Abstract
Class switch recombination (CSR) in B lymphocytes is initiated by introduction of multiple DNA double-strand breaks (DSBs) into switch (S) regions that flank immunoglobulin heavy chain (IgH) constant region exons. CSR is completed by joining a DSB in the donor Sμ to a DSB in a downstream acceptor S region (e.g., Sγ1) by end-joining. In normal cells, many CSR junctions are mediated by classical nonhomologous end-joining (C-NHEJ), which employs the Ku70/80 complex for DSB recognition and XRCC4/DNA ligase 4 for ligation. Alternative end-joining (A-EJ) mediates CSR, at reduced levels, in the absence of C-NHEJ, even in combined absence of Ku70 and ligase 4, demonstrating an A-EJ pathway totally distinct from C-NHEJ. Multiple DSBs are introduced into Sμ during CSR, with some being rejoined or joined to each other to generate internal switch deletions (ISDs). In addition, S-region DSBs can be joined to other chromosomes to generate translocations, the level of which is increased by absence of a single C-NHEJ component (e.g., XRCC4). We asked whether ISD and S-region translocations occur in the complete absence of C-NHEJ (e.g., in Ku70/ligase 4 double-deficient B cells). We found, unexpectedly, that B-cell activation for CSR generates substantial ISD in both Sμ and Sγ1 and that ISD in both is greatly increased by the absence of C-NHEJ. IgH chromosomal translocations to the c-myc oncogene also are augmented in the combined absence of Ku70 and ligase 4. We discuss the implications of these findings for A-EJ in normal and abnormal DSB repair.
Keywords: class switch recombination; DNA double-strand repair; nonhomologous end-joining, switch region internal deletions
Ig molecules are comprised of identical heavy (IgH) and light (IgL) chains. When activated via an immune response or appropriate stimulation in culture, IgM-producing B cells undergo class switch recombination (CSR) to express other IgH classes (IgG, IgE, IgA, and others). CSR changes the expressed constant- (C) region exons from Cμ to one of a set of downstream C-region exons (e.g., Cγ, Cε, Cα, among others, generically called “CH genes”) via a recombination/deletion mechanism (1, 2). CSR involves introduction of DNA double-strand breaks (DSBs) into long, repetitive switch (S) regions that lie upstream of each CH gene. Activation-induced cytidine deaminase (AID) (3) initiates the DSBs with initial AID lesions being processed into DSBs (4, 5). AID activity is targeted via transcription to the donor S region (Sμ) upstream of Cμ and to a downstream acceptor S region (2). CSR is completed when an Sμ DSB is joined to an acceptor S-region DSB, leading to deletion of Cμ and intervening DNA and thereby juxtaposing the downstream CH gene to an IgH variable region exon. AID activity also leads to somatic hypermutation (SHM) of S regions and variable region exons (3).
AID activity generates multiple DSBs within a given S region (4–6). Such DSBs might have several fates: They may be re-ligated, joined to another DSB in the same S region, or (for Sμ) joined to a DSB in a downstream S region to effect CSR (7). Rejoining DSBs within an S region, accompanied by resection or joining two DSBs within an S region, can cause internal switch deletions (ISDs), which often are large enough to be observed via Southern blotting (1, 8–11). Large ISDs typically are assayed by activating B cells in culture to switch to a particular CH and then generating hybridomas and assaying for ISD in IgM-producing hybridomas that have not undergone bona fide CSR (11, 12). Most such studies have found that large ISDs occur far more frequently in Sμ than in downstream target S regions (6, 12, 13). The almost complete absence of detectable ISDs or SHMs in downstream S regions in prior studies (6, 12) led to the proposal that AID targeting in Sμ is required to promote AID targeting of downstream S regions (6). However, it is possible that absence of downstream ISD or SHM in those studies might reflect, in part, IgM+ hybridomas arising from B cells not fully activated for CSR (2, 12).
In normal B cells, a substantial portion of CSR junctions are catalyzed by classical nonhomologous end-joining (C-NHEJ) (14). C-NHEJ ligates DNA ends with no homologies to form “direct” joins, and ends with short (1–4 nucleotide) microhomologies (MHs) to form MH-mediated joins (15); correspondingly, both types of junctions are found in CSR joins from normal B cells (14). During C-NHEJ, the Ku70/Ku80 complex binds DSBs and thereby serves as the DSB recognition component for this pathway (15). The Ku70/80 complex also serves other functions including recruiting XRCC4/ DNA ligase 4 (Lig4), which catalyze C-NHEJ DSB ligation (16). Although Ku70, Ku80, XRCC4, and Lig4 are considered core C-NHEJ factors required for all C-NHEJ repair events, other NHEJ proteins, notably DNA-PKcs and Artemis, play more limited roles that include processing a subset of DNA ends before joining (15). All known C-NHEJ components are required for completely normal CSR joining (2, 14, 17–19), although substantial CSR occurs in their absence.
End-joining in the absence of C-NHEJ has been observed in the context of transiently transfected, linearized plasmid substrates and for AID or I-SceI–generated chromosomal DSBs in cells deficient for Ku80, XRCC4, or Lig4 (20–25). In addition, XRCC4 or Lig4-deficient B cells undergo CSR at levels up to 50% those of WT B cells and, in contrast to CSR junctions in normal B cells, nearly all CSR junctions are MH-mediated (14, 19). Therefore, in the absence XRCC4 or Lig4, CSR is catalyzed by an end-joining mechanism that strongly prefers MHs and likely uses one of the other mammalian ligases (Ligase 1 or Ligase 3). Although such joining does not represent normal C-NHEJ, it might represent a variant of C-NHEJ that uses most C-NHEJ components and not a completely distinct pathway (15, 19). Recently, however, we have shown that B cells that simultaneously lack Ku70 and Lig4 still undergo substantial CSR. In these cells, CSR must be catalyzed by an A-EJ pathway that is completely independent of C-NHEJ. This pathway also appears distinct from the CSR end-joining pathway observed in the absence of XRCC4 or Lig4, because it generates a distinct junctional sequence spectrum with more direct joins (19).
Little is known about mechanisms that catalyze intra–S-region joining of AID-initiated DSBs to generate ISDs. In this context, there are notable differences between CSR and ISD. First, ISD involves rejoining of a DSB or joining of DSBs that are significantly more proximal than the DSBs involved in CSR, because the latter are separated by 100–200 kb. Second, the substrates for joining are different also. Thus, S regions are highly repetitive internally, but Sμ, although somewhat homologous to Sα and Sε, is not homologous to the various Sγ regions (26, 27). Thus, because of the repetitiveness of S regions, more and larger homologies, on average, should be present for ISD than for CSR. Earlier studies reported complete absence of ISD in activated Ku80-deficient B cells; however, the failure to detect ISD in those studies may have reflected impaired proliferation of the Ku80-deficient B cells (12). Thus, the potential for ISD to be catalyzed by pathways other than C-NHEJ is an open question.
The decrease of CSR in XRCC4-deficient cells results from an end-joining defect, because XRCC4-deficient B cells activated for CSR often harbor AID-initiated IgH locus chromosomal breaks (14, 28). Moreover, AID-dependent IgH locus breaks often are joined in normal XRCC4-deficient B cells to DSBs on other chromosomes to form translocations (14, 28). In addition, reminiscent of S-region junctions in XRCC4- or Lig4-deficient B cells, all oncogenic translocation junctions cloned from nine independent XRCC4/p53-deficient pro-B lymphomas contained junctional MH (29). These observations suggested that alternative end-joining (A-EJ), in the absence of C-NHEJ, may be prone to promoting chromosomal translocations during CSR and general DSB repair (14, 28, 29). However, as mentioned earlier, joining in the absence of XRCC4 may represent either a true A-EJ pathway or a C-NHEJ variant pathway. Therefore, the potential for a distinct A-EJ pathway to catalyze end-joining in translocations has not been addressed fully.
In this study, we addressed the question of whether true A-EJ can catalyze ISD and chromosomal translocations by assaying for these events in activated B cells that lack both Ku70 and Lig4.
Results
Ku/Lig4-Independent and Ku-Dependent/Lig4-Independent A-EJ Catalyze Robust ISD.
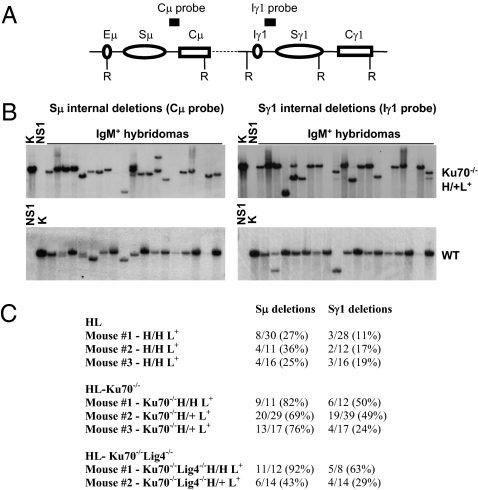
To characterize A-EJ pathways further, we assayed for ISDs within the Sμ and Sγ1 regions of Ku70-deficient, Ku80-deficient, or Ku70/Lig4 double-deficient B cells that were generated in mice harboring knock-in productive IgH and IgL variable region exons to allow generation of mature B cells (referred to as “Ku70−/−HL,” “Ku80−/−HL,” and “Ku70−/−/Lig4−/−HL” B cells, respectively) (19). We also assayed for ISD in XRCC4-deficient and Lig4-deficient B cells that were generated by conditionally inactivating Xrcc4 (14) and Lig4 in peripheral B cells. The conditional inactivation of the Lig4 gene was accomplished as described for Xrcc4 (14), by breeding mice carrying lox P sites flanking the single exon of the Lig4 gene with mice carrying the Cre transgene under the control of the CD21 promoter (Fig. S1). Deletion of the Lig4 gene and the absence of the Lig4 protein in mature B cells were verified by Southern and Western blotting (Fig. S1). Because most activated WT B cells undergo CSR on both IgH alleles, ISDs within the Sμ and Sγ1 regions can be observed most readily by Southern blotting assays of genomic DNA from IgM+ hybridomas, generated from splenic B cells activated with anti-CD40 antibody CD40 (αCD40) plus IL-4 to stimulate CSR to IgG1 and IgE (30, 31).
To assay for internal Sμ ISDs and distinguish them from CSR, EcoRI-digested genomic DNA was assayed for hybridization to probes that flank both sides of Sμ, and bands smaller than the germline Sμ were scored as ISD events (Fig. 1B and Figs. S2 and S3). Although this Southern blotting assay may miss small internal deletions and thereby underestimate the true extent of such events, we observed Sμ ISDs in 25–36% of WT HL IgM+ hybridomas (Fig. 1 B and C), a frequency somewhat higher than levels previously reported (12, 13, 32). More strikingly, we found extremely high levels of Sμ ISD in Ku70−/−HL- (70–80%) and Ku70−/−/Lig4−/−HL- (40–90%) activated B cells, as well as in XRCC4-deficient (60%) or Lig4-deficient (64%) B cells (Fig. 1 B and C and Figs. S2 and S3). Although prior studies did not detect ISD in Ku80−/− B cells activated for CSR (12), our current analyses of a very limited set of Ku80−/−HL hybridomas also revealed ISDs (Fig. S4). In this context, lack of detectable ISD in earlier studies of Ku80−/−HL B cells likely reflects the death of most Ku80−/− B cells that attempted CSR under the activation conditions used in those experiments (12). In contrast, our current activation protocols lead to better survival and much higher CSR levels than those of prior studies (19) and also allow us to observe ISDs readily even in the C-NHEJ–deficient backgrounds. Prior hybridoma studies found very few Sγ1 ISDs in WT HL B cells activated for CSR and none in activated Ku80−/−HL B cells (12). Strikingly, however, we find a substantial level of Sγ1 ISDs in WT HL B cells (11–19%) (Fig. 1C) and even greater levels in Ku70−/−HL (24–50%), Ku70−/−/Lig4−/−HL (29–63%), XRCC4-deficient (38%), and Lig4-deficient B cells (24%) (Fig. 1 B and C and Figs. S2 and S3). Together, our findings clearly demonstrate that ISD at Sμ or Sγ1 can occur robustly in the individual or combined absence of the core components of the C-NHEJ pathway.
Fig. 1.
Robust ISDs occur in Ku70- and Ku70/Lig4-deficient IgM+ B-cell hybridomas. (A) Schematic of the IgH locus with genomic probes and restriction sites used for Southern blotting. Sμ ISD events were detected with a Cμ probe and confirmed with an Iμ probe (Fig. S2). Sγ1 ISD events were detected with an Iγ1 probe and confirmed with an Sγ1 probe (Fig. S2). (B) Ku70-deficient hybridomas display frequent ISDs as shown in representative panels of IgM+ Ku70−/−H/+L+ and WT hybridomas probed with the indicated Southern probes. K, kidney DNA (indicates size of germline Sμ and Sγ1 regions); NS1, genomic DNA from NS1 hybridoma fusion partner. (C) Quantification of Sμ and Sγ1 ISD frequency in Ku70−/−HL, Ku70−/−/Lig4−/−HL, and WT HL IgM+ hybridomas.
Internal S-Region Deletions in C-NHEJ–Deficient B Cells Occur via an A-EJ Pathway.
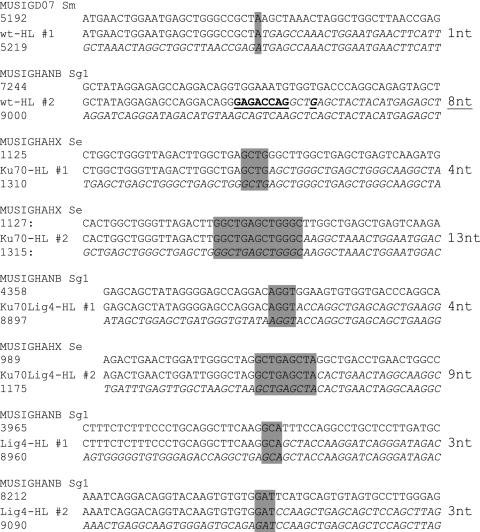
Because of the highly repetitive nature of S-region sequences, particularly Sγ sequences that harbor large numbers of 49-bp repeats (26), some ISD conceivably could be catalyzed by homologous recombination (HR) or single-strand annealing (SSA), as opposed to C-NHEJ or A-EJ end-joining (33, 34). To determine the nature of the joining pathway that mediates robust ISD in the absence of C-NHEJ, we cloned several Sμ–Sμ, Sγ1–Sγ1, and Sε–Sε ISD junctions from WT HL, Ku70-, Ku70/Lig4- and Lig4-deficient primary B cells stimulated with antiCD40 plus IL-4 in vitro. We also obtained Sγ1–Sγ1and Sε–Sε ISD junctions from IgM-expressing hybridomas derived from WT HL, Lig4-deficient, and XRCC4-deficient B cells. In agreement with prior studies performed on S-region junctions of WT cells (35), all the junctions isolated were mediated by an end-joining mechanism, with seven of nine junctions in C-NHEJ–deficient cells being MH-mediated (Fig. 2A and Table S1). In addition, some of the MHs were relatively long compared with those observed in C-NHEJ–proficient B cells (Fig. 2A) (14). Therefore, we conclude that A-EJ is the predominant pathway for ISD joining in C-NHEJ–deficient cells.
Fig. 2.
Examples of junctions from WT HL, Ku70HL, Ku70/Lig4HL, and Lig4HL primary B cells. GenBank annotated sequences for Sμ (MUSIGD07), Sγ1 (MUSIGHANB), and Sε (MUSIGHANX) were used for alignment. For each individual alignment, the annotated GenBank sequence is given at the top. The number of the nucleotides involved in the the junction is indicated on the left (top and bottom) for each alignment; between them the ISD junction sequence is shown. Junctional MHs are shaded in gray; the junctional insertion is underlined. The length of junctional MHs and insertions for each junction is indicated on the right.
Ku- and Lig4-Independent A-EJ Catalyzes Translocations.
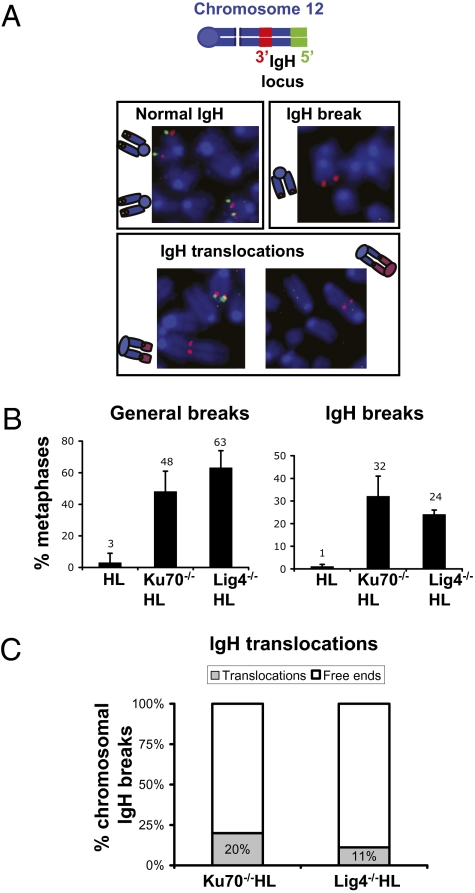
To confirm that the decreased levels of CSR observed in activated Ku70−/− and Lig4−/− splenic B cells were associated with an end-joining defect, we assayed these cells for IgH-locus chromosomal breaks. For this purpose, we used a metaphase FISH assay with BAC probes from sequences flanking the 5′ and 3′ ends of the IgH locus (Fig. 3A) (36) and found that, in contrast to WT HL B cells, a large percentage of activated Ku70−/−HL, Lig4−/−HL, or Ku70−/−/Lig4−/−HL B cells had IgH breaks (Fig. 3B and Table S2). As expected, these various C-NHEJ–deficient B cells also showed increased levels of general chr-omosome breaks and translocations, as revealed by a telomere FISH assay (Fig. 3C and Table S2). In Xrcc4-deficient B cells, a large percentage of IgH locus breaks are translocated to general breaks (14, 28). Likewise, we find that Ku70−/−HL, Lig4−/−HL, and Ku70−/−/Lig4−/−HL B cells show a dramatic increase in IgH locus translocations (Fig. 3C and Table S2); the findings in Ku70−/−/Lig4−/−HL B cells clearly indicate that a C-NHEJ–independent A-EJ pathway can catalyze translocations robustly.
Fig. 3.
Frequent IgH breaks and translocations in Ku70- and Lig4-deficient primary B cells stimulated for CSR. (A) Diagram with the 3′IgH BAC probe (green) and 5′IgH BAC probe (red) used for IgH FISH and examples of normal IgH loci, IgH breaks, and IgH translocations. Normal IgH loci retain both green and red locus-flanking signals. An IgH break results in split green and red signals. Translocations involving the IgH locus join the proximal region of chromosome12 (red signal) to other chromosomal fragments. (B) Percentage of metaphases with general breaks (Left) and IgH breaks (Right). (C) Percentage of IgH translocations.
IgH/c-myc Translocations in the Absence of C-NHEJ.
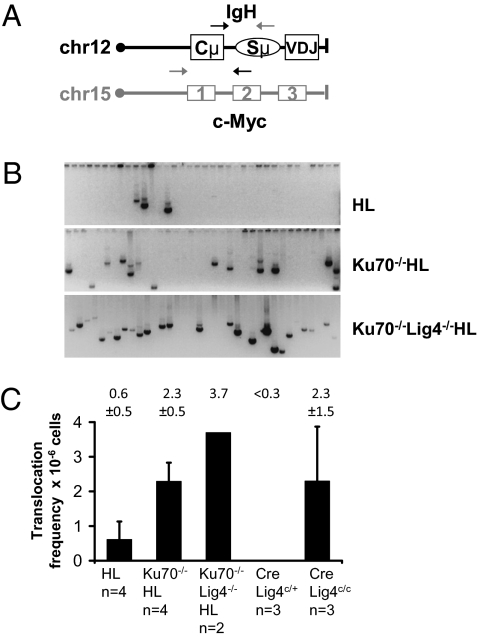
To characterize further IgH translocations in activated Ku70−/−HL, Ku70−/−/Lig4−/−HL, and Lig4-deficient cells versus WT HL B cells, we used PCR (37) to quantify the relative levels of translocations between the IgH Sμ region and the c-myc oncogene. We amplified and quantified both derivative chromosome 12 and derivative chromosome 15 IgH–c-myc translocations, using primers located just downstream of JH4 in IgH and upstream of exon 1 of c-myc (derivative chromosome 15) or downstream of c-myc exon 1 and just upstream of the Cμ IgH C region (derivative chromosome 12) (Fig. 4A) (37). In control WT HL and CD21CreLig4c/+ B cells, the frequency of observed translocations was low (less than one translocation per million B cells analyzed), as previously reported (28, 37). However, we detected a substantial increase in IgH–c-myc translocations in Ku70−/−HL, Ku70−/−/Lig4−/−HL, and Lig4-deficient B cells relative to controls (Fig. 4 B and C and Table S3). Of note, the CD21CreLig4c/+ control B cells did not have any detectable IgH–c-myc translocations (Fig. 4C), suggesting that most translocations observed in Lig4 conditionally deleted cells are caused by the end-joining defect in the absence of Lig4.
Fig. 4.
Ku70-, Ku70/Lig4- and Lig4-deficient B cells activated for CSR display increased IgH–c-myc translocations. (A) Schematic of the IgH and c-myc loci with the location of the primers used for PCR amplification indicated. Gray primers: Der15 amplification; black primers: Der12 amplification (37). (B) Examples of Der12 IgH–c-myc translocations from αCD40 plus IL-4–stimulated Ku70−/−HL, Ku70−/−/Lig4−/−HL, and control (WT HL) B cells. (C) Quantification of IgH–c-myc translocation frequency. Average and SD, genotype, and number of mice analyzed are indicated for each bar graph.
To characterize further the end-joining mechanism used to promote translocations in the absence of Ku70 or the combined absence of Ku70 and Lig4, we sequenced 11 IgH–c-myc translocation junctions from Ku70−/−HL B cells plus 6 IgH–c-myc translocation junctions from Ku70−/−/Lig4−/−HL B cells (Fig. S5). None of the 17 junctions analyzed had evidence of joining by an HR mechanism: We did not detect long junctional homologies, which are the hallmark of HR repair (34). Instead, similar to control WT HL IgH–c-myc junctions (Fig. S5), we found that all Ku70−/−HL and Ku70−/−/Lig4−/−HL translocation junctions were mediated by end-joining, with 7 of 11 Ku70−/−HL and 1 of 7 Ku70−/−/Lig4−/−HL containing MH and the remainder having insertions (Fig. S5). Thus, in activated B cells that lack Ku70 or both Ku70 and Lig4, an A-EJ pathway catalyzes increased levels of IgH chromosomal translocations to the c-myc oncogene. These findings firmly demonstrate that an A-EJ pathway that is completely independent of C-NHEJ can catalyze translocations.
Discussion
Studies of A-EJ in the context of CSR have provided major insights into the mechanism of chromosomal DNA repair by A-EJ. Little is known about the components of the A-EJ pathway that catalyzes chromosomal end-joining events. However, we recently have demonstrated that, in the absence of C-NHEJ, two forms of A-EJ catalyze the physiological process of CSR; these forms of A-EJ appear distinct based on differential reliance on Ku and different preferences for use of junctional MHs during CSR (19). In our current studies, we show that A-EJ pathways in Ku70-, Ku70/Lig4-, Lig4-, and XRCC4-deficient cells can catalyze robustly the joining of AID-initiated breaks within a given IgH S region in B cells activated for CSR. However, because some S-region repeats are long (49 bp in Sγ1), and the number of ISD junctions we obtained was limited, we cannot absolutely rule out a role for other pathways that employ longer homologies (34) in joining a fraction of ISD breaks. We also now definitively show that Ku70 and Lig4 are required for fully normal end-joining during CSR, as clearly evidenced by the increase of more than an order of magnitude in the number of unresolved IgH chromosomal breaks observed in activated Ku70- or Lig4-deficient B cells versus WT B cells (Fig. 3B and Table S2). Finally, we show that general and AID-initiated IgH breaks in Ku70-deficient, Lig4-deficient, and Ku70/Lig4-deficient B cells are frequently involved in chromosomal translocations, demonstrating that an A-EJ pathway that is independent of C-NHEJ can robustly catalyze translocations in the absence of C-NHEJ.
The finding of increased ISD but decreased CSR in the complete absence of C-NHEJ can be explained by several, not mutually exclusive, scenarios, each of which has potentially important implications for A-EJ. First, it is conceivable that A-EJ preferentially catalyzes ISD as compared with CSR because of the repetitiveness of S regions. Specifically, the repetitive nature of individual S regions might present more opportunities for the resected ends of a single DSB or the ends of two DSBs within a given S region to provide MH than found for two DSBs in different S regions. This possibility is supported by our finding that nearly all CSR joins from Ku70-, Lig4- and Ku70/Lig4-deficient cells were MH-mediated. In this regard, we note that although the CSR junctions in Lig4−/−/Ku70−/− B cells do display substantial numbers of direct junctions, they still are markedly biased toward MH-mediated junctions (19). If substrate sequences indeed influence the choice of repair pathway (e.g., by providing more MHs), A-EJ may play a substantial role in ISD in WT cells and also in CSR between S regions that have substantial homology (e.g., Sμ and Sα or Sε) (14, 38).
A second potential mechanism by which A-EJ could be biased to ISD versus CSR is based on analogy to the model proposed to explain the finding that 53BP1-deficient B cells have decreased levels of CSR but increased levels of Sμ and Sγ1 ISD. Specifically, this finding was interpreted as indicating that 53BP1 may be required for long-range S-region synapsis and that short-range DSB joining is favored in the absence of this factor (32). Correspondingly, increased ISD in core C-NHEJ–deficient B cells, coupled with their decreased CSR, might be interpreted as suggesting that A-EJ has decreased ability to perform long-range joining of DNA breaks during CSR as compared with C-NHEJ. Indeed, C-NHEJ has been shown to join DSBs on the same chromosome preferentially and thereby to suppress translocations (39). Moreover, the DSB response involving factors such as ATM, 53BP1, and H2AX has been shown to be required for normal joining of DSBs via NHEJ (25, 36, 40). In this regard, the DSB response also has been suggested as promoting long-range joining of intrachromosomal DSBs (41). Thus, among other potential mechanisms, a preferential ability of C-NHEJ versus A-EJ to promote long-range joining of spatially separated DSBs on the same chromosome theoretically might be affected by C-NHEJ rather than A-EJ being recruited preferentially by the DNA double-strand break-response complex. A corollary of this model would be that A-EJ should not be as restricted to intrachromosomal joining as C-NHEJ and thereby would be more translocation prone.
The dramatic increase in IgH translocations in Ku70-, Ku70/Lig4-, or Lig4-deficient B cells, including the significant increase in Sμ translocations to the c-myc oncogene, underscores the ability of both the Ku-dependent and Ku-independent forms of A-EJ to mediate aberrant interchromosomal joining (25). The notion that A-EJ might facilitate oncogenic translocations and tumorigenesis is supported by various studies in murine systems, which found evidence of MH in translocation junctions from tumors generated in the absence of C-NHEJ factors (e.g., 29, 42). Moreover, studies in human cell lines derived from several types of cancers indicate that DSB repair processes other than C-NHEJ are used extensively, suggesting a potential role for A-EJ in promoting and supporting DNA repair in tumor cells (43, 44). Mechanistically, an increased activity of A-EJ with respect to translocations would be consistent with the possibility that A-EJ is not so preferentially biased to joining DSBs within the same chromosome as C-NHEJ (see above). However, whether A-EJ truly catalyzes translocations more efficiently than does C-NHEJ remains to be directly demonstrated. Finally, we have shown previously that the A-EJ pathway that functions in the absence of both Ku70 and Lig4 to catalyze CSR, although biased for MH-mediated junctions, can catalyze a substantial level of direct joins (19). Our present finding that this pathway catalyzes high levels of translocations emphasizes that the absence of MH in translocation junctions does not exclude the possibility that they were catalyzed by A-EJ. Finally the relative roles of Ku-independent and Ku-dependent forms of A-EJ in catalyzing translocations remain to be determined.
Experimental Procedures
Mouse Strains.
Ku70−/−, Lig4−/−, CD21CreXrcc4c/c, and Ku80−/−HL mice were used (14, 45–47). Ku70−/−/Lig4−/−HL mice were obtained by breeding double-heterozygous Ku70+/−/Lig4+/−HL mice. The CD21CreLig4c/c mice were generated by flanking the single exon of the Lig4 gene with loxP sites (Fig. S1) and breeding with mice carrying the Cre transgene under the control of the CD21 promoter. Lig4 deletion in mature B cells results in a CSR defect similar to that in the germline-deleted Lig4HL mice (Fig. S1) (14).
Splenic B-Cell Purification and Culture.
Mature B cells were isolated and enriched from the spleen and stimulated with αCD40/IL-4 to induce CSR to IgG1 and IgE as described (14). Cytokine sources were αCD40 (Cat. # 16–0402-86; eBioscience) and IL-4 (Cat. # 14–8041; eBioscience). Hybridoma fusions were performed at day 4 of culture, and hybridomas were screened by ELISA after 1 week of selection.
Analysis of S-Region Internal Deletions.
Genomic hybridoma DNA was assayed by Southern blotting with probes for Iμ, Cμ, Iγ1, and Sγ1 as described (11, 41). To identify internal deletions in both Sμ and Sγ1, genomic DNA was digested with EcoRI. ISD junctions in primary cells were identified in some DNA sequences during isolation of CSR junctions (performed as described in ref. 14); therefore each ISD sequence also contains a CSR junction (Table S1). ISD junctions in B-cell hybridomas were obtained after PCR amplification with forward Iμ- (5′ tgtcaatcctgttcttagtcaatca3′) and reverse Sγ1-region (5′gtcgaattcccccatccctgtcacctata3′) primers. cDNA polymerase (Clontech) was used for amplification. PCR conditions were 1 min at 95 °C 1; 30 sec at 95 °C; and 12 min at 68 °C for 30 cycles.
Two-color IgH FISH and Telomere FISH.
Metaphases were prepared from B cells stimulated in vitro with αCD40 plus IL-4 and processed as described (36).
IgH-c-myc Translocations.
PCR and Southern blotting analysis of c-myc–IgH translocations were performed as described (25) on 500ng genomic DNA from αCD40 plus IL-4–stimulated splenic B cells.
Supplementary Material
Acknowledgments
We thank Klaus Rajewsky, Bjoern Schwer, Monica Gostissa, and Maria-Vivienne Boboila for stimulating discussions, and we thank Michael Lieber, Wesley Dunnick, and Kefei Yu for critical review of the manuscript. This work was supported by National Institutes of Health (NIH) Grants AI031541 and CA092625 (F.W.A.) and AI037526 (M.N.). J.H.W. was supported by a Leukemia and Lymphoma Society of America Special Fellowship and by NIH Training Grant T32CA09382-26. C.B. is supported by a Cancer Research Institute training grant
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915067107/DCSupplemental.
References
- 1.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catalan N, et al. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 6.Schrader CE, et al. Mutations occur in the Ig Smu region but rarely in Sgamma regions prior to class switch recombination. EMBO J. 2003;22:5893–5903. doi: 10.1093/emboj/cdg550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honjo T, Muramatsu M, Fagarasan S. AID: How does it aid antibody diversity? Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Alt FW, Rosenberg N, Casanova RJ, Thomas E, Baltimore D. Immunoglobulin heavy-chain expression and class switching in a murine leukaemia cell line. Nature. 1982;296:325–331. doi: 10.1038/296325a0. [DOI] [PubMed] [Google Scholar]
- 9.Iwasato T, Shimizu A, Honjo T, Yamagishi H. Circular DNA is excised by immunoglobulin class switch recombination. Cell. 1990;62:143–149. doi: 10.1016/0092-8674(90)90248-d. [DOI] [PubMed] [Google Scholar]
- 10.Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 11.Dudley DD, et al. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc Natl Acad Sci USA. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 15.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: Relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 16.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 17.Franco S, et al. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J Exp Med. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callén E, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boboila C, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2009 doi: 10.1084/jem.20092449. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 21.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stavnezer J. Antibody class switching. Adv Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri J, Alt FW. Class-switch recombination: Interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 28.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 30.Radbruch A, Müller W, Rajewsky K. Class switch recombination is IgG1 specific on active and inactive IgH loci of IgG1-secreting B-cell blasts. Proc Natl Acad Sci USA. 1986;83:3954–3957. doi: 10.1073/pnas.83.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottaro A, et al. Deletion of the IgH intronic enhancer and associated matrix-attachment regions decreases, but does not abolish, class switching at the mu locus. Int Immunol. 1998;10:799–806. doi: 10.1093/intimm/10.6.799. [DOI] [PubMed] [Google Scholar]
- 32.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur J Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 33.Yancopoulos GD, et al. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic Alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 35.Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franco S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Pan-Hammarström Q, et al. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J Exp Med. 2005;201:189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferguson DO, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jankovic M, et al. Role of the translocation partner in protection against AID-dependent chromosomal translocations. Proc Natl Acad Sci. 2009;79:7837–7841. doi: 10.1073/pnas.0908946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zarrin AA, et al. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 42.Wang JH, et al. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J Exp Med. 2008;205:3079–3090. doi: 10.1084/jem.20082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–5259. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Windhofer F, Krause S, Hader C, Schulz WA, Florl AR. Distinctive differences in DNA double-strand break repair between normal urothelial and urothelial carcinoma cells. Mutat Res. 2008;638:56–65. doi: 10.1016/j.mrfmmm.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank KM, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 47.Casellas R, et al. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.