Abstract
Corepressors play an essential role in nuclear receptor-mediated transcriptional repression. In general, corepressors directly bind to nuclear receptors via CoRNR boxes (L/I-X-X-I/V-I) in the absence of ligand and appear to act as scaffolds to further recruit chromatin remodeling complexes to specific target genes. Here, we describe the identification of the multiple LIM domain protein Ajuba as a unique corepressor for a subset of nuclear hormone receptors. Ajuba contains functional nuclear-receptor interacting motifs and selectively interacts with retinoic acid receptors (RARs) and rexinoid receptor (RXRs) subtypes in a ligand-dependent manner. Simultaneous mutation of these motifs abolishes RAR binding and concomitantly leads to loss of repression on RARE reporter genes. P19 cells depleted of Ajuba are highly sensitized to all-trans retinoic acid (atRA)-induced transcription and differentiation. In the absence of atRA, Ajuba can be readily found at the RARE control elements of RAR endogenous target genes. Stimulation of cells with atRA results in the dissociation of Ajuba from these regions. Moreover, we observed that coexpression of the known Ajuba binding partner Prmt5 (protein arginine methyltransferase-5) inhibited the Ajuba/RAR interaction. The high-affinity Ajuba-RAR/RXR interaction site overlaps the region responsible for Ajuba/Prmt5 binding, and thus binding appears to be mutually exclusive, providing a potential mechanism for these observations. Identification of Ajuba as a unique corepressor for nuclear receptors sheds new light on mechanisms for nuclear receptor-mediated repression and provides a unique target for developing more effective therapeutics to modulate this important pathway.
Keywords: hox genes, differentiation, LIM domain, retinoic acid receptors
Ajuba belongs to the Ajuba/Zyxin family of LIM proteins, which includes Ajuba, Limd1, WTIP, Zyxin, LPP, and Trip6. This family is characterized by two or three tandem C-terminal LIM domains and a unique, N-terminal preLIM region (1). This family of proteins function as scaffolds, participating in the assembly of numerous protein complexes. Consistent with this activity, Ajuba binds Grb2 to modulate serum-stimulated ERK activation (1). Ajuba also recruits the TNF receptor-associated factor 6 (TRAF6) to p62 and activates PKCζ activity (2). Ajuba interacts with α-catenin and F-actin to contribute to the formation or stabilization of adheren junctions by linking adhesive receptors to the actin cytoskeleton (3). Ajuba forms a complex with the p130Cas/Dock180 Rac guanine nucleotide-exchange factor to regulate Rac activity (4). Lastly, Ajuba binds the Aurora A kinase to regulate mitosis (5).
We recently discovered a nuclear function for Ajuba by showing that it functions as a corepressor for the zinc finger-protein Snail (6 –8). We showed that Ajuba binds to the SNAG repression domain of Snail through its LIM region. Furthermore, we identified protein arginine methyltransferase-5 (Prmt5) as an effector recruited to Snail through an interaction with Ajuba. This ternary complex functions to repress E-cadherin, a well-characterized Snail target gene. Snail, Ajuba, and Prmt5 can be found at the proximal promoter region of the E-cadherin gene concomitant with increased arginine methylation of histones at this locus (7).
In an effort to further define the Ajuba signaling system, we scanned the aminio acid sequence of the Ajuba protein for potential protein–protein interaction motifs and discovered a number of conserved NR boxes (L-X-X-L-L) and CoRNR boxes (L/I-X-X-I/V-I) within Ajuba, suggesting that Ajuba may be a potential nuclear-receptor interacting protein (9). Previous studies demonstrated that Ajuba plays a significant role in the regulation of mouse embryonic carcinoma P19 cell proliferation and differentiation, a process highly dependent upon retinoids (10). In cell culture, the addition of all-trans retinoic acid (atRA) to P19 cells results in growth inhibition and differentiation. At low doses of atRA (10–20 nM), P19 cells differentiate from an ectodermal phenotype into endodermal-like cells, whereas higher concentrations of atRA (100–200 nM) induce terminal neuroectodermal differentiation (11 –13). Collectively, these observations prompted us to hypothesize that Ajuba may interact with retinoic acid receptors to regulate retinoic acid signaling.
Here, we demonstrate that Ajuba selectively interacts with RA receptor subtypes via conserved nuclear-receptor binding motifs and functions as a corepressor to negatively regulate RA signaling.
Results
Ajuba Contains Putative Nuclear-Receptor Binding Motifs and Interacts with RARα.
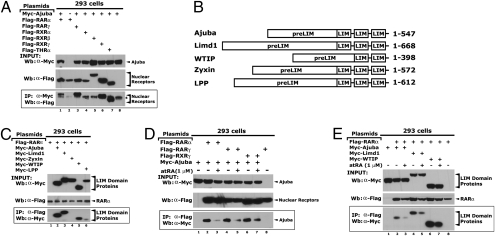
A comprehensive bioinformatic analysis of the Ajuba amino acid sequence demonstrated that Ajuba contains four putative nuclear-receptor binding motifs (NR box or CoRNR box) designated as NR1 to NR4 (Fig. 1A). NR1 and NR2 are located in the preLIM region. NR3 and NR4 are located in the LIM2 and LIM3 domains, respectively. NR1 is not conserved among species, while NR2, -3, and -4 are well conserved (see Fig. 1A). Previously, we have shown that NR2 is necessary for Prmt5 binding (7). Because retinoic acid receptor alpha (RARα) is the dominant RA receptor that mediates RA function in P19 cells (13), we speculated that Ajuba may physically interact with RARα. Subsequent testing using coimmunoprecipitation (Co-IP) assays revealed that Ajuba (Myc-tagged) interacts robustly with RARα (Flag-tagged) when transiently expressed in 293 cells (Fig. 1B). To verify an interaction between endogenous proteins, nuclear extracts were prepared from P19 cells +/− atRA (1 μM) for 8 h and RARα was immunoprecipitated from the extracts and followed by Western blotting to detect endogenous Ajuba. Consistent with the transfection results, a complex between endogenous Ajuba and RARα was readily detected in P19 cells. This interaction was inhibited by atRA (Fig. 1C).
Fig. 1.
Ajuba interacts with RARα. (A) Ajuba contains conserved nuclear-receptor binding motifs. Ajuba is characterized by the C-terminal tandem LIM domains and the glycine-proline rich N-terminal preLIM regions. The potential nuclear-receptor motifs are designated as NR1 to NR4. The second motif, NR2, serves as the Prmt5 binding site. The numbers correspond to the locations of the amino acid residues within the Ajuba protein. (B) Ajuba interacts with RARα when coexpressed in 293 cells. Myc-Ajuba and Flag-RARα were transiently expressed in 293 cells. Whole-cell lysates were immunoprecipitated with α-Myc antibody and Western blot was performed with α-Flag antibody. *, nonspecific band. (C) The endogenous Ajuba and RARα proteins interact in P19 cells. Nuclear extracts were prepared from exponentially growing P19 cells treated with/without atRA (1μM) for 8 h and were precleared with protein A/G beads. Coimmunoprecipitation assays were carried out with α-RARα antibody, and rabbit IgG was used a control. Western blot was performed with an α-Ajuba antibody. (D) In vitro interaction of Ajuba and RARα. Bacterially expressed and purified full-length GST-RARα and in vitro-translated full-length Ajuba, which was labeled with S35, were used for in vitro binding assays. atRA were added at concentration of 1 μM, 10 μM, or 100 μM into the reactions and incubated for 1 h.
To determine if Ajuba directly binds RARα, we used full-length GST-RARα recombinant protein and in vitro-translated full-length Ajuba for in vitro binding assays; Ajuba bound to GST-RARα avidly and this interaction was inhibited by atRA (Fig. 1D). Collectively, these data demonstrate a ligand-modulated binding of RARα and Ajuba in vitro and in vivo.
Ajuba Selectively Interacts with RAR and Rexinoid Receptor Subtypes.
To examine whether Ajuba can also interact with other nuclear receptors that transduce RA signaling, we coexpressed Myc-Ajuba with Flag-RARγ, rexinoid receptor (RXR)α, RXRβ, RXRγ, or thyroid hormone receptor (THR)α in 293 cells and co-IP assays were performed. We observed that in addition to RARα, Ajuba also interacted with RARγ, RXRγ, and THRα, but not with RXRα and RXRβ (Fig. 2A). Similarly, we performed co-IP assays to examine the interaction of RARα with other members of the Ajuba/Zyxin family (Fig. 2B); RARα bound to Limd1 and WTIP but not to Zyxin or LPP (Fig. 2C). These data suggest that Ajuba may broadly interact with nuclear-receptor family members and that the LIM domain proteins Limd1 and WTIP may also have the capacity to associate with nuclear receptors.
Fig. 2.
Ajuba selectively interacts with nuclear receptor subtypes. (A) Ajuba interacts with RARα, RARγ, and RXRγ when coexpressed in 293 cells. *, nonspecific band. (B) Diagram shows the Ajuba/Zyxin family members. (C) RARα interacts with Ajuba, Limd1, and WTIP. (D) The interaction between Ajuba and RARα, RARγ, and RXRγ were attenuated by atRA treatment. (E) The interaction between RARα and Ajuba, Limd1, and WTIP were also attenuated by atRA. Plasmids were transiently transfected into 293 cells. Transfected cells were treated with atRA at 1 μM for 24 h before harvest.
atRA Dissociates Ajuba from RARα, RARγ, and RXRγ.
The canonical nuclear receptor corepressors, SMRT and N-CoR, interact with nuclear receptors in a ligand-dependent manner (14). We have shown that the interaction of Ajuba and RARα is inhibited by atRA. To determine if ligand dependence extends to other RAR or RXR subtypes, Ajuba and RARα, RARγ, or RXRγ were coexpressed in 293 cells. Transfected cells were treated with either atRA (1 μM) or DMSO for 24 h and whole-cell extracts were prepared. Immunoprecipitation using α-Myc antibody from the resulting extracts showed that atRA treatment of cells dissociated Ajuba from RARα, RARγ, and RXRγ (Fig. 2D). Using this methodology, we also examined the effect of atRA on the interactions between Limd1, WTIP, and RARα. Similarly, atRA dissociated Limd1 and WTIP from RARα (Fig. 2E). Together, these data suggest that Ajuba behaves like other known corepressors for nuclear receptors with respect to ligand dependence.
Conserved Nuclear-Receptor Binding Motifs in Ajuba Are Essential for its Interaction with RARα.
To examine whether the putative nuclear receptor interacting motifs in Ajuba are essential for RARα binding, we mutated these NR motifs individually or in combination (Fig. 3A). Wild-type and mutated Ajuba constructs and RARα were transiently expressed in 293 cells and co-IP assays were performed. Mutation of the putative NR motifs individually or in the combinations of NR 2-3, NR 2-4, or NR 3-4 showed little effect on RARα binding (Fig. 3B). Strikingly, simultaneous mutation of NR2, 3, and 4 abolished the RARα binding (see Fig. 3B, Lane 11). These data suggest that NR1, which is not evolutionarily conserved, does not contribute significance to RARα binding, whereas NR2, -3, and -4 are necessary and potentially cooperate in recognizing RARα.
Fig. 3.
The putative nuclear receptor binding motifs are necessary for RARα binding. (A) A diagram shows the Ajuba mutants. Ajuba mutants were generated from full-length Ajuba cDNA tagged with 6× Myc epitopes at the N terminus using a site-directed mutagenesis method. (B) Simultaneous mutations of the nuclear receptor binding motifs NR2, -3, and -4 abolish its interaction with RARα. Plasmids were transiently transfected into 293 cells, and co-IP assays were performed using an α−Flag antibody. Western blot was performed using an α-Myc antibody. (C) Colocalization of Ajuba and RARα in P19 cells. Cells were treated with atRA at 1 μM for 2 h, fixed, and immunofluorecent assays were performed. The images were taken using confocal microscopy.
Colocalization of Ajuba and RARα in P19 Cells.
To determine the subcellular localization of the endogenous Ajuba and RARα proteins, we performed indirect immunofluorescence assays in P19 cells. The cells were treated with either atRA (1 μM) or DMSO for 2 h, and the Ajuba protein was detected by immunofluorescence and visualized by confocal microscopy. We observed that Ajuba and RARα were present in both the cytoplasm and nucleus, and were colocalized (Fig. 3C, Top). Even though the subcellular localization of RARα did not change in response to atRA treatment, the Ajuba protein accumulated in the cytoplasm (Fig. 3C, Bottom). Together, these images demonstate that in the absence of atRA, the Ajuba and RARα proteins colocalize and that atRA can disrupt this colocalization.
Ajuba Represses the Activity of a RA-Responsive Element Luciferase Reporter.
A luciferase reporter gene controlled by multiple copies of the DR5 retinoid acid-responsive element (RARE), together with plasmids encoding wild-type Ajuba, the Ajuba mutant NR 2-3-4 (see Fig. 3A), or an empty vector, were transiently transfected into P19 cells. Expression of wild-type Ajuba repressed the ability of RARα to activate the reporter in a dose-dependent manner (Fig. 4A). Conversely, the mutant NR 2-3-4, which does not interact with RARα, had no effect on DR5-Luc reporter activity (Fig. 4B). Limd1 and WTIP were also able to repress DR5-Luc activity, while Zyxin, which does not bind RARs, showed no repression (Fig. 4C). Collectively, these data indicate that Ajuba negatively regulates RAR-mediated transcription.
Fig. 4.
Ajuba negatively regulates the RA target gene in P19 cells. (A) Ajuba represses the activity of a RARE-driven promoter. The luciferase reporter construct driven by RAREs (DR5-Luc) and Ajuba were cotransfected into P19 cells and the resulting luciferase activity was normalized to β-galactosidase activity. (B) The Ajuba mutant failed to interact with RARα and loses repression to DR5-luc. (C) Similar to Ajuba, Limd1 and WTIP repress DR5-Luc activity. (D) Western blot shows shRNA-mediated depletion of Ajuba protein in P19 cells. P19 cells were infected with viral vectors containing shRNAs targeting murine Ajuba or luciferase genes and the stable cells were established using puromycin. (E) Dose-response of P19 cells to atRA. Exponentially growing cells were treated with atRA at different doses in media containing 10% FBS for 24 h and total mRNA was extracted. The expression of the RA target genes was determined using RT-PCR. The PCR-amplification for Hox genes is 30 cycles, and 24 cycles for GAPDH. (F) PCR analysis of the immunoprecipitated DNA fragments from P19 cells. ChIP assays were performed in the P19-siLuc and P19-siAjuba cells treated with atRA at 1 μM or DMSO for 8 h. The primers flanking the RAREs in Hoxa1 and Hoxb1 were used for PCR amplifications.
Ajuba Binds to the RAREs of Endogenous RA Target Genes and Negatively Regulates Their Expression in P19 Cells.
To further determine the role of Ajuba in the regulation of the RARα-mediated transcriptional regulation, we employed an RNAi knockdown approach to deplete Ajuba in P19 cells. A short interfering RNA (shRNA) targeting murine Ajuba was introduced into P19 cells via lentiviral infection. A lentivirus encoding an shRNA targeting luciferase was used as a control. We selected the cells with puromycin to establish stable cell pools and carried out Western blot assays to confirm the efficacy of the knockdown. The endogenous Ajuba protein was efficiently decreased in these mass pools (Fig. 4D). We stimulated these cells with atRA for 24 h and well-defined RARα target genes were examined using semiquantitative RT-PCR. Treatment of cells with atRA induced the expression of hoxa1, hoxa4, hoxb1, and hoxb4 in both cell pools but with different magnitudes: At all of the chosen doses, atRA induced stronger RA target gene expression in the P19-siAjuba cells than that in the P19-siLuc cells, with the most striking difference seen in the hoxb1 gene (Fig. 4E). Notably, the kinetics of hox gene expression resulting from atRA stimulation was not altered in the absence of Ajuba.
To examine whether Ajuba directly occupies the chromatin of RA target genes in living cells, we applied a ChIP analysis to the endogenous RA target genes hoxa1 and hoxb1. ChIP assays were performed in P19-siAjuba cells and P19-siLuc cells treated with either atRA (1 μM) or DMSO for 8 h using antibodies specific to Ajuba and SMRT. Rabbit IgG was used as negative control. The immunoprecipitated DNA fragments were examined by PCR amplification using primer sets flanking the well-defined RAREs in the hoxa1 and hoxb1 loci. DNA fragments generated by ChIP with Ajuba, SMRT, or RARa antibodies were highly enriched for hoxa1 and hoxb1 RAREs in untreated P19-siLuc cells (Fig. 4F). However, enrichment was not observed in the P19-siLuc cells treated with atRA (see Fig. 4F). In the P19-siAjuba cells, the antibody to Ajuba failed to immunoprecipitate these DNA fragments from either hoxa1 or hoxb1 genes. Depletion of Ajuba in P19 cells slightly decreased RARα binding to the RARE of the hoxa1 gene, while occpancy of hoxb1 was unaffected (see Fig. 4F). Interestingly, we observed that RARα was dissociated from RAREs of hoxa1 and hoxb1 genes in the P19-siLuc cells treated with atRA, suggesting a receptor shift may take place in these loci. Taken together, these data strongly suggest that endogenous Ajuba occupies RAREs in bona fide RA target genes.
P19-siAjuba Cells Are More Sensitized to atRA-Induced Differentiation.
TROMA-1 (also known as cytokeratin A) is an intermediate filament protein and a well-established marker for early differentiation of embryonic stem cells (15). Retinoic acid triggers F9 and P19 embryonic stem cells to differentiate into endoderm-like cells and induces the synthesis of TROMA-1 (10, 16). In cell culture, low doses of atRA induce P19 cells to differentiate along the endodermal lineage, whereas at higher concentrations atRA induces terminal neuroectodermal differentiation (16). To examine the role of Ajuba in the regulation of atRA-induced differentiation in these lineages, we used the P19-siAjuba and the P19-siLuc cells described above. As expected, atRA treatment at either 10 or 50 nM induced profound morphological changes in these cells (Fig. 5A). Analysis of TROMA-1 induction via immunofluorescence and Western blot revealed a dramatically higher level of TROMA-1 expression in the P19-siAjuba cells compared to the P19-siLuc cells (Fig. 5 B and C). We next determined the time course for atRA to induce differentiation in the P19-siLuc and -siAjuba cells. We treated these cells with atRA at 50 nM. The expression of TROMA-1 could be detected as early as day 2, and peaked on day 5 in both cell lines. Strikingly, no TROMA-1 expression was detected after day 6 (Fig. 5D). Collectively, these data suggest that decreasing the level of Ajuba protein in P19 cells sensitizes them to atRA stimulation but does not affect the timing of their responsiveness to atRA.
Fig. 5.
Modulation of Ajuba in P19 cells results in the cells sensitized to atRA-induced differentiation. (A) atRA induces differentiation in P19-siLuc and P19-siAjuba cells. (B) Immunofluorecent assays show the expression of TROMA-1 in P19 cells. (C) Western blot shows the induction of TROMA-1 in P19 cells in the presence or absence of Ajuba. (D) Temporal expression of TROMA-1 in P19 cell variants stimulated with atRA.
Prmt5 Inhibits the Interaction Between Ajuba and RARα.
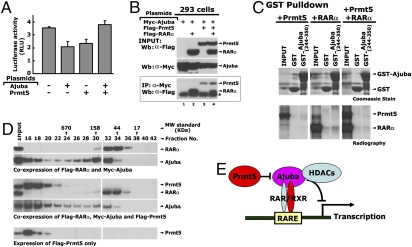
We have recently shown that Ajuba recruits the arginine methyltransferase Prmt5 to the Snail transcription factor to repress Snail target genes (7). This prompted us to examine whether Prmt5 could be recruited to RARα via Ajuba. We first examined the effect of Prmt5 on Ajuba-mediated repression of the DR5-Luc reporter activity. Interestingly, Prmt5 by itself repressed DR5-Luc activity. However, when Ajuba and Prmt5 were coexpressed, no repression of DR5-Luc activity was seen (Fig. 6A). To examine the effect of Prmt5 on the interaction between Ajuba and RARα, these proteins were coexpressed in 293 cells via transient transfection and whole-cell extracts were used for co-IP assays. Strikingly, we observed that Prmt5 attenuated the interaction between Ajuba and RARα (Fig. 6B, Lanes 2 and 4). In contrast, RARα did not affect the interaction between Ajuba and Prmt5 (see Fig. 6B, Lanes 2 and 3). To further confirm these observations, we performed in vitro binding-competition assays using a GST-Ajuba (244–350 aa) recombinant protein and in vitro-translated, full-length RARα and Prmt5 proteins. The truncated Ajuba (244–350 aa) protein contains the NR2 motif, which encodes both the Prmt5 and the RARα binding sites. When Prmt5, RARα, and GST-Ajuba (244–350 aa) proteins were added together, the amount of RARα bound to GST-Ajuba was reduced (Fig. 6C), suggesting a direct competition for the same binding site exists between Prmt5 and RARα proteins. To further strengthen the observation that Prmt5 is not a component of the Ajuba/RARα complex, we performed size-exclusion fractionation of whole-cell extracts prepared from 293 cells expressing Myc-Ajuba, Flag- RARα, or Flag-Prmt5. The Ajuba protein eluted in two well-defined peaks (Fig. 6D, fractions 16–18 and 30–32), whereas RARα eluted in a single peak (fractions 32–34), well seperated from Prmt5, which eluted in one peak (fractions 16–18). These observations suggest that Ajuba and RARα can be coeluted in fractions 30 to 34. However, Prmt5 appeared to be excluded from this complex (see Fig. 6D). Together, these data suggest that Ajuba, Prmt5, and RARα do not coexist in the same protein complex under these conditions. However, Prmt5 could potentially act as a “switch” to toggle Ajuba function toward either Snail or RARs.
Fig. 6.
Prmt5 inhibits the interaction between Ajuba and RARα. (A) Prmt5 inhibits the repression of Ajuba on DR5-Luc activity. (B) Prmt5 inhibits the interaction between Ajuba and RARα when coexpressed in 293 cells. (C) In vitro competition assays between Prmt5 and RARα. Bacterially expressed and purified GST-Ajuba (244–350aa) and in vitro-translated Prmt5 and RARα were used for in vitro binding assays. (D) Western blots show the patterns of the RARα, Ajuba or Prmt5 proteins eluted from superose-6 sizing column. Whole-cell extract (8 mg) was prepared from 293 cells overexpressing Myc-Ajuba, Flag- RARα, and Flag-Prmt5 and was loaded onto a superpose-6 gel-filtration column. (E) Model illustrating the roles of Ajuba in retinoic acid-mediated gene expression.
Discussion
Ajuba is a cytoplasmic protein that can shuttle into the nucleus. Ajuba functions as a scaffold protein participating in assembly of numerous protein complexes in both cell compartments, and has been found to bind Grb2, TRAF6, the p130Cas/Dock180 Rac guanine nucleotide-exchange factor, α-catenin, F-actin, and Aurora A kinase to regulate multiple cellular activities, including cell adhesion, migration, wound healing, and mitosis (1 –5). In the nucleus, one function of Ajuba is to act as a corepressor for the zinc finger-protein Snail, recruiting Prmt5 to repress Snail target genes and function in the epithelial-mesenchymal transition differentiation process (6 –8).
In this study, we have discovered that Ajuba modulates RA receptor signaling, probably as a classic corepressor. We demonstrated that Ajuba directly interacts with RARα via its conserved nuclear-receptor binding motifs in a ligand-dependent manner. Furthermore, we showed that depletion of Ajuba in P19 cells results in the cells being more sensitized to atRA-stimulated transcription and differentiation. ChIP assays showed that Ajuba occupies RARE loci of endogenous genes, but is released upon stimulation with atRA and concomitantly active transcription was noted from these loci. Collectively, these findings suggest that Ajuba functions as a previously unrecorded corepressor for RARα and as a unique modulator of RA signaling.
In addition to RARα, we also demonstrated that Ajuba can interact with RARγ, RXRγ, and THRα. These data suggest that Ajuba may broadly interact with a variety of nuclear hormone receptors to regulate their transcriptional activities. We are currently examining the biological significance of these findings. We also observed that the Ajuba/Zyxin family members Limd1 and WTIP can interact with RARs and RXRs in a manner similar to that of Ajuba and repress RARE-controlled luciferase activity, suggesting Limd1 and WTIP may be potential corepressors for nuclear receptors.
Because Ajuba itself does not contain any apparent enzymatic activity, it likely functions as a transcriptional coregulator by recruiting other chromatin remodeling factors. Previously, we demonstrated that the Snail protein functions as a nuclear anchor to retain Ajuba in the nucleus. In addition, Ajuba recruits Prmt5 as an effector to repress Snail target genes (7). We found that Prmt5 binds to the NR2 motif in the preLIM region of Ajuba and is translocated into the nucleus in a Snail- and Ajuba-dependent manner. Here, our data suggest that RA receptors also function as nuclear anchors for Ajuba via direct interactions. Surprisingly, we demonstrated that Ajuba, Prmt5, and RARα do not coexist in the same protein complex, rather Prmt5 inhibits the interaction between Ajuba and RARα via a direct competition for the NR2 binding site on Ajuba (see Fig. 6D). Binding of Prmt5 to the NR2 motif, which also serves as the RARα binding site, may further affect other protein–protein interactions mediated by the C-terminal LIM domains. It is also possible that Prmt5 methylates Ajuba, RARα, or other cofactors in the RARα/Ajuba complex and that this may result in the dissociation of this complex (17, 18).
Recently, the class I of histone deacetylases (HDACs) were identified as repressors recruited to GFI1 (a SNAG family of transcription factors) via an interaction with Ajuba to repress its target gene (19). Whether HDACs are also recruited to RARs and RXRs by Ajuba remains an intriguing question and needs further study. A potential model for this complex can be found in Fig. 6E.
Ajuba is expressed in totipotent embryonic stem cells, in the embryonic yolk sac, and in the E12.5 fetal liver (1). In early postimplantation embryos (E7.5–8.5), Ajuba is expressed in all embryonic germ layers, in extraembryonic yolk-sac blood islands, and in the fetal components of the developing placenta. As development progressed, the Ajuba expression pattern becomes dramatically restricted, similar to maturing embryos (post-E12.5). The temporal and spatial expression pattern of Ajuba in developing embryos suggests that Ajuba may play a regulatory role in the early embryonic events. Here we demonstrated that in P19 cells, Ajuba functions as a gauge to regulate the magnitude of atRA-induced differentiation. Hox genes are critical regulators of embryonic patterning and organogenesis (20, 21). In P19 cells, atRA can induce the expression of a number of hox genes, including hoxa1, hoxa4, hoxb1, hoxb4, and hoxd4, which parallel atRA-induced differentiation. It has been shown that hoxa1 is essential for atRA to induce P19 cells to undergo neuronal differentiation (22, 23). Consistent with this finding, we observed that depletion of Ajuba in P19 cells amplified hox gene expression, and potentiated robust differentiation in response to atRA stimulation. Interestingly, TROMA-1 expression was quickly lost in P19 cells treated with atRA for 6 days and longer. This sudden decrease in TROMA-1 expression may be a result of P19 cells differentiating into other cell types that do not express this marker, or the activation of a protein-degradation pathway resulting in the quick degradation of the TROMA-1 protein.
In addition to hox genes, oct4, sox2, nanog3, GATA4, and GATA6 are identified as key regulators of the p19 cell differentiation (13). Our preliminary studies suggest that oct4 and GATA4 may be the downstream targets of Ajuba in P19 cells. These data suggest an even broader role for Ajuba in endodermal differentiaion.
In summary, we have provided evidence that Ajuba is a corepressor to repress RA signaling via interactions with RARs and RXRs. Whether Ajuba functions as a corepressor to other members of the nuclear receptor family remains yet to be determined. Although HDACs are suggested to be downstream effectors for Ajuba-mediated repression, the exact mechanism by which Ajuba represses RAR signaling is not clear and needs to be examined further. Nevertheless, identification of Ajuba as a unique corepressor for a subset of nuclear receptors sheds light on a mechanism for nuclear receptor-mediated repression involving LIM-dimain containing proteins. Moreover, this work provides another potential target for developing more effective cancer therapeutics.
Materials and Methods
Plasmids.
The Myc epitope-tagged Ajuba plasmids pMEX-myc-Ajuba have been described (1). All Ajuba mutants were made using the QuikChange Site-Directed Mutagenesis procedures following the manufacturer’s protocol (Stratagene), and all mutants were confirmed by DNA sequencing. The pcDNA-Flag-Prmt5 and pGL2-DR5 luciferase reporter plasmids have been described (7, 24). The full-length cDNAs encoding human RARα, murine RARγ, and THRα was kindly provided by M. Lazar at the University of Pennsylvania. Human RXRα, RXRβ, and RXRγ were obtained from Open Biosystems. To make Flag-epitope tagged proteins, we subcloned these cDNAs into pcDNA3.1-Flag plasmids via PCR cloning.
Cell Culture, Transfections, and Luciferase Reporter Assays.
293 and P19 cells were maintained in DMEM containing 10% FBS, 2-mM L-glutamine, and penicillin (50 U/mL)/streptomycin (50 μg/mL) at 370 C under 5% CO2 in a humidified chamber. Transfection and luciferase reporter assays in P19 cells and 293 cells were essentially the same as that described (3).
Coimmunoprecipitation, Western Blot, Immunofluorescence, and Antibodies.
Plasmids encoding Myc-Ajuba, Flag-RARα, RARγ, RXRα, RXRβ, and RXRγ were transiently transfected into 293 cells and, 24 h after transfection, cells were lysed in the cell lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 2.5 mM EDTA, DTT, and protease inhibitor mixture. The whole-cell extracts were precleared with protein A/G beads and co-IP were performed with either α-Myc or α-Flag antibodies. The Western blot and immunofluorescent assays were described previously (7). Mouse monoclonal α-Myc (Zymed), α-Flag (Sigma), rabbit polyclonal α-RARα (Santa Cruz), mouse monoclonal α-RARα (Millipore), and α-SMRT (Abcam) antibodies were purchased. The rabbit polyclonal α-Ajuba antibody was raised by immunizing rabbits with a bacterial expressed 6-His fusion protein of murine Ajuba (aa 1–216) as the antigen.
ShRNA Knockdown Ajuba in P19 Cells and RT-PCR.
The retroviral shRNA vectors targeting the murine Ajuba gene and the luciferase gene were described previously (7, 8). The viruses were generated in 293T cells, and were infected into P19 cells. The resulting cells were selected with puromycin to create the P19-siAjuba and P19-siLuc cells. For atRA stimulation, the resulting cells were seeded in 6-well plates at 2× 105 cells/well on day 0. On day 1, various doses of atRA (10nM–10μM) or DMSO were added into the culture media for 24 h. Total RNA from P19 cells was isolated with the RNeasy Kit (Qiagen). RNA was treated with RQ DNase I to remove any genomic DNA contamination (Promega Corp.). Two micrograms of the treated total RNA were used for cDNA synthesis in a 20-μl reaction using SuperScript II reverse transcriptase (Invitrogen). The primer pairs used for PCR amplification will be available upon request. PCR amplification was carried out using Taq DNA Polymerase (Promega Corp.) at 94 C for 15 s, at 60 C for 15 s, and at 72 C for 60 s for 30 or 35 cycles in PCR Express Thermal Cycler (HYBAID).
Chromatin Immunoprecipitation.
ChIP experiments were carried out in the P19-siAjuba cells and the P19-siLuc cells. To prepare cells for ChIP assays, the P19-siAjuba cells and the P19-siLuc cells were grown in 150-mm plates to 70 to 90% confluency and atRA (1 μM) or DMSO were added into the culture media for 8 h. These cells were processed as previously described (7). The immunoprecipitated DNAs were amplified with the primer sets which amply the DNA fragements flanking the known RAREs in the hoxa1 and hoxb1 genes. The PCR fragments were cloned and confirmed by DNA sequencing. For quantification, the PCR products were resolved on 2% agarose gels and visualized with ethidium bromide.
Cell Differentiation Assays.
The procedure for atRA induction of P19 cell differentiation have been described previously (10, 11). In brief, 1 × 104 of the P19-siLuc cells and P19-siAjuba cells were seeded in 10-cm plates, respectively, in normal cell culture media. The next day, the media was replaced with the induction media containing atRA at designated doses. The media were changed every other day and morphological changes of these cells were examined daily under microscopy. These cells were harvested on designated time points and cell extracts were prepared in a lysis buffer containing 2 μM of the N-ethylmaleimide inhibitor.
Acknowledgments
We thank Dr. Gideon Dreyfuss for the gift of the Prmt5 plasmid and Dr. Mitchell A. Lazar for the RARE reporter and RA receptor plasmids, and Drs. David C. Schultz and Alexey V. Ivanov for helpful discussion and critical reading the manuscript. We thank the National Cancer Institute-Supported Wistar Institute Cancer Center Shared Facilities: Genomics, Protein Expression, Proteomics, and Hybridoma; and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. This work is supported in part by National Institute of Health (NIH) Grants CA095561 and CA092088 (to F.J.R.), the Samuel Waxman Cancer Research Foundation (F.J.R), NIH Training Grant T32, CA09171-31A1 (to Z.H.), NIH Grants CA85839 and GM080673 (to G.D.L.), and the Leukemia Research Fund of Great Britain (A.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Goyal RK, et al. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol Cell Biol. 1999;19:4379–4389. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, Longmore GD. The LIM protein Ajuba influences interleukin-1-induced NF-kappaB activation by affecting the assembly and activity of the protein kinase Czeta/p62/TRAF6 signaling complex. Mol Cell Biol. 2005;25:4010–4022. doi: 10.1128/MCB.25.10.4010-4022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marie H, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- 4.Pratt SJ, et al. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J Cell Biol. 2005;168:813–824. doi: 10.1083/jcb.200406083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirota T, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 6.Ayyanathan K, et al. The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic Shuttling. Cancer Res. 2007;67:9097–9106. doi: 10.1158/0008-5472.CAN-07-2987. [DOI] [PubMed] [Google Scholar]
- 7.Hou Z, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langer EM, et al. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 10.Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol Biol Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bain G, Ray WJ, Yao M, Gottlieb DI. From embryonal carcinoma cells to neurons: the P19 pathway. Bioessays. 1994;16:343–348. doi: 10.1002/bies.950160509. [DOI] [PubMed] [Google Scholar]
- 12.McBurney MW. P19 embryonal carcinoma cells. Int J Dev Biol. 1993;37(1):135–140. [PubMed] [Google Scholar]
- 13.Soprano DR, Teets BW, Soprano KJ. Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam Horm. 2007;75:69–95. doi: 10.1016/S0083-6729(06)75003-8. [DOI] [PubMed] [Google Scholar]
- 14.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Brûlet P, Jacob F. Molecular cloning of a cDNA sequence encoding a trophectoderm-specific marker during mouse blastocyst formation. Proc Natl Acad Sci USA. 1982;79:2328–2332. doi: 10.1073/pnas.79.7.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy B, Taneja R, Chambon P. Synergistic activation of retinoic acid (RA)-responsive genes and induction of embryonal carcinoma cell differentiation by an RA receptor alpha (RAR alpha)-, RAR beta-, or RAR gamma-selective ligand in combination with a retinoid X receptor-specific ligand. Mol Cell Biol. 1995;15:6481–6487. doi: 10.1128/mcb.15.12.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Guezennec X, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostaqul Huq MD, et al. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya-Durango DE, et al. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. J Biol Chem. 2008;283:32056–32065. doi: 10.1074/jbc.M802320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 21.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Gillespie RF, Gudas LJ. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J Mol Biol. 2007;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Ceballos E, Gudas LJ. Hoxa1 is required for the retinoic acid-induced differentiation of embryonic stem cells into neurons. J Neurosci Res. 2008;86:2809–2819. doi: 10.1002/jnr.21729. [DOI] [PubMed] [Google Scholar]
- 24.Shulemovich K, Dimaculangan DD, Katz D, Lazar MA. DNA bending by thyroid hormone receptor: influence of half-site spacing and RXR. Nucleic Acids Res. 1995;23:811–818. doi: 10.1093/nar/23.5.811. [DOI] [PMC free article] [PubMed] [Google Scholar]