Abstract
Heterozygous coding mutations in the INS gene that encodes preproinsulin were recently shown to be an important cause of permanent neonatal diabetes. These dominantly acting mutations prevent normal folding of proinsulin, which leads to beta-cell death through endoplasmic reticulum stress and apoptosis. We now report 10 different recessive INS mutations in 15 probands with neonatal diabetes. Functional studies showed that recessive mutations resulted in diabetes because of decreased insulin biosynthesis through distinct mechanisms, including gene deletion, lack of the translation initiation signal, and altered mRNA stability because of the disruption of a polyadenylation signal. A subset of recessive mutations caused abnormal INS transcription, including the deletion of the C1 and E1 cis regulatory elements, or three different single base-pair substitutions in a CC dinucleotide sequence located between E1 and A1 elements. In keeping with an earlier and more severe beta-cell defect, patients with recessive INS mutations had a lower birth weight (−3.2 SD score vs. −2.0 SD score) and were diagnosed earlier (median 1 week vs. 10 weeks) compared to those with dominant INS mutations. Mutations in the insulin gene can therefore result in neonatal diabetes as a result of two contrasting pathogenic mechanisms. Moreover, the recessively inherited mutations provide a genetic demonstration of the essential role of multiple sequence elements that regulate the biosynthesis of insulin in man.
Keywords: gene regulation, genetic testing, gene expression regulation, RNA instability, promoter regions
Neonatal diabetes is diagnosed within the first 6 months of life (1, 2) and there are two main clinical subtypes: the persistent, permanent neonatal diabetes (PNDM) and the remitting and frequently relapsing, transient neonatal diabetes (TNDM). Recently there have been considerable advances in the understanding of the genetics of neonatal diabetes (3). Most patients with PNDM have activating mutations in KCNJ11 or ABCC8, the genes encoding the potassium ATP-sensitive (KATP) channel subunits Kir6.2 (4) and SUR1 (5 –7), or heterozygous mutations in the preproinsulin (INS) gene (8 –12). In contrast, abnormalities in chromosome 6q24 are the most common cause of TNDM (13), followed by mutations in the KCNJ11 and ABCC8 genes (14). Despite these advances, the etiology of neonatal diabetes is still not known in at least 30% of patients with PNDM, suggesting other genetic causes are still to be found (9).
Insulin is secreted from islet beta cells of the pancreas. Insufficient secretion of insulin results in hyperglycemia and diabetes, whereas excessive secretion results in hypoglycemia. Insulin biosynthesis and secretion are therefore tightly regulated to maintain blood glucose levels within a narrow physiological range. Extensive studies have dissected an array of cis sequence elements in the INS promoter region and their cognate DNA binding factors, which together ensure the cellular specificity and rate of INS transcription (15 –22). In addition, insulin biosynthesis is strongly dependent on posttranscriptional regulatory mechanisms, including the modulation of translation and stability (23 –25). The latter is largely mediated through sequences located in the untranslated regions of INS transcripts (26 –28).
Heterozygous missense mutations in the coding region of the INS gene have recently been described as a cause of neonatal diabetes (8 –12). Most of the reported mutations are predicted to disrupt the folding of the proinsulin molecule. The resulting misfolded protein accumulates in the endoplasmic reticulum (ER), resulting in ER stress and beta-cell apoptosis (29, 30). An alternative potential genetic mechanism would be reduced insulin secretion because of a disruption of the INS coding sequence, as seen in the double Ins1 and Ins2 knockout mouse (31), or of the sequences that regulate insulin biosynthesis. However, as yet this has not been demonstrated in humans.
We now report recessively acting mutations within the INS gene in a series of patients with neonatal diabetes. In contrast to the previously described dominant mutations, these mutations reduce insulin synthesis and thus represent a unique pathogenic mechanism for human diabetes. These mutations also provide genetic evidence for the essential role of distinct nucleotide sequences in the regulation of the human preproinsulin gene.
Results
Recessive INS Mutations Cause Neonatal Diabetes.
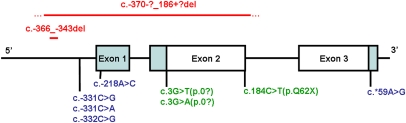
We sequenced 117 unrelated probands with diabetes diagnosed before 6 months (13 offspring of consanguineous parents) in whom the known common genetic causes had been excluded. We identified 10 different INS recessive mutations in 15 unrelated families (Figs. 1 and 2). Four homozygous mutations affected the coding region: c.184C > T (p.Q62X), c.3G > T (p.0?), c.3G > A (p.0?), and a large deletion that removes a segment of the promoter, exon 1 and coding exon 2 of INS (c.-370-?_186+?del). Five homozygous mutations were found in regulatory regions: c.-331C > A (2 families), c.-331C > G (5 families), c.-218A > C, and a 24-base pair deletion (c.-366_-343del) are located in the promoter region, whereas c.*59A > G is within the 3′ untranslated region. One proband was a compound heterozygote for two regulatory region mutations, c.-331C > G and c.-332C > G.
Fig. 1.
A schematic of the INS gene showing the 10 mutations identified in 15 families. Positions of point mutations are indicated below the exons, while deletions are shown above the gene. The blue shaded regions are noncoding, the red text indicates a deletion, the blue text are noncoding mutations, and the green are coding mutations. The precise breakpoints of the multiexonic deletion are not known; the solid line represents the minimal deleted region. Mutation nomenclature is based on the coding sequence where nucleotide 1 represents translational start site.
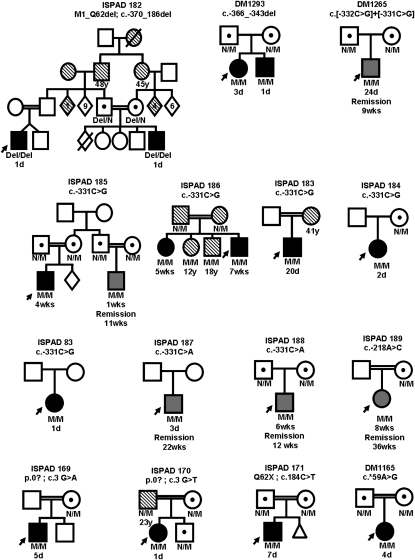
Fig. 2.
Partial pedigrees of the 15 families with recessive INS mutations. (Del, deletion; n, Normal allele; M, mutation). Solid black-filled shapes represent patients with permanent neonatal diabetes, gray filled shapes represent patients with transient neonatal diabetes, and shapes filled with diagonal lines represent those patients diagnosed with diabetes after 6 months of age. Age at diagnosis and remission (where applicable) are shown below the symbols.
The mutations were inherited in a recessive manner either homozygous or compound heterozygous, with heterozygous carrier parents being unaffected with neonatal diabetes (Fig. 2). Pathogenicity of mutations was suggested by conservation across species and absence of variants in controls (Table S1).
Recessive INS Mutations Uncover Essential Regulatory Sequences in Humans.
Further support for the pathogenicity of mutations came from known function of mutated residues and functional studies (Figs. 3 and Figs. 4; see also SI Results). Multiple mutation mechanisms were involved in the recessive INS mutations, which are described briefly below.
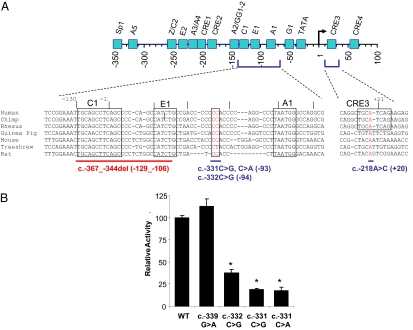
Fig. 3.
Functional evidence for the pathogenicity of recessive promoter INS mutations. (A) Schematic of the genomic sequence of the INS promoter structure with major cis regulatory elements, and the sequence context of mutated elements in several mammalian species that do not exhibit major divergence in these regions. Mutated bases are highlighted in red. The numbering of promoter landmarks is relative to the transcription start site (genomic numbering, where g.1 is equivalent to c.-238) consistent with the convention used in previous studies. Mutations are described according to Human Genome Variation Society guidelines (http://www.hgvs.org/mutnomen/) (cDNA numbering according to the translational start site where c.1 is equivalent to g.238), and distance to the conventional INS transcriptional start site is shown in parenthesis. (B) Evidence for loss-of-function of the c.-331(C > G, C > A) and c.-332C > G mutations. Firefly luciferase expression is compared in constructs containing the wild-type (WT) INS promoter sequence (INS WT), or c.-331 C > G, c.-331 C > A, c.-332 C > G, c.-339G > A mutations, after transfection in MIN6 β-cells. Data shown are means (+/−SE) from three independent constructs for each mutation (n = 3 replicates). c.-339G > A is a control mutation that does not impair INS transcription. Results are corrected for transfection efficiency using a vector that constitutively expresses Renilla luciferase, and are expressed relative to the INS WT results. The asterisks denote P < 0.0001 in ANOVA for the difference between INS WT and mutant constructs.
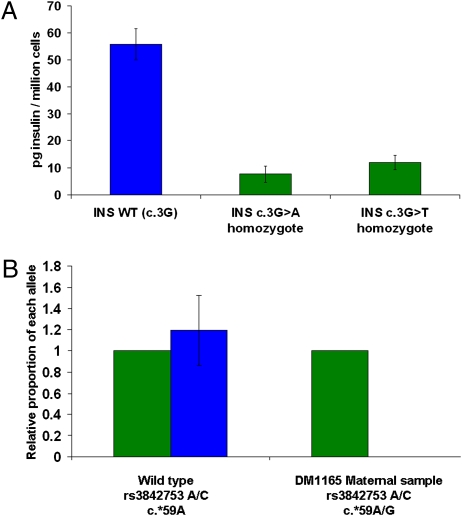
Fig. 4.
Functional evidence for the pathogenicity of recessive INS mutations affecting translation and mRNA stability (A) Homozygous mutations in the translation initiation codon of the INS gene result in reduced insulin content of transfected HeLa cells. The insulin content of HeLa cells was measured by RIA after transfection with wild-type insulin (INS WT) or either of two INS mutant constructs, as shown. Both nucleotide changes were identified in patients with permanent neonatal diabetes. Nonspecific values obtained with HeLa cells transfected with empty vector were subtracted from all samples and those data are presented as mean +/− SE (n = 3 replicates). (B) Allele-specific quantitative real-time PCR of c.*59A > G and normal transcripts. The graph shows the relative abundance of the wild-type and mutant RNA transcripts in mutant and normal cell lines. The rs3842753 A allele tags the c.*59A (wild type, shown in green); the c.*59G (mutant) was tagged by rs3842753 C allele (blue). The graph shows the level of transcripts in the control sample heterozygous only for rs3842753 and in the maternal sample (family DM1165), which is heterozygous for both rs3842753 and c.*59A/G. The level of the mutant transcript is reduced to less than 3 × 10−4 percent compared with the normal transcript in the heterozygous c.*59A > G cell line. Experimental error as calculated from the standard deviation of the replicate experiments is indicated. The standard deviation for the quantification of the c.*59G allele in the maternal sample is 3 × 10−6, and thus the experimental error is not visible in the figure.
Truncated proteins.
The nonsense mutation (p.Q62X) is predicted to give rise to a mutant protein that is truncated within the C-peptide region and will lack the insulin-A chain.
Promoter mutations.
The (c.-366_-343del) 24-base pair deletion abolishes the INS promoter evolutionary conserved C1 and E1 elements, where MAFA and NEUROD1 bind, respectively (16, 20, 32) (Fig. 3A). The c.-218A > C mutation disrupts the CRE3 site that interacts with multiple DNA binding proteins in vitro (22) (Fig. 3A). All of these elements have been previously shown to be critically important for the INS promoter activity in transient transfection studies (15, 18, 33 –35). The c.-331(C > G, C > A) and c.-332C > G mutations were located between the E1 and A1 elements (Fig. 3A). This sequence is conserved among a subset of mammalian species (Fig. 3A) and mutagenesis of multiple bases neighboring this dinucleotide impairs INS promoter activity (36). We constructed insulin promoter fragments carrying the point mutations c.-331(C > G, C > A) and c.-332C > G. The point mutations induced up to 90% reduction in transcriptional activity, while a control mutation, c.-339G > A, did not alter the transcriptional activity in pancreatic beta-cell lines (Fig. 3B). Thus, the CC dinucleotide that is mutated in eight unrelated probands with neonatal diabetes forms part of an essential positive cis regulatory sequence of the INS promoter.
Mutated or absent translational start site.
The two point mutations (c.3G > A and c.3G > T) at the first methionine residue (p.Met1) abolish the native translation initiation site for the preproinsulin protein. Quantification of total INS mRNA levels by real-time PCR revealed no differences in mRNA abundance for c.3G > A or c.3G > T mutations compared with the wild type. The insulin content of HeLa cells transfected with these mutations was reduced by 86% and 79% for c.3G > A and c.3G > T, respectively, compared to cells transfected with the wild-type sequence (Fig. 4A and SI Results). The multiexon deletion (exons 1 and 2) removes over half the coding region including the translational start site and is expected to be a null mutation.
Altered mRNA stability through a mutation in the 3′ untranslated region.
The c.*59A > G mutation is located in the polyadenylation signal of the 3′ untranslated region and potentially impairs mRNA stability. In a heterozygous lymphoblastoid cell line generated from the proband’s mother, the mutant mRNA transcript was present at a very low level compared to the wild-type allele. This is consistent with reduced mRNA stability (Fig. 4B and SI Results).
Clinical Phenotype of Patients with Recessive INS Mutations.
The clinical characteristics of patients with recessive INS mutations are shown in Table 1 (Table S1 and S2). In keeping with the known actions of insulin before and after birth, the phenotype was limited to markedly reduced fetal growth and diabetes.
Table 1.
Comparison of clinical characteristics in patients with isolated neonatal diabetes with recessive and dominant INS mutations
| Characteristic | INS recessive | INS dominant | P-value |
| n | 19 | 46 | NA |
| Sex, % male | 63.2 | 47.8 | 0.3 |
| Birth weight, g | 1,680 (1,410; 2,050) | 2,530 (2,350; 2,900) | <0.001 |
| Gestational age, wk | 37.5 (36, 40) | 40 (38.5, 40) | 0.008 |
| Birth weight, SD score | −3.2 (−4.1, −2.6) | −2.0 (−2.5, −1.0) | <0.001 |
| Age at diagnosis, wk | 1 (0, 3) | 10 (5, 22) | <0.001 |
| Remission, % | 26 | 0 | 0.001 |
| Age at remission, wk | 12 (11, 22) | NA | NA |
| Age at relapse, yr | 1 (only 1 case) | NA | NA |
| Current age, yr | 5 (2, 14) | 11 (4, 23) | 0.2 |
Data are median (interquartile range). NA, not applicable.
The diabetes phenotype within the families is shown in Fig. 2. Nineteen patients had neonatal diabetes (15 probands and 4 family members); 14 had PNDM and were treated with insulin from diagnosis, and 5 patients had TNDM, having gone into remission at a median age of 12 weeks [interquartile range (IQR) 11, 22]. Birth weight was markedly reduced in all patients with neonatal diabetes resulting from recessive mutations [median birth weight 1,680 g (1,420; 2,050), which is −3.2 SD score (−4.1, −2.6)]. In keeping with more severe insulin deficiency, patients with PNDM had a more severe intrauterine growth retardation [median SD score for birth weight −3.9 (−4.4, −2.8) vs. −1.8 (−3.4, −0.9) in TNDM, P = 0.03] and diabetes was diagnosed earlier [2 days (1, 9.5) vs. 24 days (5, 62), P = 0.04] (Table S3). All patients with mutations that altered the coding region or mRNA stability had PNDM. The noncoding promoter mutations were associated with both PNDM and TNDM. A summary of the remaining 98 patients with neonatal diabetes of unknown origin is given in Table S4.
Differences in the Clinical Phenotype with Recessive and Dominant INS Mutations.
To identify whether the different mutation mechanisms in the same gene resulted in phenotypic differences, we compared the clinical characteristics of patients with neonatal diabetes as a result of recessive INS mutations with patients with the previously identified dominant mutations in INS (Table 1). Patients with neonatal diabetes resulting from recessive INS mutations had a markedly different phenotype, with lower birth weight [median SD score −3.2 (IQR −4.1, −2.6) vs. −2.0 (−2.5, −1.0), P < 0.001] and an earlier age of diagnosis [median age in weeks 1 (0, 3) vs. 10 (5, 22), P < 0.001]. TNDM is only seen in patients with recessive mutations (26 vs. 0%, P = 0.001). Overall, recessive INS mutations accounted for 3.7% PNDM (n = 300) and 2.2% TNDM (n = 134) in a consecutive series of patients with isolated neonatal diabetes referred to the two laboratories for genetic testing.
Discussion
We have shown that recessively acting mutations in the preproinsulin gene (INS) are a cause of neonatal diabetes. They act by reducing synthesis of the preproinsulin peptide because of a truncated protein, abnormal transcription, reduced mRNA stability, or disrupted translation. These mutations usually cause PNDM but may manifest as TNDM or diabetes outside the neonatal period. In keeping with the recessive inheritance, many probands (60%) were the offspring of consanguineous parents.
The clinical manifestations of recessive INS mutations reflect the consequences of insulin deficiency in humans during pre- and postnatal life. The birth weight was markedly reduced [median SD score −3.2 (−4.1, −2.6)], consistent with the major role of insulin in fetal growth. The early onset of neonatal diabetes (median 1 week) reflects severe insulin deficiency postnatally. In contrast to many other subtypes of neonatal diabetes, there are no extrapancreatic features.
Differences in the underlying pathophysiology explain why patients with recessive INS mutations are diagnosed earlier and have a lower birth weight than patients with heterozygous INS mutations (8 –12). The disrupted insulin synthesis seen with recessive mutations occurs as soon as the fetal beta cell starts to secrete insulin. In contrast, insulin secretion is required before beta-cell dysfunction develops in patients with heterozygous mutations, which result in misfolding of the preproinsulin peptide, accumulation of the misfolded protein in the ER, and hence the destruction of the beta cell through ER stress. These two distinct disease mechanisms are supported by phenotypic studies in mouse models, where reduced insulin secretion at birth or progressive ER stress and cell death have been described in mice carrying analogous recessive or dominant mutations, respectively (30, 31, 37).
The majority of the patients with neonatal diabetes have PNDM, but 26% (5 of 19) have TNDM. TNDM is only found in patients with noncoding mutations and they have a higher birth weight and are diagnosed later. This finding is consistent with TNDM resulting from a less severe insulin deficiency, and is comparable to the situation with mutations in the Kir6.2 subunit of the KATP mutations (38), where TNDM mutations have less severe functional consequences. The mechanism of remission in recessive INS mutation carriers is not understood but is likely to reflect a variation in demand or the ability of the beta cell to meet this demand, as a similar timing of remission is seen in some patients with less severe mutations, resulting in channelopathies (14, 38) and pancreatic developmental defects (39, 40).
The mutations identified in this study illustrate multiple mechanisms by which insulin biosynthesis can be disrupted. These include absent or altered translation because of coding-sequence deletions or mutations, reduced transcription because of mutations of the promoter, or abnormal mRNA stability. Our functional studies established that the 3′ UTR mutation that abolishes the polyadenylation signal results in severe RNA instability and that the initiation codon mutations result in reduced transcription of the preproinsulin gene. The promoter mutations are highly informative because they provide human genetic evidence that discrete INS cis regulatory elements are essential. Numerous studies have demonstrated that multiple cis elements are required for the activity of episomal INS reporter constructs in cultured cells (15, 17 –19, 21, 22). However, it is not known if each of those cis elements is truly necessary in vivo, because such studies can only partially predict their function in the integrated chromatin environment of bona fide differentiated cells. Studies in other selected genes have addressed this by targeted deletion of transcriptional regulatory elements in mice (41). Our findings now reveal that the C1/E1, CC, and CRE3 elements are essential for INS gene transcription in humans. The discovery of three separate mutations that target the CC dinucleotide sequence is particularly significant. This element forms part of a canonical CCACC binding-site motif for Kruppel-like zinc-finger proteins. Two recent studies have shown that artificial mutations of this CCACC element lead to decreased INS promoter activity (36, 42). Although previous studies failed to identify protein complexes interacting with this region in beta-cell nuclear extracts (17, 36), a recent study showed that in vitro translated GLIS3, a zinc-finger transcription factor that is mutated in patients with neonatal diabetes and congenital hypothyroidism (43), exhibits sequence-specific binding to this region (42). Multiple DNA binding factors may bind to CCACC elements in vitro, and thus future studies are warranted to determine if additional factors act through this element in vivo in beta cells. Our findings, therefore, demonstrate that the natural CC element mutations that cause diabetes disrupt INS gene activity and establish the importance of this cis regulatory element.
In conclusion, we have shown that homozygous INS mutations are a unique cause of neonatal diabetes. The mutations result in reduced synthesis of the insulin polypeptide through a variety of mechanisms and may yield further insights into the regulation of insulin biosynthesis.
Materials and Methods
Cohort Characteristics.
We studied an international cohort of 117 unrelated patients (67 males) with diabetes diagnosed before 6 months (median age 4 weeks) and without a known genetic etiology, which were referred to the Exeter (n = 105) or Bilbao laboratories (n = 12). Thirteen patients were offspring of consanguineous parents (second-degree relatives or closer). In the 100 probands with PNDM, we excluded mutations in KCNJ11, ABCC8, GCK, and previously described heterozygous coding mutations in INS (9). In the 17 patients with TNDM, we excluded 6q24 anomalies, KCNJ11, and ABBC8 mutations. Remission was defined as the disappearance of clinical symptoms with normalization of blood glucose or HbA1c for a period longer than 15 days after withdrawal of insulin therapy. Babies born before 33 weeks of gestation were excluded to avoid hyperglycemia of prematurity. Studies were approved by Cruces Hospital committee and North and East Devon Research Ethics Committee. Informed consent was obtained from all patients or their parents and the studies were conducted in line with the Declaration of Helsinki. Clinical data were obtained from the patients’ clinical records. We calculated standard deviation scores for birth weight (44).
Molecular Genetic Analysis.
Genomic DNA was extracted from peripheral leukocytes using standard procedures. Regulatory elements up to 450 bp upstream of the transcriptional start site and exons 1 to 3 of the INS gene (Fig. 1) were amplified by the PCR in three amplicons (primers and conditions available on request). Unidirectional sequencing was carried out on an ABI3730 (Applied Biosystems) and analyzed using Mutation Surveyor v3.20. Sequences were compared with the published sequence (Ensembl sequence ENSG00000129965) and published polymorphisms. The genomic reference sequence nucleotide 1 is the transcriptional start site (g.1A or c.-238A), whereas the translational start site is located at g.238 (c.1). Mutation nomenclature is shown in compliance with the Human Genome Variation Society, where nucleotide 1 represents the A of the translational start-site codon ATG (c.1). Suspected mutations were tested for conservation across species and cosegregation within families. Putative gene deletions were investigated using multiplex ligation-dependent probe amplification assay oligonucleotide probes specific for the three exons of INS (see SI Materials and Methods).
Functional Studies.
Investigating the effect of INS promoter mutations on transcriptional activity.
To determine the functional impact of the c.-331(C > G, C > A) and c.-332C > G mutations we performed site-directed mutagenesis of an INS promoter firefly luciferase reporter construct (pSOUAPRL-251hINS-Luc), and compared the activity of control and mutated promoters in MIN6 β-cells, using a Renilla luciferase promoter (pGL4.75) to correct for differences in transfection efficiency (see SI Materials and Methods).
Investigating the effect of the translation initiation mutations (c.3G > T and c.3G > A).
To determine the effect of these mutations on insulin production, we transfected HeLa cells, which do not express insulin, with wild-type or mutant INS and analyzed intracellular insulin content using radio-immunoassay (SI Materials and Methods).
Investigating the effect of the c.*59A > G mutation on mRNA stability.
We determined the effect of the c.*59A > G mutation on insulin mRNA stability using real time PCR to measure the relative levels of the INS mRNA transcripts in a heterozygous lymphoblastoid cell line derived from the proband’s mother. We used a heterozygous SNP, rs3842753, to identify the mutation bearing allele (SI Materials and Methods).
Statistical Analysis.
Clinical numeric data are given as median (IRQ range). Functional data are given as mean (SE). The clinical features of patients were compared using Kruskal-Wallis, χ2 (Fisher’s exact) tests or Mann-Whitney U in the statistical package SPSS version 13. Student’s t-test or analysis of variance was used for expression studies.
Supplementary Material
Acknowledgments
We thank Chris Boustred, Piers Fulton, Amna Khamis, Andrew Parrish, Gustavo Pérez-Nanclares, Xavi Garcia, and Eleftheria Eiakogiannaki for technical assistance; Ann Kelly, Anthony Dixon, Srikanth Bellary, and Abigail Britten for supplying the Pakistani control DNAs; Dr Roland Stein (Vanderbilt University) for providing a human insulin promoter plasmid; and Dr Jun-ichi Miyazaki for the MIN6 cells. This work was funded by the European Union (Integrated Project EURODIA LSHM-CT-2006-518153 in the Framework Programme 6 and CEED3 RTD REG/F.2(2009)D/518731 in the Framework Programme) To A.T.H. and J.F, Wellcome Trust Grants 067463/Z/2/Z (to A.T.H.) and 081278/Z/06/Z (to L.W.H.); the Centro de Investigación Biomédica en Red de Diabetes y Enfermedades Metabólicas and PI06/0690 from the Instituto de Salud Carlos III of the Spanish Ministry of Health (to G.P.d.N.); Grant BFI06.266 from the Basque Department of Education (to I.G.); an “Ayuda para contratos post-Formación Sanitaria Especializada” from the “Instituto de Salud Carlos III” (FIS CM06/00013) (to O.R.-C.); FIS researcher Grant CP03/0064 from the Instituto de Salud Carlos III of the Spanish Ministry of Health (to G.P.d.N.); and Grants 021620819 from the Czech Ministry of Education and 64203 from the Czech Ministry of Health (to Z.S.). Work in Poland is supported by the Polish Diabetes Association. A.T.H. is a Wellcome Trust Research Leave Fellow, L.W.H. is a Research Councils United Kingdom Academic Fellow, and J.M.L. is funded by a Peninsula Medical School PhD studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910533107/DCSupplemental.
References
- 1.Iafusco D, et al. Early Onset Diabetes Study Group of the Italian Society of Paediatric Endocrinology and Diabetology. Permanent diabetes mellitus in the first year of life. Diabetologia. 2002;45:798–804. doi: 10.1007/s00125-002-0837-2. [DOI] [PubMed] [Google Scholar]
- 2.Edghill EL, et al. HLA genotyping supports a nonautoimmune etiology in patients diagnosed with diabetes under the age of 6 months. Diabetes. 2006;55:1895–1898. doi: 10.2337/db06-0094. [DOI] [PubMed] [Google Scholar]
- 3.Aguilar-Bryan L, Bryan J. Neonatal diabetes mellitus. Endocr Rev. 2008;29:265–291. doi: 10.1210/er.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gloyn AL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 5.Proks P, et al. A heterozygous activating mutation in the sulphonylurea receptor SUR1 (ABCC8) causes neonatal diabetes. Hum Mol Genet. 2006;15:1793–1800. doi: 10.1093/hmg/ddl101. [DOI] [PubMed] [Google Scholar]
- 6.Ellard S, et al. Permanent neonatal diabetes caused by dominant, recessive, or compound heterozygous SUR1 mutations with opposite functional effects. Am J Hum Genet. 2007;81:375–382. doi: 10.1086/519174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babenko AP, et al. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N Engl J Med. 2006;355:456–466. doi: 10.1056/NEJMoa055068. [DOI] [PubMed] [Google Scholar]
- 8.Støy J, et al. Neonatal Diabetes International Collaborative Group. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edghill EL, et al. Neonatal Diabetes International Collaborative Group. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57:1034–1042. doi: 10.2337/db07-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo C, et al. Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes (SIEDP) Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J Clin Invest. 2008;118:2148–2156. doi: 10.1172/JCI33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molven A, et al. Norwegian Childhood Diabetes Study Group. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57:1131–1135. doi: 10.2337/db07-1467. [DOI] [PubMed] [Google Scholar]
- 12.Polak M, et al. French ND (Neonatal Diabetes) Study Group. Heterozygous missense mutations in the insulin gene are linked to permanent diabetes appearing in the neonatal period or in early infancy: a report from the French ND (Neonatal Diabetes) Study Group. Diabetes. 2008;57:1115–1119. doi: 10.2337/db07-1358. [DOI] [PubMed] [Google Scholar]
- 13.Temple IK, et al. Transient neonatal diabetes: widening the understanding of the etiopathogenesis of diabetes. Diabetes. 2000;49:1359–1366. doi: 10.2337/diabetes.49.8.1359. [DOI] [PubMed] [Google Scholar]
- 14.Flanagan SE, et al. Mutations in ATP-sensitive K+ channel genes cause transient neonatal diabetes and permanent diabetes in childhood or adulthood. Diabetes. 2007;56:1930–1937. doi: 10.2337/db07-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson O, Edlund T, Moss JB, Rutter WJ, Walker MD. A mutational analysis of the insulin gene transcription control region: expression in beta cells is dependent on two related sequences within the enhancer. Proc Natl Acad Sci USA. 1987;84:8819–8823. doi: 10.1073/pnas.84.24.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 17.Boam DS, Clark AR, Docherty K. Positive and negative regulation of the human insulin gene by multiple trans-acting factors. J Biol Chem. 1990;265:8285–8296. [PubMed] [Google Scholar]
- 18.Sharma A, Stein R. Glucose-induced transcription of the insulin gene is mediated by factors required for beta-cell-type-specific expression. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.German M, et al. The insulin gene promoter. A simplified nomenclature. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 20.Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci USA. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 22.Hay CW, Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006;55:3201–3213. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- 23.Permutt MA, Kipnis DM. Insulin biosynthesis. I. On the mechanism of glucose stimulation. J Biol Chem. 1972;247:1194–1199. [PubMed] [Google Scholar]
- 24.Wicksteed B, et al. A cis-element in the 5′ untranslated region of the preproinsulin mRNA (ppIGE) is required for glucose regulation of proinsulin translation. Cell Metab. 2007;5:221–227. doi: 10.1016/j.cmet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Goodge KA, Hutton JC. Translational regulation of proinsulin biosynthesis and proinsulin conversion in the pancreatic beta-cell. Semin Cell Dev Biol. 2000;11:235–242. doi: 10.1006/scdb.2000.0172. [DOI] [PubMed] [Google Scholar]
- 26.Sander M, Griffen SC, Huang J, German MS. A novel glucose-responsive element in the human insulin gene functions uniquely in primary cultured islets. Proc Natl Acad Sci USA. 1998;95:11572–11577. doi: 10.1073/pnas.95.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wicksteed B, et al. Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. J Biol Chem. 2001;276:22553–22558. doi: 10.1074/jbc.M011214200. [DOI] [PubMed] [Google Scholar]
- 28.Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci USA. 2007;104:15841–15846. doi: 10.1073/pnas.0702697104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbach N, et al. Dominant-negative effects of a novel mutated Ins2 allele causes early-onset diabetes and severe beta-cell loss in Munich Ins2C95S mutant mice. Diabetes. 2007;56:1268–1276. doi: 10.2337/db06-0658. [DOI] [PubMed] [Google Scholar]
- 31.Duvillié B, et al. Phenotypic alterations in insulin-deficient mutant mice. Proc Natl Acad Sci USA. 1997;94:5137–5140. doi: 10.1073/pnas.94.10.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuoka TA, et al. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Docherty HM, et al. Relative contribution of PDX-1, MafA and E47/beta2 to the regulation of the human insulin promoter. Biochem J. 2005;389:813–820. doi: 10.1042/BJ20041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inagaki N, et al. c-Jun represses the human insulin promoter activity that depends on multiple cAMP response elements. Proc Natl Acad Sci USA. 1992;89:1045–1049. doi: 10.1073/pnas.89.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hay CW, et al. Glucagon-like peptide-1 stimulates human insulin promoter activity in part through cAMP-responsive elements that lie upstream and downstream of the transcription start site. J Endocrinol. 2005;186:353–365. doi: 10.1677/joe.1.06205. [DOI] [PubMed] [Google Scholar]
- 36.Niu X, et al. Human Krüppel-like factor 11 inhibits human proinsulin promoter activity in pancreatic beta cells. Diabetologia. 2007;50:1433–1441. doi: 10.1007/s00125-007-0667-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gloyn AL, et al. Relapsing diabetes can result from moderately activating mutations in KCNJ11. Hum Mol Genet. 2005;14:925–934. doi: 10.1093/hmg/ddi086. [DOI] [PubMed] [Google Scholar]
- 39.Yorifuji T, et al. Neonatal diabetes mellitus and neonatal polycystic, dysplastic kidneys: Phenotypically discordant recurrence of a mutation in the hepatocyte nuclear factor-1beta gene due to germline mosaicism. J Clin Endocrinol Metab. 2004;89:2905–2908. doi: 10.1210/jc.2003-031828. [DOI] [PubMed] [Google Scholar]
- 40.Edghill EL, et al. Hepatocyte nuclear factor-1 beta mutations cause neonatal diabetes and intrauterine growth retardation: support for a critical role of HNF-1beta in human pancreatic development. Diabet Med. 2006;23:1301–1306. doi: 10.1111/j.1464-5491.2006.01999.x. [DOI] [PubMed] [Google Scholar]
- 41.Fujitani Y, et al. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang HS, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senée V, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 44.Freeman JV, et al. Cross sectional stature and weight reference curves for the UK, 1990. Arch Dis Child. 1995;73:17–24. doi: 10.1136/adc.73.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.