Abstract
Recent genomewide association studies have found multiple genetic variants on chromosome 8q24 that are significantly associated with an increased susceptibility to prostate, colorectal, and breast cancer. These risk loci are located in a ”gene desert,” a few hundred kilobases telomeric to the Myc gene. To date, the biological mechanism(s) underlying these associations remain unclear. It has been speculated that these 8q24 genetic variant(s) might affect Myc expression by altering its regulation or amplification status. Here, we show that multiple enhancer elements are present within this region and that they can regulate transcription of Myc. We also demonstrate that one such enhancer element physically interacts with the Myc promoter via transcription factor Tcf-4 binding and acts in an allele specific manner to regulate Myc expression.
Frequent rearrangements and amplifications on the long arm of human chromosome 8 have long been linked to cancer (1–6). More recently, several genome-wide association studies (GWAS) have found multiple single nucleotide polymorphisms (SNPs) on 8q24 that are associated with an increased risk for cancer (7–14). After compiling these data, three adjacent genomic blocks on 8q24 were identified with an increased risk for prostate cancer (15). These three areas were termed regions 1, 2, and 3, based on their time of discovery, with the most significant SNPs being rs1447295 in region 1, rs16901979 in region 2, and rs6983267 in region 3 (15). In addition to an increased susceptibility to prostate cancer, SNP rs6983267 in region 3 has also been found to be associated with an increased colorectal cancer risk (12–14), whereas SNP rs13281615 in region 3 has been found to be associated with an increased breast cancer risk (16). Together, regions 1, 2, and 3 span ≈600 kbp. This area has been termed a “gene desert,” with relatively few predicted genes, including DQ515897, DQ486513, CB104826, and pseudogene POU5F1P1, which is homologous to transcription factor POU5F1. The closest cancer associated gene, the proto-oncogene Myc, is located ≈263 kb telomeric to rs1447295 in region 1 and 624 kb telomeric to rs16901979 in region 2, respectively. It has been speculated that the associated genetic variants identified in these studies could affect genomic instability, or alter the transcriptional regulation of causal genes located outside of the region (17). Although the associated variants are far away from Myc, it is possible that they could regulate Myc expression as enhancers, and other regulatory elements have been demonstrated to operate as far as 1 Mb away from their target genes (18, 19). Consistent with these data, it has been reported that the in vivo expression of the Myc gene depends on distal enhancer elements (20) and that some of these enhancer elements have been found up to 37 kb upstream of Myc gene (21). It has also been determined that long-range chromosome and chromatin integrity are essential for proper control of Myc transcription and that sequences in excess of 50 kbp upstream of Myc are necessary for this control (22). Even today, transcriptional regulation of Myc remains enigmatic, with the Myc promoter and upstream regulatory elements remaining poorly understood (22). What is clear, however, is that Myc regulation is extremely complex, as would be expected from a gene whose expression is capable of inducing either the proliferative or apoptotic/senesce response (23). Here, we show that multiple enhancer elements are present within the genetically identified cancer-associated regions on chromosome 8q24 and that they can regulate transcription of Myc. We also demonstrate that one such enhancer element physically interacts with the Myc promoter via transcription factor Tcf-4 binding and acts in an allele specific manner to regulate Myc expression.
Results and Discussion
Computational Prediction of Enhancer Elements in 8q24 Cancer-Associated Regions.
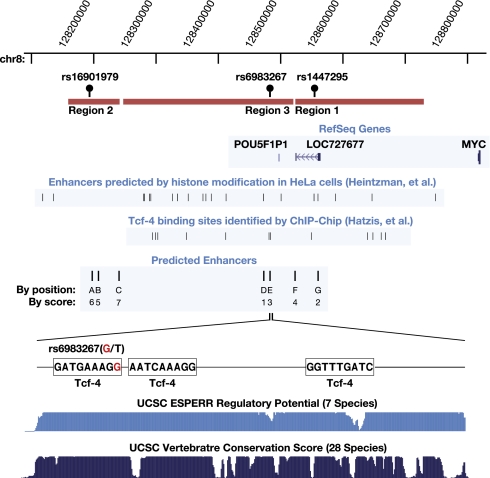
To identify evolutionarily conserved regulatory elements in the genetically identified cancer-associated regions on 8q24, we aligned the human genome sequence to dog and mouse orthologs. We then used computational tools, including enhancer element locator (EEL) to screen for potential evolutionarily conserved enhancer elements (21). EEL computes enhancer module scores based on the alignment of transcription factor (TF) binding sites instead of direct DNA sequence alignment, in a manner similar to aligning peptide sequences instead of the corresponding DNA sequence to find conserved protein regions. Such a strategy has been successfully employed in the past to identify enhancers for both the c-Myc and n-Myc genes (21). We chose the top seven enhancer candidates based in regions 1, 2, and 3, of the broad genetically identified prostate cancer region, for follow-up. Other computational information was also incorporated, including the conserved blocks predicted by the PhastCons mammalian conservation score and the ESPERR seven species regulatory potential, as described on the University of California, Santa Cruz (UCSC) genome website. Most of the seven enhancers are located within these highly conserved blocks. The EEL predicted enhancer modules were extended to include the whole conserved block. The locations of these enhancer modules, termed (A–G) are shown in Fig. 1. Many of the cancer-associated SNPs are located within, or in close proximity to, these predicted enhancer modules. For example, rs6983267, the SNP most strongly associated with prostate and colon cancer in region 3, is located within enhancer E (Fig. 1). The importance of this enhancer is underscored by its evolutionary conservation (Fig. 1), including 100% sequence conservation among greater primates (SI Text). The presence and location of these predicted enhancer elements is consistent with recent data from Heintzman et al. (24), who used histone modification patterns to predict the location of cell-type specific enhancers. Analysis of their data reveals partial overlap between the enhancers predicted by their method with those identified by our algorithm (Fig. 1).
Fig. 1.
Computationally predicted enhancers in the 8q24 cancer-associated gene desert region. Enhancers were predicted by aligning the human 8q24 genomic sequence with dog and mouse orthologs by using EEL. A physical map of 8q24, including Myc and 800 kb enveloping the 8q24 cancer associated region, are shown. The three prostate cancer-associated regions not in LD are shown in orange. The three most significant SNPs (rs16901979, rs6983267, and rs1447295) are illustrated above the region. Enhancers predicted by Heintzman et al. (24) and Tcf-4 binding sequences identified by Hatzis et al. (25) are indicated. The top seven predicted enhancers (along with their relative scores) are indicated as A–G on the map. Enhancer E is shown in detail together with its three predicted Tcf-4 binding sites, conservation score, and regulatory potential score from UCSC genome database.
A Luciferase Reporter Assay Demonstrates Enhancer Functionality in Both Prostate and Colon Cells.
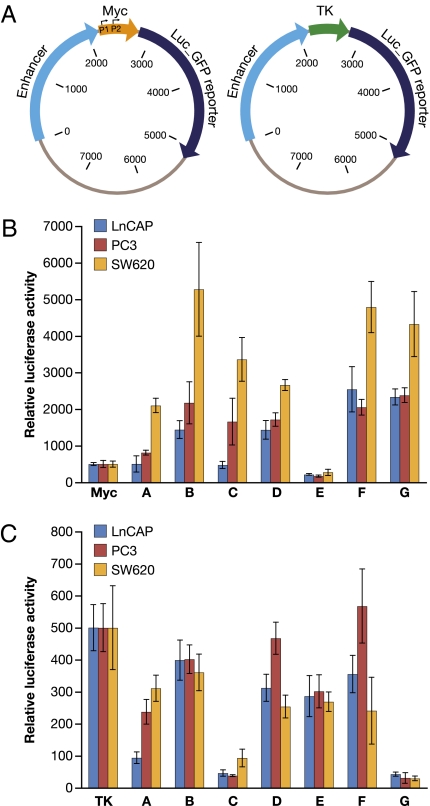
To determine whether the computationally identified enhancers possess biological functionality, they were cloned upstream of either a basal thymidine kinase (TK) promoter-driven luciferase reporter construct or a native Myc promoter driven luciferase (Luc) reporter construct (Fig. 2A). Although not as strong as the SV40 or the CMV promoter, the TK promoter is well characterized and has been used extensively in similar reporter assays (26–28). The Myc proximal promoter is less well-defined (22, 29). For the purpose of these experiments, we used a basal Myc promoter that starts 55 bp upstream of exon 1 and includes the dual P1/P2 promoters, but not many of the transcription factor binding sites are located more upstream such as the Tcf-4 binding elements (TBE1, TBE2, and TBE3) (30). Two random DNAs from 8q24, and one other random DNA from elsewhere in the genome, were used as negative controls and to determine the baseline for enhancer activity. A CMV enhancer construct was used as a positive control for enhancer activity. Reporter constructs were transfected into prostate cancer cell lines LnCAP, PC3, and colorectal cell line SW620. As readout of enhancer activity, luciferase activity was measured 24 h after transfection (Fig. 2 B and C). As expected, the CMV enhancer greatly increases both Myc promoter-Luc-reporter and TK promoter-Luc-reporter activities ≈100-fold (Fig. S1). Most of the putative enhancers stimulate Myc promoter-Luc-reporter activity as shown in Fig. 2B, except for enhancer E, which produced little to no Myc promoter-Luc-reporter activity in any of the three cell lines. Most of the enhancers increased Myc promoter-Luc-reporter activity to a greater degree in SW620 cells than in LnCAP and PC3 cells, possibly due to an APC mutation in SW620 that leads to constitutive activation of the Myc pathway (30, 31). Surprisingly, most of the enhancers did not increase TK promoter-Luc-reporter activity, with some (enhancer C and G) significantly repressing reporter activity (Fig. 2C). These results suggest selective interaction of the enhancers with basal core or proximal promoter elements. However, regardless of stimulation or repression, the change in reporter activity indicates that these putative enhancers may have regulatory potential.
Fig. 2.
Enhancer reporter assay. (A) The enhancers were cloned in front of a Myc promoter-driven or TK promoter-driven luciferase-GFP reporter. (B) Luciferase activity of enhancer-Myc-luciferase constructs in prostate cell line LnCAP, PC3, and colorectal cell line SW620. (C) Luciferase activity of enhancer-TK-luciferase constructs in prostate cell line LnCAP, PC3, and colorectal cell line SW620. Enhancer reporter plasmids were cotransfected into cell lines as described in Materials and Methods. Luciferase activity was measured 24 h after transfection by using the Dual-Glo Luciferae assay system.
Enhancer E Activity Is Mediated by β-Catenin/Tcf-4.
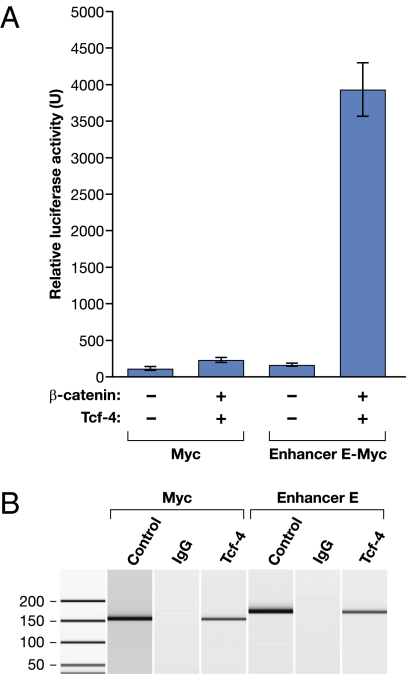
The SNP most strongly associated with prostate and colon cancer susceptibility is contained within the computationally identified enhancer E module and, as such, warranted further investigation. Regulation of Myc expression has been shown to be mediated via the Wnt signaling pathway through the binding of β-catenin and the transcription factor Tcf-4 (30). Enhancer E contains three conserved Tcf-4 binding elements (TBEs). Therefore, we reasoned that enhancer E may function through β-catenin/Tcf-4 interaction. To test this hypothesis, we measured the ability of enhancer E to stimulate luciferase activity directed by a Myc promoter-Luc-reporter in LnCAP cells cotransfected with Tcf-4 and/or β-catenin expression construct(s). We found that enhancer activity was slightly enhanced by β-catenin or Tcf-4 alone (Fig. S2) but was significantly elevated by the coexpression of both β-catenin and Tcf-4 (Fig. 3A).
Fig. 3.
Enhancer E interacts with Tcf-4 in vivo. (A) β-catenin/Tcf-4 can stimulate enhancer E activity in the luciferase reporter assay. Luciferase activity of Myc-luciferase constructs (with and without enhancer E) in prostate cell line LnCAP with (+)/without (−) β-catenin and/or Tcf-4. (B) Tcf-4 binds to enhancer E in LnCAP cells in a ChIP assay. ChIP was performed (lanes 2, 3, 5, and 6) by using genomic LnCAP DNA, a Tcf-4 monoclonal antibody (Tcf-4), or total mouse IgG (IgG), and PCR primers directed against the Myc promoter (Myc) or enhancer E as indicated. Lanes 1 and 4 contain positive control PCR products generated from input genomic DNAs.
Tcf-4 Binds to Enhancer E.
To further confirm that enhancer E functions through Tcf-4 interaction, we performed the chromatin immunoprecipitation (ChIP) assay in LnCAP cells by using an anti-Tcf-4 antibody. The Myc proximal promoter is known to contain three TBEs and therefore was used as a positive control in this assay (22). Anti Tcf-4-immunoprecipitated DNAs were PCR-amplified by using primers directed against either the Myc promoter or enhancer E. Fig. 3B shows that Myc and enhancer E DNAs are both enriched after Tcf-4 immunoprecipitation, suggesting physical binding of Tcf-4 to enhancer E. These findings are consistent with those reported by Hatzis et al. (25), who reported 6,868 high confidence Tcf-4 binding sites in a Tcf-4 ChIP-on-Chip study of colorectal cell line LS174T. We have analyzed these data and determined that one of the identified sites locates within the 8q24 region of interest and overlaps with enhancer E (Fig. 1). These results suggest that Tcf-4 binds to enhancer E in both prostate and colorectal cell lines.
Enhancer E Comes in Close Physical Contact with the Myc Promoter.
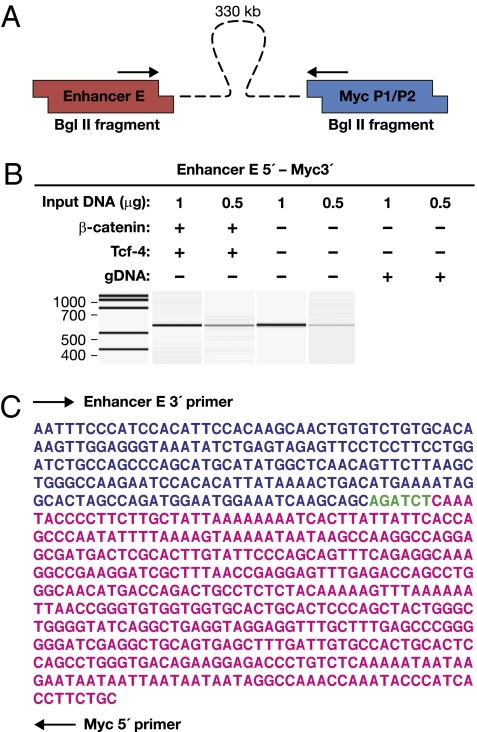
An important question, related to the understanding of long-range enhancer function, is in what manner do enhancers physically communicate or interact with promoters. The prevailing model is that enhancer-bound protein factors interact with promoter-bound factors, bringing the enhancers in close physical contact with the promoter, and as a result, causing the intervening DNA to “loop out” (17). In an effort to demonstrate physical interaction between enhancer E and the Myc promoter, we employed the chromosome conformation capture (3C) assay (17). In this assay, physical interaction involving disparate genomic regions are first preserved by formaldehyde cross-linking as in the ChIP assay. The cross-linked chromatin is then digested with a restriction enzyme and ligated under dilute conditions to promote ligation of fragments within close proximity. A novel ligation product will form between two previously faraway regions if the interactions bring the two regions together. The presence of these product(s) can be assayed by PCR. We used BglII as the restriction enzyme in this study. PCR primers were designed near the end of BglII fragments that include the Myc promoter or enhancer E (Fig. 4A). The 3C assay was performed by using LnCAP cells both with and without β-catenin/Tcf-4 transfection. Fig. 4B shows that a ligation product was detected between enhancer E and the Myc promoter, which are located >340 kb apart. A genomic DNA control did not yield such a band (Fig. 4B). The PCR products were sequenced and confirmed to be the expected Myc promoter-enhancer E junction products (Fig. 4C). Further, using the same amount of input DNA, more such PCR product was generated in the β-catenin/Tcf-4 transfected cells as compared to untransfected cells, suggesting that the interaction is strengthened by β-catenin/Tcf-4 interaction (Fig. 4B).
Fig. 4.
Enhancer E forms close contact with Myc promoter in 3C assay. (A) The 3C primer locations. (B) PCR results in the presence or absence of β-catenin and/or Tcf-4 by using the indicated amounts of LnCAP BglII digested/ligated genomic DNA or nondigested/ligated genomic DNA (gDNA). The concentration of DNA templates for PCR amplifications was determined by UV absorbance and confirmed by qPCR. (C) Sequence of the recovered PCR product with primer locations and BglII site are indicated.
SNP rs6983267 Is a Functional Variant of Enhancer E Activity.
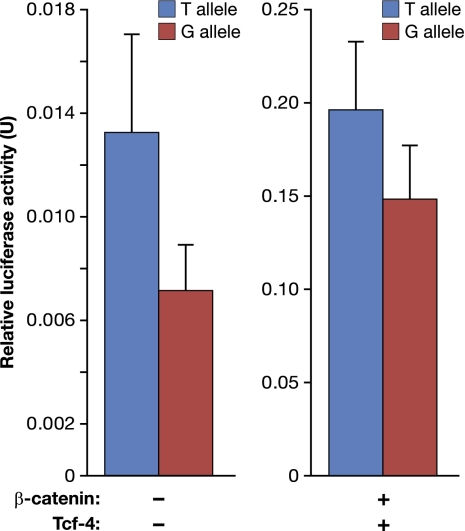
Although the majority of genetic variants identified as a part of GWAS association studies serve to merely indicate the general physical location of important causative loci, some may be true functional variants. This is much easier to decipher if the variants are located within protein coding regions or well characterized regulatory elements. SNP rs6983267, the marker most strongly associated with prostate and colon cancer in region 3, is located within enhancer E with the polymorphic locus (G > T) overlapping a conserved TBE (Fig. 1). We reasoned that SNP rs6983267 may influence enhancer E activity. To directly test this hypothesis, we constructed enhancer E-Myc-Luc luciferase reporter constructs, similar to those described in Figs. 2 and 3, containing either the T or G allele of SNP rs6983267. As shown in Fig. 5, after transfection into LnCAP cells, the T allele consistently stimulated luciferase activity to a greater extent than the G allele in both the presence and absence of supplemental β-catenin/Tcf-4. Although the effect of SNP rs6983267 is not large, it was found to be both highly reproducible and significant through multiple repetitions, with P < 0.0022 (Datasets S1–S4).
Fig. 5.
SNP rs6893267 influences enhancer E activity. Luciferase activity of enhancer-Myc-luciferase constructs in prostate cell line LnCAP with or without β-catenin/Tcf-4 (25 ng of each plasmid). The G and T allele variants of enhancer E are indicated. Multiple repeats were performed for each allele with statistically significant P values of P < 0.0022 (Datasets S1–Dataset S4).
Concluding Remarks.
These data demonstrate that there are enhancer elements located within the cancer-associated regions on chromosome 8q24 and that these elements can regulate Myc promoter activity in a reporter assay (Fig. 2). One of these enhancers showed responsiveness to stimulation with a known Myc transcription factor (Tcf-4) (Fig. 3) and was demonstrated to form close physical contact with Myc promoters in vivo via interaction Tcf-4 (Fig. 4). These data are supported by reports from other laboratories that: (i) predict the presence of enhancer elements which overlap with those presented here (ref. 24; Fig. 1), and (ii) demonstrate, by ChIP-on-Chip experiments, that Tcf-4 binds to DNA sequences that include one of the enhancers which we describe (ref. 31; Fig. 1). Finally, we have shown that a previously identified cancer risk locus, SNP rs6983267, located within this enhancer, acts as a functional variant in the regulation of Myc transcription (Fig. 5).
At first glance, the functional variation demonstrated for SNP rs6983267 may appear contradictory. The G allele is associated with increased cancer risk (12–15), yet the T allele confers ≈2-fold increased Myc transcription (Fig. 5). This apparent paradox can be understood when viewed in the light of Myc being a prototypical example of a phenomenon termed “intrinsic tumor suppression” (23, 32). The term intrinsic tumor suppression was coined by Lowe et al. (32) to describe the observed tight coupling of cellular proliferative programs to tumor suppression (i.e., senescence/apoptosis). A recent publication by Murphy et al. (23) provides an excellent illustration of this phenomenon with respect to Myc. They have directly demonstrated, in an in vivo mouse model, that low levels of Myc transcription drives cell proliferation, whereas subtly higher levels induces ARF-mediated senescence/apoptosis. In light of these data, we propose that it may be more appropriate to view SNP rs6983267 as less of a cancer “susceptibility” locus and more of a cancer “protective” locus. These data provide a biological link to the observed association of common genetic variation on chromosome 8q24 and the risk of cancer. It seems likely that one or more enhancer elements are located in this region and that they function to regulate Myc transcription. Furthermore, given that the G allele of SNP rs6983267 is found to be significantly predominant in the African-American population, whereas the T allele is significantly predominant in the European-American population (www.ncbi.nlm.nih.gov), it is possible that the functional variation found to be associated with this locus could contribute to the higher incidence rate of prostate cancer in African-Americans as compared to European-Americans. However, further studies will be needed to evaluate how much the genetic variation located within and around these enhancer elements actually contributes to cancer development and progression.
Materials and Methods
Computational Prediction of Enhancers in 8q24.
The genomic DNA sequence of the human 8q24 cancer-associated regions was downloaded from the UCSC genome web site. The corresponding mouse and dog genomic regions were extracted according to the blastz pairwise alignment tables available from UCSC genome web site. The sequences for prostate cancer-associated region1 and region3 are in close physically proximity and were processed together with the EEL program. For region 1/3, ≈150 kb region of human genome sequence covering SNP rs1562871 to rs4407842 (hg18 chr8:128470954–128619305) and its corresponding dog and mouse genomic sequences were used as input for the EEL program. For prostate cancer-associated region 2, ≈100 kb of human genome sequence surrounding rs16901979 (hg18 chr8:128144098–128244098) and its corresponding dog and mouse genomic sequences were processed by using EEL. Enhancer predictions were performed with human-dog and human-mouse pair-wise alignment separately. The top enhancers predicted in each region were compared across all three species. The enhancer modules were extended to include the most adjacent blocks with high conservation scores by using PhastCons conservation track and 7× regulatory potential track at the UCSC genome web site. The top four enhancers in region 1/3 and top three enhancers in region 2 were chosen for experimental validation. Sequence and detailed alignments are described in SI Text.
Construction of Enhancer Reporter Plasmids.
The top seven enhancers were PCR amplified from human genomic DNA (Promega) and cloned. Primer sequences are listed in SI Text. The enhancers were placed in front of a proximal Myc-promoter driven- or TK-promoter driven-luciferase reporter gene. The Myc promoter used in our study is a 766-bp fragment starting 55 bp upstream of exon1 and includes the alternative promoters P1 and P2. This promoter does not contain the 3 Tcf-4 binding elements (TBE1, TBE2, and TBE3) described by He et al. (30). The promoter sequence was amplified from a human genomic DNA library. The TK promoter was derived from a commercial vector, pRL-TK (Promega). Promoters and enhancers were cloned into Multisite Gateway Entry clones by PCR amplification using Gateway attB site-containing primers. Enhancers were flanked by att4 and att1 sites, whereas the promoter-reporter fusions were flanked by att1 and att2 sites. The promoter-reporter fusions were constructed by overlap PCR, which merged the Myc or TK promoters with the firefly luciferase (ffLuc2)-eGFP fusion reporter. The PCR products were cloned by using the Gateway BP reaction (Invitrogen) and final Entry clones were fully sequence verified. To generate the final expression clones, combinations of enhancers and promoter-reporters were merged by using LR Clonase Plus (Invitrogen) into pDest-302, a minimal Gateway Multisite Destination vector that contains no eukaryotic promoter elements, and only a downstream poly(A) sequence and bacterial origin/antibiotic resistance markers. Thus, final expression clones contain only the enhancer-promoter-reporter-poly(A) region for mammalian expression. As controls, clones were constructed, which contained an out-of-frame CAT gene in place of the enhancer or “random” DNA sequences from regions of 8q24, which did not show predicted enhancers.
In the case of enhancer E, site-directed mutagenesis was performed to create the alternative SNP allele, and to knock out predicted Tcf4 binding sites. Mutagenesis was carried out by using the QuikChange kit (Stratagene) on the Entry clones, and mutant clones were fully sequence verified before subcloning. A full list of Entry and Expression clones can be found in the SI Text. All expression clones were prepared for transfection by using the GenElute HQ Maxiprep kit (Sigma).
Cell Culture, Transfections and Luciferase/Renilla Quantification.
LNCAP, PC3, and SW620 were plated in 24-well plates and were used at 50–70% confluence. Firefly luciferase plasmids (250 ng) were transfected with Transit Prostate transfection kit (Mirus) for prostate cell lines or lipofectamine for SW620 according to manufacturer recommendations. Cotransfection of a CMV-driven Renilla luciferease construct was used to normalize for cell content in every well. In all cases 24 h of incubation after transfection was used. Detection of the activity of both luciferases was carried out sequentially with the Dual-Glo Luciferae assay system (Promega) by using a Tecan luminometer. Data were analyzed by using Excel (Microsoft).
Chromosome Conformation Capture-PCR.
Chromosome conformation capture to detect interaction between enhancer areas and myc promoter in vivo was performed according to Hagège et al. (33). Two different conditions were analyzed: control and cells transfected with TCF-4 and β-catenin at the conditions specified previously. Two million cells were cross-linked with 1% formaldehyde in the presence of 10% FBS for 10 min at room temperature. Quenching was done with ice-cold glycine, and the cells were lysed on ice. Collected nuclei were digested with 400 U of BglII restriction enzyme overnight at 37 °C with rotation. Ligation was performed for 4 h at 16 °C and 30 min at room temp with 100 U of T4 DNA ligase. Decross-linking in the presence of protenase K was done overnight at 65 °C. After RNase digestion the DNA was purified by using phenol-chloroform. Concentration of template DNA was assessed with SYBR green qPCR by using GAPDH primers internal to BglII sites in comparison with Roche human DNA.
PCR was performed by using 0.5 μg to 5 μg of template DNA using a touchdown protocol, lowering the annealing temperature from 60 °C to 55 °C in two cycle steps, followed by 25 cycles of amplification. Semiquantitative analysis of ligation products (1 μl) was assessed by capillary electrophoresis on an Agilent Bioanalyzer using a DNA1000 DNA chip. The products with expected size were sequenced to determine the exact location of the ligation event. Undigested DNA and primers directed to random genomic locations in proximity to a BglII restriction site were used as controls of nonspecific ligations.
Chromatin Immunoprecipitation for Tcf-4.
LnCap cells were grown to subconfluency to reach a total of 2 × 107 cells. ChIP was performed by using the EZ-ChIP kit from Upstate (Millipore) according to manufacturer specifications. The protein/DNA complexes were cross-linked with 1% formaldehyde in growth media for 10 min at room temperature. After quenching with glycine, cells were lysed in SDS buffer in the presence of protease inhibitor mixture. Shearing of DNA by sonication was performed at conditions that yielded a DNA smear on agarose gels ranging from 100 to 1,000 bp in size. Aliquots containing 2 million cell equivalents were stored at −80 °C to use in each immunoprecipitation. After preclearing of Protein G agarose, a 1% aliquot of supernatant was collected as input DNA. Immunoprecipitation was carried out in the presence of protease inhibitors in a rocker overnight at 4 °C. One microgram of mouse IgG was used as negative control and 1 μg of anti-PolII as positive control. Tcf4 ChIP grade monoclonal antibodies (Upstate) were used at two different concentrations (3 and 6 μg) (13). Syber green qPCR was used to quantify the presence of GAPDH, SP5, MYC, and enhancer E in each immunoprecipitate.
Supplementary Material
Acknowledgments
The authors thank Dr. B. Vogelstein for sharing the Tcf4 β-catenin plasmids and Drs. Robert Eisenman and David Levens for helpful comments and suggestions. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906067107/DCSupplemental.
References
- 1.Cher ML, et al. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996;56:3091–3102. [PubMed] [Google Scholar]
- 2.Visakorpi T, et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- 3.Korsmeyer SJ. Chromosomal translocations in lymphoid malignancies reveal novel proto-oncogenes. Annu Rev Immunol. 1992;10:785–807. doi: 10.1146/annurev.iy.10.040192.004033. [DOI] [PubMed] [Google Scholar]
- 4.Douglas EJ, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- 5.Ribeiro FR, et al. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–3970. doi: 10.1158/1078-0432.CCR-05-1977. [DOI] [PubMed] [Google Scholar]
- 6.Ried T, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 8.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 10.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlinson I, et al. CORGI Consortium. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 13.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 14.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witte JS. Multiple prostate cancer risk variants on 8q24. Nat Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 16.Easton DF, et al. SEARCH collaborators kConFab AOCS Management Group. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 18.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 19.Qin Y, et al. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- 20.Lavenu A, Pournin S, Babinet C, Morello D. The cis-acting elements known to regulate c-myc expression ex vivo are not sufficient for correct transcription in vivo. Oncogene. 1994;9:527–536. [PubMed] [Google Scholar]
- 21.Hallikas O, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Wierstra I, Alves J. The c-myc promoter: Still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- 23.Murphy DJ, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzis P, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger A, Salazar G, Le Cam A, Kervran A. Characterization of an enhancer element in the proximal promoter of the mouse glucagon receptor gene. Biochim Biophys Acta. 2001;1517:236–242. doi: 10.1016/s0167-4781(00)00279-7. [DOI] [PubMed] [Google Scholar]
- 27.Stewart CL, Schuetze S, Vanek M, Wagner EF. Expression of retroviral vectors in transgenic mice obtained by embryo infection. EMBO J. 1987;6:383–388. doi: 10.1002/j.1460-2075.1987.tb04766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner EF, Vanek M, Vennström B. Transfer of genes into embryonal carcinoma cells by retrovirus infection: efficient expression from an internal promoter. EMBO J. 1985;4:663–666. doi: 10.1002/j.1460-2075.1985.tb03680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Levens D. Making myc. Curr Top Microbiol Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- 30.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 31.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 32.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 33.Hagège H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.