Abstract
Hepatitis C virus (HCV) remains a major public health problem, affecting approximately 130 million people worldwide. HCV infection can lead to cirrhosis, hepatocellular carcinoma, and end-stage liver disease, as well as extrahepatic complications such as cryoglobulinemia and lymphoma. Preventative and therapeutic options are severely limited; there is no HCV vaccine available, and nonspecific, IFN-based treatments are frequently ineffective. Development of targeted antivirals has been hampered by the lack of robust HCV cell culture systems that reliably predict human responses. Here, we show the entire HCV life cycle recapitulated in micropatterned cocultures (MPCCs) of primary human hepatocytes and supportive stroma in a multiwell format. MPCCs form polarized cell layers expressing all known HCV entry factors and sustain viral replication for several weeks. When coupled with highly sensitive fluorescence- and luminescence-based reporter systems, MPCCs have potential as a high-throughput platform for simultaneous assessment of in vitro efficacy and toxicity profiles of anti-HCV therapeutics.
Keywords: viral hepatitis, liver, tissue engineering, drug development, infection
Recent advances have allowed HCV to be propagated in human hepatoma cells (HCV cell culture system, HCVcc). These cell lines, however, display abnormal proliferation, deregulated gene expression, as well as aberrant signaling and endocytic functions (1 –4). Consequently, neither the perturbation of normal hepatocyte biology by infection, nor authentic host responses to HCV, can be studied accurately in culture (1). Primary hepatocytes are considered a more physiologically relevant system, but are notoriously difficult to maintain in culture as they precipitously decline in viability and phenotype upon isolation from their in vivo microenvironment (6). This rapid deterioration, as well as the lack of HCV detection methods with high specificity and sensitivity, has made it difficult to assess viral replication in primary human cell cultures (5, 7 –11). Over the last few decades, investigators have employed a plethora of different strategies to preserve liver-specific functions in vitro and to extend the lifetime of the model systems (12). These strategies typically include extracellular matrix manipulations, defined culture media, fluid flow using bioreactors, or alteration of cell–cell interactions by forming 3D spheroidal aggregates or cocultivation with nonparenchymal cell types (6, 12, 13, 14). Although some of these models provide necessary extracellular matrix cues, they lack crucial heterotypic cell−cell interactions or control over tissue architecture, known to affect liver-specific functions (6, 12). In culture techniques using fragile extracellular matrix gels, 3D aggregates, and/or continuous perfusion, scaling down to 96-well and smaller formats appropriate for drug screening remains challenging. Most importantly, it is unclear whether any of these model systems supports persistent HCV infection.
Results
Primary Human Hepatocytes in Micropatterned Cocultures Form Polarized Cell Layers and Support HCV Glycoprotein-Mediated Entry.
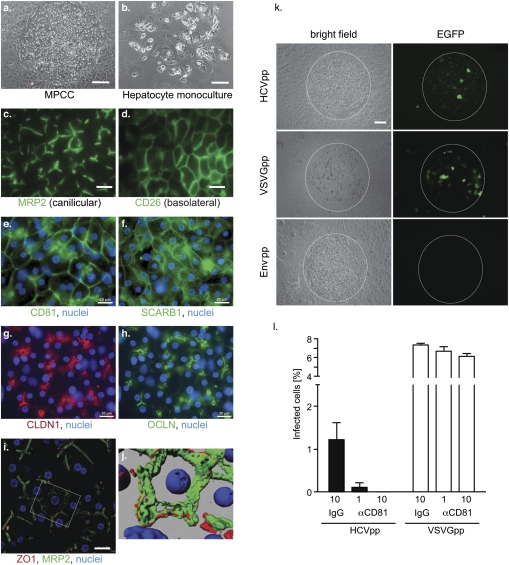
We have recently developed a miniaturized, multiwell model of human liver tissue with optimized microscale architecture that maintains phenotypic functions for several weeks in vitro (12). In this cell culture system, primary adult human hepatocytes do not seem to proliferate. Our system is comprised of primary hepatocytes organized in micropatterned colonies of empirically optimized dimensions and subsequently surrounded by supportive stroma (micro-patterned cocultures, MPCC; Fig. 1 A and B). Here, we show that primary human hepatocytes form polarized cell layers in MPCCs. Multidrug resistant protein 2 (MRP2), zona occludens protein 1 (ZO1), and HCV entry factors claudin-1 (CLDN1) (15) and occludin (OCLN) (16, 17), were located in tight junction (TJ)-like structures (canalicular domain), whereas CD26 was localized on the basolateral domain (Fig. 1). The presence of bile canalicular structures between adjacent hepatocytes was confirmed via 3D renderings reconstructing ZO1, MRP2, and nuclear staining (Fig. 1 I and J). Compared to human liver tissue, primary hepatocytes in MPCCs expressed similar patterns of the other known HCV entry factors, CD81 (18), scavenger receptor class B type 1 (SCARB1) (19), and CLDN1 (15) (Fig. 1 and Fig. S1).
Fig. 1.
Primary human hepatocytes in MPCCs form polarized cell layers, express HCV entry factors, and support HCV glycoprotein-mediated entry. Bright field images of primary hepatocytes in MPCCs (A) and in monocultures (B). Wide-field fluorescence images of fixed MPCCs stained for the canilicular marker MRP2 (C), and the basolateral marker CD26 (D). Nuclear (blue) and antigen-specific staining (green) for CD81 (E), SCARB1 (near edge of hepatocyte island) (F), CLDN1 (red) (G), OCLN (H) in MPCCs. (I) Merged image of primary hepatocytes stained for MRP2 (green), ZO1 (red), and nuclei (blue). (J) 3D rendering of boxed area in I. (K) Infection of MPCCs with retroviral pseudoparticles bearing HCV glycoproteins (HCVpp), vesicular stomatitis virus glycoprotein (VSVGpp), or no glycoproteins (Env-pp) and containing an EGFP reporter gene. Representative images are shown for all experiments. (L) Anti-CD81 antibody blocks entry of HCVpp (dark bars), but not VSVGpp (white bars). Concentrations of antibody (μg/mL) are noted. Mean and SD are shown. Scale bars: 100 μm (a, b, k), 50 μm (c, d), 20 μm (e-i).
To test whether MPCC hepatocytes can support HCV glycoprotein-mediated entry, we infected cultures with HCV pseudoparticles (HCVpps). HCVpp, which are defective lentiviral particles that display the HCV glycoproteins (E1 and E2) and encode a reporter gene (here EGFP), allow rapid quantitation of infection in the absence of replication. Approximately 1–3% of the human hepatocytes in MPCCs, but none of the supporting murine embryonic fibroblasts (3T3-J2), could be infected with HCVpp (Fig. 1 K and L). Pseudoparticles lacking glycoproteins did not infect the cultures, although MPCCs were readily infected by pseudoparticles displaying the pan-tropic vesicular stomatitis virus glycoprotein (VSVGpp). A blocking antibody targeting CD81 completely abrogated HCVpp, but not VSVGpp, infection (Fig. 1L).
HCV Persistently Replicates in Primary Human Hepatocyte MPCCs
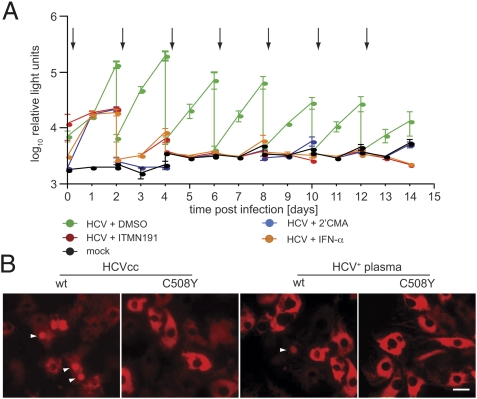
Despite considerable effort, it has not been possible to unequivocally demonstrate HCV replication in primary hepatocyte cultures over prolonged periods of time. Previous studies have relied on quantifying HCV RNA by quantitative reverse transcription-polymerase chain reaction (qRT-PCR), a technique inappropriate for detecting rare infectious events due to high background of nonspecifically bound viral RNA. We instead employed a highly sensitive HCVcc reporter virus expressing secreted Gaussia luciferase (Gluc), Jc1FLAG2(p7-nsGluc2A) (20). After inoculation, cultures were washed to remove Gluc carryover, and luciferase secretion was monitored as an indicator of viral replication. We found that several conventional culture systems (i.e., collagen gel sandwich, Matrigel overlay, and randomly distributed cocultures) could not sustain HCV replication, presumably due to a decline in liver-specific phenotype (12) (Fig. S2). In contrast, MPCCs in multiwell formats supported HCV replication for at least two weeks (Fig. 2 and Fig. S3). Treatment with HCV NS3-4A protease (ITMN191) or NS5B polymerase inhibitors (2’CMA), or IFN alpha (IFN-α), reduced luciferase activity to background levels (Fig. 2A), indicating that persistent signal was indeed due to ongoing viral replication. Persistent HCV infection was achieved in MPCCs created from freshly isolated or cryopreserved human hepatocytes from several donors, reflecting the reproducibility of the optimized microscale architecture and the concomitant phenotypic stability. We next attempted to quantify HCV RNA and proteins in MPCCs by qRT-PCR, Western blot, and immunofluorescence. In contrast to recent reports (10), we were unable to obtain specific signals above background, reminiscent of failed attempts to detect HCV proteins in infected liver biopsies - probably due to the low number of HCV RNA copies per cell (21).
Fig. 2.
Primary human hepatocyte MPCCs are susceptible to HCV. (A) Persistent infection of primary human hepatocytes with HCVcc. Primary hepatocytes in MPCC were infected with Jc1FLAG2(p7-nsGluc2A). After 24 h, virus was removed and MPCC medium containing DMSO (0.1%) or the indicated inhibitors was added. All inhibitors were used at approximately 50× IC50 (polymerase inhibitor 2’CMA = 2.16 mM, protease inhibitor ITMN191 = 0.16 mM, IFN-α = 500 U/mL). Samples were taken daily and the media replaced with washing every 48 h. Accumulated luciferase activity in the supernatants is plotted. Arrows indicate the addition of fresh inhibitor. (B) Visualization of HCV infection in primary human hepatocytes. MPCCs were transduced with lentiviruses expressing wild-type (wt) or mutant (C508Y) RFP-NLS-IPS HCV reporter. Twenty-four hours after transduction, MPCCs were infected with Jc1FLAG2(p7-nsGluc2A) or plasma from HCV-infected patients in the presence of heparin (5 IU), CaCl2 (9 mM), and MgCl2 (6 mM). Twelve hours following infection, virus was removed and MPCC medium was added. Unfixed MPCCs were imaged by wide-field fluorescence microscopy at 48 h postinfection. Representative pseudocolored fluorescent images are shown; white arrow heads show nuclear RFP, indicative of HCV infection. Scale bar: 20 μm.
To demonstrate that HCV actively replicates in the primary hepatocyte component of the MPCCs, we made use of a recently developed fluorescence-based live cell reporter (22). This system uses a reporter (RFP-NLS-IPS) composed of a red fluorescent protein (RFP), an SV40 nuclear localization sequence (NLS), and a C-terminal mitochondrial-targeting domain (IPS) derived from the IFN-β promoter stimulator 1 protein (IPS-1), a known cellular substrate for the HCV NS3-4A protease. The RFP-NLS-IPS substrate was stably expressed in primary hepatocyte MPCCs (Fig. 2B). In HCV-infected cells, RFP-NLS-IPS processing by NS3-4A results in translocation of the cleavage product, RFP-NLS, from the mitochondria to the nucleus. This redistribution of fluorescence was detected in approximately 1–5% of HCVcc-infected MPCC hepatocytes transduced with wild-type RFP-NLS-IPS; no relocalization was detected after infection of MPCCs harboring a cleavage-resistant reporter (C508Y) or in supporting fibroblasts (Fig. 2B). At very low frequency (approximately 1 in 30,000 cells), we also detected RFP-NLS-IPS cleavage in MPCC cultures infected with plasma or sera from HCV-infected patients (Fig. 2B).
Primary Hepatocytes in MPCCs Produce Infectious Virus.
To determine whether primary hepatocytes in MPCCs are capable of producing infectious virions, filtered culture supernatants were used to inoculate naïve Huh-7.5 cells, followed by staining for HCV protein (NS5A) at 72 h postinfection. Infectious virus was detected in MPCC supernatants harvested at day 4 postinfection and for all time points measured up to day 12 (Fig. S4). Supernatants from MPCCs infected in the presence of specific antiviral inhibitors did not yield NS5A-positive foci in Huh-7.5 cells, indicating that de novo virus production, rather than carry over of the inoculum, was detected. Attempts to passage MPCC-produced virus onto naïve MPCCs were unsuccessful due to the low titers produced by the primary cells. The low titers also precluded further biophysical analysis of the virus.
Proof of Principle for Preclinical Screening of Anti-HCV Therapeutics in MPCCs.
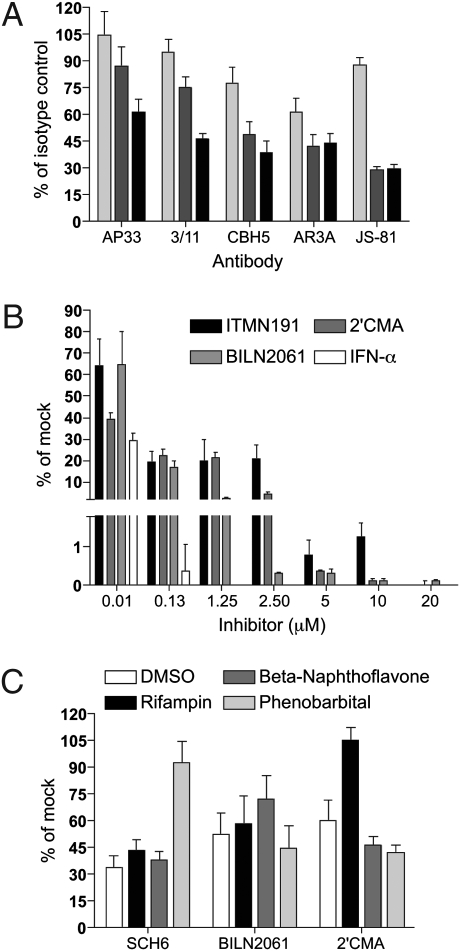
Persistently infected MPCCs may be a viable and relevant platform for preclinical screening of anti-HCV therapeutics. Antibodies blocking HCV entry factors, in particular CD81 and SCARB1, have been shown to be effective in vitro (23, 24) and in small animal models (25). We tested the ability of monoclonal antibodies against these cellular targets, as well as four antibodies specific for HCV E2, to inhibit HCVcc entry in MPCCs; none of these reagents had previously been tested in primary cell cultures. Anti-CD81 (JS-81) blocked HCVcc entry very efficiently (IC50 < 1 μg/mL), whereas anti-SCARB1 (C167) did not effectively inhibit viral uptake. All antibodies against E2 were able to inhibit HCVcc infection, although with varying efficiencies (IC50 for AP33 > 3/11 > CBH5 > AR3A) (Fig. 3A). A variety of specific antivirals targeting HCV enzymes are also under preclinical development. Currently, however, in vitro platforms capable of simultaneously assessing drug toxicity and efficacy are not widely available. We have previously shown the utility of MPCCs in drug metabolism and toxicity screening via assessment of gene expression profiles, phase I/II metabolism, canalicular transport, secretion of liver-specific products, and susceptibility to hepatotoxins (12). Here, we examined the use of MPCCs in evaluating antiviral efficacy (Fig. 3B). We measured HCV replication by luciferase activity at 4 days posttreatment with protease inhibitors (BILN2061 and ITMN191), polymerase inhibitor (2’CMA), or IFN-α. These compounds inhibited HCV replication in the submicromolar range, indicating the relevance of MPCCs for monitoring HCV inhibition. We then evaluated the efficacy of protease inhibitors (SCH-6 and BILN2061) and polymerase inhibitor (2’CMA) in HCVcc-infected MPCCs pretreated for 3 days with compounds known to modulate drug metabolism and other cellular functions in vivo (12) (Fig. 3C). We found that the addition of certain drugs severely reduced the efficacy of SCH-6 and 2’CMA, as compared to DMSO solvent control. Although the mechanisms underlying these adverse drug interactions remain unknown, these observations demonstrate the importance of conducting drug combination studies during in vitro efficacy assessment. These studies indicate that MPCCs may be well suited as a metabolically competent in vitro model of the liver, allowing HCV replication to be studied over several days to weeks, and a variety of intervention strategies to be tested for efficacy and toxicity.
Fig. 3.
Utility of primary human hepatocyte MPCCs in antibody and small molecule screening. (A) Dose-dependent inhibition of HCVcc replication in MPCCs treated with antibodies against HCV glycoproteins (AP33, 3/11, CBH5, AR3A) or cellular CD81 (JS-81). Antibody concentrations are 0.1 (light gray), 1 (dark gray), and 10 (black) μg/mL (B) Dose-dependent inhibition of HCVcc replication in MPCCs treated with IFN-α (up to 0.13 μM) or small molecules (NS3-4A protease inhibitors, BILN2061 and ITMN191, or polymerase inhibitor, 2’CMA). HCVcc-infected MPCCs were pulse-treated for 2 days with compounds and supernatants were collected at days 2 and 4 (shown) postinhibitor treatment. (C) Drug-drug interactions lead to reduced efficacy of small molecules in HCVcc-infected MPCCs. Infected MPCCs were treated for 3 days with prototypical inducers of drug metabolism enzymes (12, 32), followed by treatment of cultures with small molecules for 2 days. In all experiments, HCVcc replication was monitored by luciferase secretion into the supernatants. Mean and standard error of the mean are shown.
Discussion
Here we have described a microscale primary human hepatocyte in vitro culture platform that supports the entire HCV life cycle. Primary hepatocyte MPCCs are stable for several weeks and therefore allow monitoring of human hepatotropic infections over extended periods of time. Although this is an important step forward, limitations remain. Entry of HCVpp and tissue culture-derived virus into MPCC hepatocytes was inefficient. Although the four critical viral entry factors are present on these cells, it is possible that differences in their spatial distribution might account for the low uptake efficiency. Indeed, antibodies block entry into primary hepatocytes at different efficacies than previously reported (26 –28), possibly due to limited accessibility of the HCV entry factors on polarized cells. Furthermore, following isolation from the liver and disruption of hepatic polarity, it may be that in some hepatocytes in MPCCs, expression levels of critical viral entry factors on polarized membranes do not reach the threshold required with proper spatial localization for efficient viral uptake.
We also did not observe any increase in the number of infected cells over time, arguing for limited spread of HCV in the cultures. Several factors could contribute to this phenomenon, including limited numbers of infectious particles, heterogeneous polarity, or an inherent or acquired refractory nature of a proportion of cells. Furthermore, certain critical host factors may be heterogeneously expressed and therefore limiting in some cells, rendering them resistant to infection or unable to sustain HCV RNA replication. Although our data demonstrate that primary hepatocytes in MPCCs can produce infectious virus, the titers are low and few infectious virions are available for spread. In those cells that do become infected, HCV is capable of interfering with innate antiviral immunity via NS3-4A-mediated cleavage of critical signaling molecules, including IPS-1, TRIF, and IRF3 (29). Although this mechanism is presumably sufficient to blunt antiviral signaling and sustain replication, IFN production may not be entirely prevented, rendering adjacent cells nonpermissive. The low permissivity of MPCC hepatocytes to HCV may also reflect the in vivo reality of chronic hepatitis. Technical challenges have traditionally made it difficult to estimate the number of infected cells in an HCV-positive liver. Recently, however, two-photon microscopy methods have been used to determine that only a low proportion (7–20%) of ex vivo patient hepatocytes express viral antigens (30).
Although further improvements in infection efficiency may be possible, our system lays the foundation for preclinical assessment of antiviral therapeutics against human hepatotropic pathogens in a more physiologically relevant microenvironment. Importantly, due to the phenotypic stability of the MPCCs, infection processes can be monitored longitudinally, potentially allowing the kinetics of viral spread and antiviral signaling to be characterized at the single cell level. The polarized nature of the MPCC hepatocytes allows HCV entry and uptake inhibitors to be studied in the context of intact tight junction structures. Furthermore, using sera from HCV-infected patients and a very sensitive fluorescent reporter system (22), we were able to detect an extremely low frequency of productive infection—suggesting that a combination of authentic virus and host cells may be achievable. Proof of principle studies reported here also demonstrate the value of MPCCs in drug studies. The high baseline activities of drug metabolism enzymes (i.e., cytochrome P450s) and their drug-mediated induction/inhibition in MPCCs (12) allows for simultaneous measurements of drug efficacy, drug–drug interactions, and drug toxicity, thereby providing critical preclinical parameters. These advantages combine to make MPCCs a highly valuable system for studies of HCV biology.
Methods
Virus Genomes and Stocks.
Jc1FLAG2(p7-nsGluc2A) is a fully-infectious HCVcc reporter virus encoding Gaussia luciferase between p7 and NS2 (20). Virus stocks were created by electroporation and titered by limiting dilution as previously described (23).
Liver Sections and Hepatocytes.
Primary human hepatocytes were purchased from vendors permitted to sell products derived from human organs procured in the United States by federally designated Organ Procurement Organizations. Vendors included: Celsis In vitro Technologies, BD-Gentest and CellzDirect. Human hepatocytes were pelleted by centrifugation at 50–100 xg for 5–10 min at 4 °C, resuspended in hepatocyte culture medium, and assessed for viability using Trypan blue exclusion (typically 70–90%). Liver-derived nonparenchymal cells, as judged by size (<10 μm diameter) and morphology (nonpolygonal), were consistently found to be less than 1% in these preparations. Human liver sections were obtained from the NewYork-Presbyterian Hospital from uninfected donor tissue deemed unacceptable for liver transplantation. Tissue was processed by immediately freezing in OCT compound at -80°C or by fixation in 10% formalin solution for 24 h followed by paraffin embedding. Tissue sections were cut (∼5-6 μm) on poly-L-lysine coated slides. Human serum and plasma samples were obtained at Weill Cornell Medical Center. All protocols for human primary material procurement were approved by the Committee on Use of Human Experimental Subjects, MIT, or by the IRB, Rockefeller University and Weill Cornell Medical Center.
Micropatterned Co-cultures of Primary Human Hepatocytes and Supportive Stromal Cells.
Off-the-shelf tissue culture polystyrene (24- and 96-) or glass bottom (24-) multi-well plates, coated homogenously with rat tail type I collagen (25–50 μg/ml), were subjected to soft-lithographic techniques (12) to pattern the collagen into micro-domains (islands of 500 μm in diameter with 1200 μm center-to-center spacing). To create MPCCs, hepatocytes were seeded on collagen-patterned plates that mediate selective cell adhesion. The cells were washed with medium 2–3 h later (∼3x104 adherent hepatocytes in 96 collagen-coated islands in 24-well plate and ∼4.5x103 hepatocytes in 14 islands in 96-well plate) and incubated in hepatocyte medium overnight. Hepatocyte culture medium was DMEM with high glucose, 10% FBS, 0.5 U/ml insulin, 7 ng/ml glucagon, 7.5 μg/ml hydrocortisone and 1% penicillin-streptomycin. 3T3-J2 murine embryonic fibroblasts were seeded (9×104 cells in each well of 24-well plate and 1.4×104 cells in each well of 96-well plate) in fibroblast medium (DMEM with high glucose, 10% bovine serum and 1% penicillin-streptomycin) 12–24 h later. Fibroblast-to-hepatocyte ratio was estimated to be 4:1, once the fibroblasts reached confluency. Fibroblast culture medium was replaced with hepatocyte culture medium 24 h after fibroblast seeding and subsequently replaced daily. Control conventionally-plated pure hepatocyte cultures (Collagen gel sandwich, Matrigel overlay, Matrigel substratum, and randomly distributed co-cultures of hepatocytes and murine fibroblasts) were created as described previously (12).
Pseudoparticle Generation and Infection Assays.
Pseudoparticles were generated by cotransfection of plasmids encoding (1) an EGFP-encoding provirus (CSGW) or provirus encoding transgene (pTRIP) (2) HIV gag-pol and (3) envelope glycoprotein(s), as previously described (17). HCVpp were generated using H77 E1E2 (residues 170–746).
Antibodies, Immunostaining, and Blocking.
For immunostaining, cells or tissue sections were fixed in 1% paraformaldehyde and/or −20 °C methanol. Following washing and blocking in 1% BSA/0.2% milk or 1% BSA/0.3% Triton X-100 in PBS, cells were incubated in primary antibody overnight at 4 °C: mouse anti-human CD81 (clone JS-81, BD Pharmingen; 1:200), rabbit anti-SCARB1 (NB110-57591, Novus Biologicals; 1:100), rabbit anti-CLDN1 (51-9000, Zymed; 1:200), rabbit anti-ZO1 (61-7300, Zymed; 1:200), mouse anti-OCLN (33-1500, Zymed; 1:200), mouse anti-MRP2 (Clone M2III-6, Kamiya Biomedical; 1:50), mouse anti-EEA1 (clone 14, BD Biosciences; 1:100), mouse anti-NS5A (9E10 (23), 1:2000). Secondary antibodies were goat-anti-mouse or goat-anti-rabbit AlexaFluor488- or AlexaFluor594-conjugates (Invitrogen; 1:1000) for immunofluorescence, or goat-anti-mouse HRP (ImmPress kit, Vector Labs) with DAB+ substrate (Dako) for immunohistochemistry. Nuclei were detected using Hoechst dye (500 ng/mL in PBS, Invitrogen). Images were captured on a Nikon inverted microscope using SPOT image analysis software. Confocal images were captured on a Zeiss LSM 510 inverted microscope at 0.3 μm optical slices using Zeiss software (v3.2). Z-stack files were uploaded into ImageJ64 software with images generated using a “Sum Slices” projection and 3D renderings were done using Imaris software. Final images were assembled using Adobe Photoshop CS3 software. Blocking experiments used human anti-SCARB1 antibody C167 (24), anti-CD81 (clone JS-81; BD Pharmingen), anti-E2 3/11 (31), and antibodies kindly provided by A.H. Patel (University of Glasgow, Scotland) (AP33) (26), S.K. Foung (Stanford University, Palo Alto, CA) (CBH5) (27), and D.R. Burton (The Scripps Research Institute, La Jolla, CA) (AR3A) (28). Human IgG1 (clone MOPC-21), IgG4 (MOR6391), and rat IgG2a (MCA1124R) isotype control antibodies were purchased from AbD Serotec.
Supplementary Material
Acknowledgments
The authors thank Amelie Forest, Megan Holz, Michelle Hunter, Maryline Panis, Jodie Tassello, and Anesta Webson for excellent technical assistance, and Catherine Murray for superbly editing the manuscript. Alison North and The Rockefeller University Bioimaging Core Facility provided outstanding technical support. The authors also thank Ira Jacobson, Queeny Brown, Ray Peterson, and Steve Gonzales (Weill Cornell Medical College), Luis Chiriboga, and Herman Yee (New York University) for assistance with the histological analysis, as well as Robert Brown and Raghu Varadarajan (Columbia University), who kindly provided primary liver tissue for histological analysis. This work was supported by the Greenberg Medical Research Institute, the Ellison Medical Foundation, the Starr Foundation, the Ronald A. Shellow Memorial Fund, the Richard Salomon Family Foundation, and funded in part by a Grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative (grants to C.M.R.), and the National Institutes of Health (grants R01 AI075099 to C.M.R, R01 DK56966 to S.N.B., and NRSA DK081193 to C.T.J.). C.M.R. is an Ellison Medical Foundation Senior Scholar in Global Infectious Diseases. This work was funded by the National Institutes of Health through the National Institutes of Health Roadmap for Medical Research, Grant 1 R01 DK085713-01 (to C.M.R., S.N.B., and A.P.). Information on this Roadmap Transformative R01 Program can be found at http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-08-029.html. A.P. is a recipient of a Kimberly Lawrence-Netter Cancer Research Discovery Fund Award. S.N.B. is an HHMI investigator.
Footnotes
The authors declare no conflict of interest. Materials used as controls in this study, the HCVcc cell culture virus system and Huh-7.5 hepatoma cells and reporter derivatives, were created at Washington University or Rockefeller University. These were then licensed to a commercial entity, Apath LLC, in which C.M.R. holds equity.
S.R.K and S.N.B have equity in Hepregen Corporation, which holds commercial licenses from the Massachusetts Institute of Technology for micropatterned co-cultures (MPCCs) and other related microscale liver technologies.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915130107/DCSupplemental.
References
- 1.Durantel D, Zoulim F. Going towards more relevant cell culture models to study the in vitro replication of serum-derived hepatitis C virus and virus/host cell interactions? J Hepatol. 2007;46:1–5. doi: 10.1016/j.jhep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Hsu IC, et al. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis. 1993;14:987–992. doi: 10.1093/carcin/14.5.987. [DOI] [PubMed] [Google Scholar]
- 3.Nagao K, et al. Expression of hTERT mRNA in a mortal liver cell line during S phase without detectable telomerase activity. Int J Mol Med. 2005;15:683–688. [PubMed] [Google Scholar]
- 4.Yokoo H, et al. Proteomic signature corresponding to alpha fetoprotein expression in liver cancer cells. Hepatology. 2004;40:609–617. doi: 10.1002/hep.20372. [DOI] [PubMed] [Google Scholar]
- 5.Fournier C, et al. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J Gen Virol. 1998;79:2367–2374. doi: 10.1099/0022-1317-79-10-2367. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: Cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 7.Carloni G, et al. Susceptibility of human liver cell cultures to hepatitis C virus infection. Arch Virol Suppl. 1993;8:31–39. doi: 10.1007/978-3-7091-9312-9_4. [DOI] [PubMed] [Google Scholar]
- 8.Iacovacci S, et al. Replication and multiplication of hepatitis C virus genome in human foetal liver cells. Res Virol. 1993;144:275–279. doi: 10.1016/s0923-2516(06)80040-3. [DOI] [PubMed] [Google Scholar]
- 9.Rumin S, et al. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro . J Gen Virol. 1999;80:3007–3018. doi: 10.1099/0022-1317-80-11-3007. [DOI] [PubMed] [Google Scholar]
- 10.Buck M. Direct infection and replication of naturally occurring hepatitis C virus genotypes 1, 2, 3 and 4 in normal human hepatocyte cultures. PLoS One. 2008;3:e2660. doi: 10.1371/journal.pone.0002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lázaro CA, et al. Hepatitis C virus replication in transfected and serum-infected cultured human fetal hepatocytes. Am J Pathol. 2007;170:478–489. doi: 10.2353/ajpath.2007.060789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 13.Sivaraman A, et al. A microscale in vitro physiological model of the liver: Predictive screens for drug metabolism and enzyme induction. Curr Drug Metab. 2005;6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 14.LeCluyse EL. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur J Pharm Sci. 2001;13:343–368. doi: 10.1016/s0928-0987(01)00135-x. [DOI] [PubMed] [Google Scholar]
- 15.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, et al. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 19.Scarselli E, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marukian S, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiffler JD, et al. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010 doi: 10.1038/nbt.1604. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 24.Catanese MT, et al. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. J Virol. 2007;81:8063–8071. doi: 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meuleman P, et al. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 26.Owsianka A, et al. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol. 2005;79:11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadlock KG, et al. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74:10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 29.Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 30.Liang Y, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 31.Flint M, et al. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madan A, et al. Effects of prototypical microsomal enzyme inducers on cytochrome P450 expression in cultured human hepatocytes. Drug Metab Dispos. 2003;31:421–431. doi: 10.1124/dmd.31.4.421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.