GA1, GA2, and LacCer (Fig. 1B). Serial digests with ABS and bovine testes β-galactosidase (BTG) further confirmed the identified peaks with the complete loss of the GA1 and a marked increase in the GA2 peak (Fig. 1C). Additional digests with Jack bean β-N-acetyl-hexosaminidase and coffee bean α-galactosidase were also performed to confirm the identities of specific peaks.
Table 1.
GSL carbohydrate headgroup structures and NP-HPLC retention GU values
| GSL species | Carbohydrate headgroup structure | 2AA GU value |
| LacCer | Galβ1–4Glc | 2.04 |
| GA2 | GalNAcβ1–4Galβ1–4Glc | 2.57 |
| ceramide trihexoside (Gb3) | Galα1–4Galβ1–4Glc | 2.82 |
| isoglobohexosylceramide (iGb3) | Galα1–3Galβ1–4Glc | 2.83 |
| GM3 | NeuNAcα2–3Galβ1–4Glc | 2.93 |
| GM2 | GalNAcβ1–4(NeuNAcα2–3)Galβ1–4Glc | 3.29 |
| GA1 | Galβ1–3GalNAcβ1–4Galβ1–4Glc | 3.72 |
| GM1a | Galβ1–3GalNAcβ1–4(NeuNAcα2–3)Galβ1–4Glc | 4.12 |
| αGal GSL | Uncharacterized | 4.22 |
| GD1a | NeuNAcα2–3Galβ1–3GalNAcβ1–4(NeuNAcα2–3)Galβ1–4Glc | 4.96 |
Species with the hydroxylated analogs of N-acetylneuraminic acid (NeuNAc) and N-glycolylneuraminic acid (NeuGc) had an increase in GU value of 0.4 per sialic acid residue. 2AA, anthranilic acid.
Fig. 1.
GSL profile of RAW cells. Lipids were extracted from RAW cell pellets and characterized by labeling with 2AA and NP-HPLC analysis. RAW cellular GSL glycans were characterized (A) before and after digest with the exoglycosidases (B) ABS and (C) ABS + BTG. Arrows indicate the shift in retention time of various species after exoglycosidase digest. These are representative profiles from 15 (GSL glycan analysis) and 5 (exoglycosidase) sample sets.
Table 2.
Relative abundance of selected GSL in RAW cells and GSL bound to mCD1d-Fc expressed at steady state and in the setting of activation (LPS)
| GSL species |
Untreated RAW cells |
LPS-treated cells |
||
| Cells | mCD1d-Fc | Cells | mCD1d-Fc | |
| GA2 | 174 (2.4%) | 1,199 (18.1%) | 257 (9.3%) | 1,535 (12.9%) |
| GM3 | 100 (1.4%) | 140 (2.1%) | 94 (3.4%) | 1,265 (10.7%) |
| GM2 | 359 (5.0%) | 672 (10.2%) | 190 (6.8%) | 1,022 (8.6%) |
| GA1 | 69 (1.0%) | 0 (0%) | 39 (1.4%) | 223 (1.9%) |
| GM1a | 2,596 (36.3%) | 781 (11.9%) | 1,007 (36.3%) | 2,404 (20.3%) |
| αGal GSL | ND | 86 (1.3%) | ND | 556 (4.7%) |
| GM1a (NeuGc) | 320 (4.5%) | 45 (0.7%) | 75 (2.7%) | 596 (2.5%) |
| GD1a | 3,158 (44.2%) | 1,181 (17.9%) | 646 (23.3%) | 2,581 (21.7%) |
GSL species abundance was quantified by calculating peak areas (mV × seconds) in the NP-HPLC chromatograms of 2AA labeled glycans released from untreated or LPS-treated RAW cells or eluted from mCD1d-Fc expressed in these cells. Relative abundance for each GSL species is also shown as a percentage of total GSL (in brackets). ND, not detected.
GSL Repertoire Eluted from Soluble Murine CD1d.
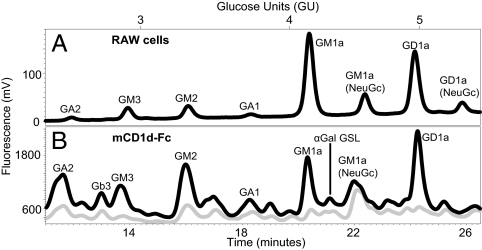
Next, the GSL bound to mCD1d fusion protein (mCD1d-Fc), which is secreted as a soluble CD1 molecule, were identified. Soluble mCD1d-Fc and MHC class I-Fc expressed by RAW cells were isolated from the culture supernatants using protein A beads that were washed and directly extracted with increasingly polar organic solvent conditions to isolate GSL. At no time were the beads or attached proteins exposed to harsh salt or acidic conditions. Control samples, including protein A beads and protein A beads exposed to FCS-containing culture media extracted with the same series of organic solvents, demonstrated the absence of GSL similar to any of the peaks observed in the mCD1d samples. The mCD1d-Fc and MHC class I-Fc eluted lipids were subsequently digested with ceramide glycanase, and the released glycans were fluorescently labeled to allow for their identification. The GSL glycan profile eluted from mCD1d-Fc produced in the RAW murine monocyte/macrophage cell line showed several glycan peaks such as GD1a (17.9%), GM1a (11.9%), and GM2 (10.2%), in order of relative abundance. Additional GSL glycans eluted included GM3, GA1, GM1a (NeuGc), and LacCer. A significant amount of GA2 was eluted in all sample sets, although the relative abundance was variable (Fig. 2B). These peaks were not observed in the ceramide glycanase-digested eluates of the MHC I-Fc control (Fig. 2B).
Fig. 2.
Identification of GSL eluted from soluble mCD1d-Fc expressed in RAW cells and comparison with total cellular RAW cell GSL. Lipids were extracted from (A) RAW cells or eluted from (B) mCD1d-Fc (black) or MHC class 1-Fc (gray) expressed in these cells. GSL glycan headgroups were characterized by labeling with 2AA and NP-HPLC analysis. Results are representative of six sample sets.
Comparison of mCD1d-Fc Eluted Lipids with RAW GSL Profile.
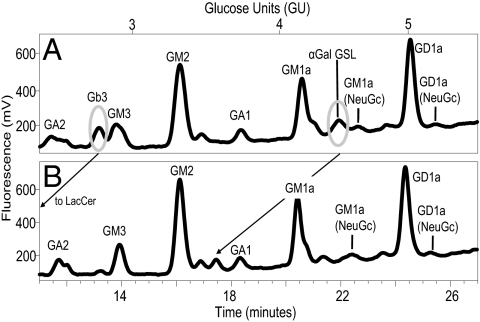
GD1a and GM1a, the most predominant RAW cell GSL, were eluted from soluble mCD1d-Fc but were not the most predominant GSL species eluted (Fig. 2). Soluble mCD1d preferentially bound GD1a, GM1a, GM2, and GA2 (Fig. 2B; Table 2). These findings suggest that the GSL bound to soluble mCD1d do not solely reflect the GSL content of the RAW cells. Instead, it reveals preferential binding of specific GSL to mCD1d. In addition, finer mapping of smaller peaks was also performed. GSL glycan analysis with this methodolgy is capable of identifying GSL glycans that are present in as low as 1% of the total sample (29). Given the importance of α-galactose (Gal)-expressing GSL as CD1d antigens, exoglycosidase digests were performed to identify the α-Gal-terminating GSL glycans bound to mCD1d (Fig. 3). These analyses also allow for differentiation between Gb3 and iGb3. We were able to confirm the presence of the Gb3 peak and the absence of iGb3 in the lipid eluates from soluble murine CD1d-Fc. The retention times for Gb3 and iGb3 were clearly distinct and confirmed with known standards (29). An additional α-galactose–bearing GSL glycan at 4.22 glucose units (GU) that will be referred to as α-Gal-GSL was also identified. This GSL glycan was not observed in the total RAW cell GSL glycan analysis and suggests preferential enrichment of this GSL by mCD1d (Table 2; Fig. 3A). However, the GU and retention times of the α-Gal-GSL peak and exoglycosidase digest peaks did not correspond directly to any GSL in our glycan database, and thus, its identity could not be determined.
Fig. 3.
Identification and confirmation of α-galactose–expressing GSL eluted from mCD1d-Fc. mCD1d-Fc eluted GSL glycans (A) before and (B) after digest with coffee bean α-galactosidase (cbag) were analyzed by NP-HPLC. Encircled peaks represent α-Gal–terminating glycan species that are lost after digest with cbag. Arrows indicate the shift in retention time of various species after exoglycosidase digest. Results are representative of three sample sets.
GSL Profile of Cell Surface-Cleavable mCD1d.
The ability of GSL to be loaded and exchanged on mCD1d is likely influenced by many factors including cellular localization, GSL availability, and accessory proteins, such as microsomal triglyceride transfer protein (MTP), saposins, and GM2 activator. Because soluble mCD1d-Fc is directly secreted from the cell, it would only have exposure to the GSL available in the ER and along the secretory routes. To analyze intact mCD1d that traffics through the lysosome and recycling pathways, a cytoplasmic tail (CT)-containing, cell surface-cleavable construct was developed. This cell surface-cleavable construct had the added advantage of not necessitating detergent cell lysis to release mCD1d.
The cell surface-cleavable mCD1d Tobacco Etch Virus (mCD-1d-TEV) CT construct was designed as a single chain construct with β2-microglobulin. The TEV cleavage sequence was inserted into the connecting peptide region of mCD1d so that the wild-type mCD1d transmembrane and cytoplasmic regions remained intact. Importantly, this construct was shown to traffic properly by confocal microscopy as demonstrated by marked colocalization with lysosomal-associated membrane protein-1 (LAMP-1), which was also observed for wild-type, full-length CD1d (Fig. S2). The proper intracellular trafficking and folding of mCD1d-TEV-CT molecules is further supported by the ability of RAW cell mCD1d-TEV-CT transfectants to present Gal(α1–2)galactosyl-ceramide (α-GalGalCer), which requires lysosomal processing to activate NKT cells (Fig. S3). These findings are consistent with a fully functional mCD1d-TEV-CT molecule that traffics through the ER, Golgi, secretory, recycling, and endocytic cellular networks.
RAW mCD1d wild-type and mCD1d-TEV-CT stable transfectants were incubated with TEV buffer. The amount of intact mCD1d-TEV cleaved was determined by ELISA. Intact, soluble mCD1d was captured and detected from the RAW mCD1d-TEV-CT transfectants. This was not observed with the full-length mCD1d RAW transfectants (Fig. S4). The cleaved mCD1d-TEV sample was passed over anti-CD1d mAb-conjugated beads or anti-MHC class I mAb-conjugated control beads; then, the beads were directly extracted, the total lipids were digested with ceramide glycanase, and the released glycans were labeled and analyzed as described above. The GSL profile for mCD1d-TEV showed clear GM1a and GD1a glycan peaks. However, the GM2 peak that was a prominent GSL glycan eluted from soluble mCD1d was not observed in the mCD1d-TEV eluates (Figs. 2A and 4; Table 2). These findings suggest that GM1a and GD1a are prominent mCD1d ligands in both the ER/secretory and lysosomal routes and that the GM2 acquired in the ER/secretory pathway may be exchanged or processed in the lysosomal microenvironment. Given that the amount of cleaved mCD1d TEV protein analyzed was not as abundant as in the soluble mCD1d-Fc studies, changes in the smaller, less-prominent GSL glycan peaks could not be fully assessed.
Fig. 4.
Characterization of cell surface-cleaved mCD1d-TEV eluted GSL glycans. mCD1d-TEV cleaved from RAW cell transfectants was isolated using anti-CD1d mAb (black) or control mAb (gray) conjugated beads. GSL eluted from mCD1d-TEV were characterized by labeling with 2AA and NP-HPLC analysis. Results are representative of two sample sets.
Impact of APC Activation on the mCD1d and Cellular GSL Repertoire.
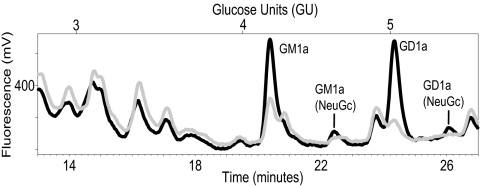
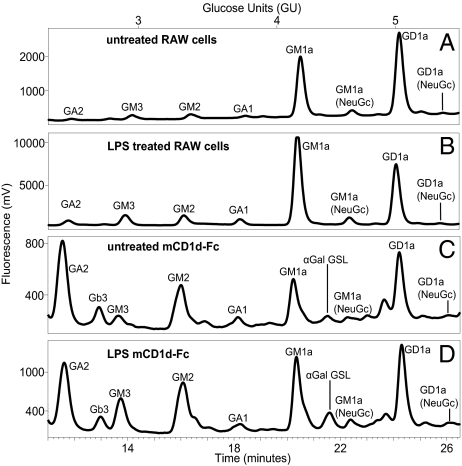
We next examined the impact of RAW cell activation on the GSL content and mCD1d GSL ligands. We first examined the impact of LPS on the GSL profile of RAW cells. On activation with LPS, there was a marked decrease in the GD1a content of the RAW cells (Fig. 5; Table 2). The relative abundance of GA2 increased 3-fold. In contrast, little to no change was observed for GM2, GA1, and the prominent GM1a peak (Fig. 5; Table 2). Next, the impact of RAW cell activation by LPS on lipids bound by soluble mCD1d was examined. There was an overall increase in the relative abundance of several of the GSL eluted. This was most notable for GM3, α-Gal GSL, and GM1a (NeuGc), which increased 5-, 3.6- and 3-fold, respectively, and it was also observed for GM1a. The relative abundance of GA2 bound to soluble mCD1d decreased (Fig. 5; Table 2). As was observed with analyses at steady state, the GSL species bound to soluble mCD1d in activated RAW cells do not mirror the GSL content of the activated total RAW cell GSL. In fact, the GSL that increased most prominently in the cellular GSL pool with LPS-mediated activation such as GA2 were less prominently represented in the eluates from soluble mCD1d from LPS-activated RAW cells compared with the GSL eluted from mCD1d-Fc generated by unstimulated RAW transfectants (Table 2; Fig. 5).
Fig. 5.
Profiles of cellular GSL and GSL eluted from soluble mCD1d-Fc isolated from untreated and LPS-activated RAW cells. GSL glycan headgroups from lipids extracted from (A) untreated or (B) LPS-treated RAW cells and from GSL eluted from mCD1d-Fc expressed in (C) untreated or (D) LPS-treated RAW cells were characterized by labeling with 2AA and NP-HPLC analysis. Profiles are representative of five sample sets.
Discussion
This study describes the range of GSL bound to mCD1d in a myeloid lineage APC using a well-characterized and sensitive method. The number and diversity of GSL eluted from mCD1d is significantly broader than the GSL identified in prior studies that to date have only identified GM3 (30). In fact, despite the well-documented importance of GSL as CD1 antigens, they have not been well-characterized or identified in prior elution studies (20, 30, 31) that used B cell transfectants, different lipid-identification methods, CD1 protein purification and elution conditions. Given the significant differences that have been described in GSL content for different leukocyte subsets, these myeloid APC are likely more reflective of myeloid DC, which are a major subset of APC that expresses CD1 molecules in vivo.
Soluble mCD1d was observed to bind a broad array of GSL that did not solely reflect the GSL content of the RAW cells. This suggests that soluble mCD1d molecules that survey the ER/secretory routes preferentially bind specific GSL, such as GM2 and GA2, in addition to GM1a and GD1a, which are the most abundant cellular GSL in RAW cells. In addition, secreted soluble CD1d significantly bound a unique GSL with an α-Gal epitope. The subcellular distribution of GSL and contributions of lipid-transfer proteins may be important factors that enhance the binding of specific GSL to mCD1d. Prior studies have suggested that the lipid classes bound to human CD1d are altered by their trafficking patterns. However, GSL as a class were not identified or isolated in those studies (31). Using cell surface-cleaved mCD1d-TEV-CT that, in addition to being assembled in the ER, also surveys the lysosomal routes, we found that GM1a and GD1a were the most abundant GSL bound. The GM2 glycan peak eluted from soluble mCD1d was not identified as bound to mCD1d-TEV-CT, suggesting that GM2 and possibly other less highly expressed GSL may be edited or exchanged in the lysosomal microenvironment. The contribution of lysosomal lipid-transfer proteins, such as saposin and GM2 activator protein that have been shown to facilitate the binding of α-GalCer to mCD1d (36), may also play a critical role in the determining the endogenous GSL bound to mCD1d.
This work highlights that TLR activation of myeloid APC has a marked effect on the GSL repertoire of cells, and it revealed that the GSL eluted from all forms of CD1d did not mirror endogenous GSL or the marked changes to the cellular GSL pool after LPS activation. Instead, there were specific changes to the GSL bound to mCD1d on LPS activation. For example, although the most dramatic change to the RAW cell GSL pool was an almost 50% decrease in GD1a, the relative abundance of GD1a eluted from soluble mCD1d expressed by RAW transfectants stimulated with LPS was essentially unchanged (Table 2). In contrast, the relative expression of GM1a in the cellular GSL pool was unchanged with LPS stimulation, but it increased in the eluates of mCD1d-Fc expressed in LPS-stimulated RAW transfectants. These findings highlight the dichotomy between cellular GSL expression and the GSL bound to mCD1d. Changes to total cellular GSL content, subcellular GSL localization, lipid transfer, and GSL synthetic enzyme activity may all be contributors to alterations to the repertoire of endogenous GSL bound to CD1d in activated APC.
Although the focus of this report is mCD1d, modification of the endogenous GSL repertoire by activation and changes observed in GSL loading and editing for molecules that traffic through the lysosome are likely important for other CD1 molecules. These findings may be relevant in the case of human CD1b where the lysosomal microenvironment has been shown to be important in facilitating the loading of lipids (37) and possibly, with specific endogenous GSL. In addition, CD1e has been shown to play important roles in CD1b antigen loading and to rapidly redistribute from the Golgi network to the lysosome in myeloid DC matured by LPS (38); additionally, CD1e could play a role in selecting the endogenous GSL that are bound to CD1b in both the steady state and in the setting of myeloid DC activation.
This report does not address the phospholipids, sphingolipids, or lipopeptides that are bound to mCD1d. In addition, our methods do not identify sulfatide or GSL with glycan head groups smaller than lactose. The GSL that were identified in this study may only partially overlap with the GSL bound to mCD1d in other APC, which may have a different or more limited GSL repertoire as discussed above. It is interesting to speculate that the differences observed in the autoreactivity of NKT cells to specialized murine tissues, such as the spleen and thymus, may reflect differences in the endogenous pool of GSL or other lipid classes (39).
These analyses describe the repertoire of GSL bound to mCD1d and show that the endogenous GSL bound can be altered by intracellular trafficking, cellular GSL content, and changes to the activation state of APC. The GSL identified define the main GSL antigen complexes available for TCR recognition. This may be compared with the repertoire of peptides bound by MHC molecules, which include hundreds of distinct peptides containing common anchor residues specific for each MHC allele (40). In contrast, the repertoire of bound GSL reveals that a limited number of specific GSL dominate and that the pattern results from CD1d preferentially selecting only certain species. Further, because polymorphisms of CD1d molecules are very limited, this repertoire of antigens is likely to be very similar among individual animals or humans. Together, changes in GSL along with inflammatory cytokines work together to elicit NKT cell activation. These studies describe this CD1d GSL repertoire and provide a window into its changes under physiological conditions that alter T cell activation.
Materials and Methods
Soluble mCD1d-Fc and Cell Surface-Cleavable mCD1d Constructs.
The generation of the soluble, single-chain β2m-mCD1d-mouse IgG2a constant fragment mCD1d-Fc (21) has been previously described. In the case of the MHC class I Fc control, the extracellular domain of mCD1d was replaced by the extracellular domain of MHC class I. The extracellular domain of MHC class I was cloned using full-length HLA-B27 cDNA in pSRα-neo, generously provided by Masahiko Sugita (Kyoto University, Kyoto, Japan), as a template. A single-chain, β2m-mCD1d cell surface-cleavable construct was generated by inserting the seven amino acid protease cleavage sequence for TEV (Glu-Asn-Leu-Tyr-Phe-Gln-Gly) into the connecting peptide sequence of mCD1d with minimal disruption of the wild-type mCD1d sequence and without changing the number of amino acid residues in the connecting peptide segment (..ILYWDARQAPVG.. was changed to ..ILYWDENLYFQG..). This generated a single-chain β2m-mCD1d TEV (mCD1d-TEV-CT) construct with β2m covalently linked to the extracellular domain of mCD1d followed by the connecting peptide region containing the TEV cleavage site and the complete wild-type mCD1d transmembrane region and CT. All PCR primer sequences used to generate the constructs are available on request.
Cell Lines and Transfectants.
The murine myeloid lineage monocyte/macrophage RAW 264.7 and HeLa cell lines were obtained from the ATCC. The mCD1d-Fc, MHC class I Fc, wild-type mCD1d full-length and mCD1d-TEV constructs were transfected into RAW cells by electroporation, and stable clones were isolated by limiting dilution under selection with G418 sulfate (Invitrogen). The Fc constructs were screened by ELISA as described below. The mCD1d-TEV constructs were screened by cell-surface staining and flow cytometry. For the confocal microscopy experiments, full-length, wild-type mCD1d and mCD1d-TEV constructs were transiently transfected into HeLa cells with FuGENE 6 (Roche). When the soluble Fc transfectants were propagated for the production of soluble Fc, they were switched to complete media in which the standard FCS was replaced with 10% Ultra Low IgG FCS (Invitrogen). For the LPS-stimulation experiments, identical numbers of RAW cells and RAW transfectants were incubated in media or media containing LPS at 25 μg/mL (Escherichia coli; Sigma) for 48 h.
Affinity Purification and Lipid Extraction.
The soluble fusion proteins were purified from equal volumes of culture supernatants by affinity chromatography using recombinant rmp protein A Sepharose Fast Flow agarose (GE Healthcare). The cell surface-cleaved mCD1d-TEV protein was purified by affinity chromatography using anti-mCD1d monoclonal antibody mAb (19G11; provided by Albert Bendelac, Chicago, IL) or control anti-MHC class I (W6/32) mAb (ATCC) coupled to agarose by the Aminolink Plus immobilization kit (Thermo Scientific). The soluble Fc bound to protein A Sepharose and the cleaved mCD1d TEV proteins bound to anti-mCD1d or control MHC class I mAb-conjugated beads were washed with at least 20 volumes of PBS and drained before their direct extraction with chloroform/methanol (1:1 vol/vol) for 3 h at room temperature. The beads were pelleted by centrifugation, and the supernatants were aspirated and stored. The beads were then incubated in chloroform/methanol (1:2 vol/vol). The final extraction was performed with chloroform/methanol/water (4.8:3.5:1 vol/vol/v). The extractions were pooled and stored at −80 °C pending analysis. All of the solvents, including the water, were HPLC grade. Analysis of total cellular GSL was performed on the lipids extracted from RAW or LPS-treated RAW cell pellets using the same solvent protocol described above.
GSL Glycan Isolation, Fluorescent Labeling, and HPLC Analysis.
The GSL glycan analyses were carried out as previously described (41). Briefly, lipids were purified from nonhydrophobic contaminants by passing the eluted mCD1d or MHC class I organic extracts over a SepPak C18 column solid-phase extraction column. The glycan headgroups of the GSL were cleaved from the ceramide backbone using ceramide glycanase (Macrobdella decora ceramide glycanase; Europa Bioproducts) or purified from Hirudo medicinalis (D. Neville and T. Butters). The released glycans were labeled with the fluorescent marker anthranilic acid (2AA) and were analyzed by normal phase-HPLC (NP-HPLC) as described in ref. 41. Retention times of different species were converted to GU values, which were determined with reference to an external standard, a ladder of glucose oligomers obtained from a partial hydrolysate of dextran resolved with each sample set. The number of glucose residues in each dextran peak was plotted against the retention times of each of the peaks to obtain a standard curve using a fifth-order polynomial line fit. Preliminary identification of GSL glycans was done by comparison of GU values from published database of known standards (41). This method was found to be both sensitive and specific with standard deviations from the GU values listed in Table 1 in the range of 0.006–0.021 (41). Glycan identities were confirmed after digest with various exoglycosidases. A digest blank control was analyzed with each sample. Each lipid sample was split into two equal aliquots. One aliquot was treated with ceramide glycanase and fluorescently labeled before HPLC analysis. The other aliquot, designated digest blank, was not treated with ceramide glycanase before fluorescent labeling and HPLC analysis. This digest blank contained only non GSL-derived sugars. The chromatogram peak areas of various species were normalized by subtracting the digest-blank peak area to distinguish GSL and non–GSL-derived sugars.
Exoglycosidase Digests.
Additional characterization of the fluorescently labeled glycans was performed by exoglycosidase digestion. The fluorescently labeled glycans were dried down and resuspended in enzyme digest buffer (sodium acetate 100 mM at pH 6) and one or more of the following: Athrobacter ureafaciens sialidase (ABS; Roche), bovine testes β-galactosidase (BTG; Prozyme), Jack bean β-N-acetyl-hexosaminidase (JBH; Prozyme), or coffee-bean α-galactosidase (Boehringer). The samples were incubated overnight at 37 °C. Proteins were removed by centrifugation through a 10-KDa molecular weight cut-off filter before HPLC analysis.
Cleavage of Cell-Surface mCD1d-TEV.
Full-length, wild-type mCD1d and cell surface-cleavable mCD1d-TEV-CT RAW transfectants were detached by brief incubation with 0.5% trypsin-EDTA, washed with PBS, and incubated in TEV buffer (50 mM Tris at pH 8.0, 0.5 mM EDTA, and 0.01% Triton X-100) for 2 h at 4 °C. AcTEV protease (Invitrogen), an enhanced form of TEV protease, was added as per manufacturer’s instructions in several of the assays. After incubation, the cells were pelleted, and the supernatants frozen at −20 °C. Equal amounts of the supernatants were passed over anti-mCD1d or MHC class I control mAb conjugated agarose beads as described above.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants AI028973 and AI063428 (to M.B.B.). G.S.B. acknowledges support as a Personal Research Chair from Mr. James Badrick (Royal Society Wolfson Research Merit Award) and a former Lister Institute-Jenner Research Fellow as well as from the Medical Research Council and The Wellcome Trust (084923/B/08/7). K.M., D.C.A.N, and T.D.B acknowledge support from the Oxford Glycobiology Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915056107/DCSupplemental.
References
- 1.Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol. 2003;4:1230–1237. doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- 2.Kinjo Y, Kronenberg M. V alpha14 i NKT cells are innate lymphocytes that participate in the immune response to diverse microbes. J Clin Immunol. 2005;25:522–533. doi: 10.1007/s10875-005-8064-5. [DOI] [PubMed] [Google Scholar]
- 3.Brigl M, Brenner MB. CD1: Antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 4.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novak J, Griseri T, Beaudoin L, Lehuen A. Regulation of type 1 diabetes by NKT cells. Int Rev Immunol. 2007;26:49–72. doi: 10.1080/08830180601070229. [DOI] [PubMed] [Google Scholar]
- 6.Terabe M, Berzofsky JA. NKT cells in immunoregulation of tumor immunity: A new immunoregulatory axis. Trends Immunol. 2007;28:491–496. doi: 10.1016/j.it.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Shamshiev A, et al. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Wu L, Van Kaer L. Natural killer T cells and autoimmune disease. Curr Mol Med. 2009;9:4–14. doi: 10.2174/156652409787314534. [DOI] [PubMed] [Google Scholar]
- 9.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 10.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 11.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 12.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 13.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 15.Mallevaey T, et al. Activation of invariant NKT cells by the helminth parasite schistosoma mansoni. J Immunol. 2006;176:2476–2485. doi: 10.4049/jimmunol.176.4.2476. [DOI] [PubMed] [Google Scholar]
- 16.Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol. 2007;178:2706–2713. doi: 10.4049/jimmunol.178.5.2706. [DOI] [PubMed] [Google Scholar]
- 17.Paget C, et al. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity. 2007;27:597–609. doi: 10.1016/j.immuni.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Salio M, et al. Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc Natl Acad Sci USA. 2007;104:20490–20495. doi: 10.1073/pnas.0710145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Libero G, et al. Bacterial infections promote T cell recognition of self-glycolipids. Immunity. 2005;22:763–772. doi: 10.1016/j.immuni.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Joyce S, et al. Natural ligand of mouse CD1d1: Cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 21.Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 22.Fox LM, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shamshiev A, et al. The alphabeta T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity. 2000;13:255–264. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 24.Wu D, et al. Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc Natl Acad Sci USA. 2005;102:1351–1356. doi: 10.1073/pnas.0408696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamshiev A, et al. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahng A, et al. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 28.Porubsky S, et al. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci USA. 2007;104:5977–5982. doi: 10.1073/pnas.0611139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speak AO, et al. Implications for invariant natural killer T cell ligands due to the restricted presence of isoglobotrihexosylceramide in mammals. Proc Natl Acad Sci USA. 2007;104:5971–5976. doi: 10.1073/pnas.0607285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox D, et al. Determination of cellular lipids bound to human CD1d molecules. PLoS One. 2009;4:e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiels J, Mangeney M, Tétaud C, Tursz T. Sequential shifts in the three major glycosphingolipid series are associated with B cell differentiation. Int Immunol. 1991;3:1289–1300. doi: 10.1093/intimm/3.12.1289. [DOI] [PubMed] [Google Scholar]
- 33.Taga S, Tétaud C, Mangeney M, Tursz T, Wiels J. Sequential changes in glycolipid expression during human B cell differentiation: Enzymatic bases. Biochim Biophys Acta. 1995;1254:56–65. doi: 10.1016/0005-2760(94)00167-w. [DOI] [PubMed] [Google Scholar]
- 34.Kiguchi K, Henning-Chubb CB, Huberman E. Glycosphingolipid patterns of peripheral blood lymphocytes, monocytes, and granulocytes are cell specific. J Biochem. 1990;107:8–14. doi: 10.1093/oxfordjournals.jbchem.a123016. [DOI] [PubMed] [Google Scholar]
- 35.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307:1630–1634. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 36.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 38.Angénieux C, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 39.Park SH, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- 40.Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 41.Neville DC, et al. Analysis of fluorescently labeled glycosphingolipid-derived oligosaccharides following ceramide glycanase digestion and anthranilic acid labeling. Anal Biochem. 2004;331:275–282. doi: 10.1016/j.ab.2004.03.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.