Abstract
Gene and genome duplications create novel genetic material on which evolution can work and have therefore been recognized as a major source of innovation for many eukaryotic lineages. Following duplication, the most likely fate is gene loss; however, a considerable fraction of duplicated genes survive. Not all genes have the same probability of survival, but it is not fully understood what evolutionary forces determine the pattern of gene retention. Here, we use genome sequence data as well as large-scale phosphoproteomics data from the baker’s yeast Saccharomyces cerevisiae, which underwent a whole-genome duplication ∼100 mya, and show that the number of phosphorylation sites on the proteins they encode is a major determinant of gene retention. Protein phosphorylation motifs are short amino acid sequences that are usually embedded within unstructured and rapidly evolving protein regions. Reciprocal loss of those ancestral sites and the gain of new ones are major drivers in the retention of the two surviving duplicates and in their acquisition of distinct functions. This way, small changes in the sequences of unstructured regions in proteins can contribute to the rapid rewiring and adaptation of regulatory networks.
Keywords: gene duplication, whole-genome duplication, gene retention, phosphorylation, posttranslational modification
Whole-genome duplications (WGDs) have occurred repeatedly in eukaryotic evolution and have been linked to genetic innovation, adaptation, speciation, and survival (1 –4). Following a WGD, most duplicate copies are lost (5, 6), but a considerable fraction survive, with either selection or genetic drift accounting for the pattern of duplicate gene retention. Interestingly, this pattern of retention is not random, but, rather, biased to certain functional categories. In particular, genes involved in regulation are preferentially retained (2, 6, 7) and it is this preferential retention that likely predetermines the future of a lineage. However, the mechanisms that determine which genes are maintained in duplicate and which return to a single-copy state are largely unknown.
After a WGD, there is a relatively short period of genome instability, extensive gene loss, and elevated levels of nucleotide substitution (8). During that period, regulatory networks must be rapidly rewired to integrate the newly duplicated (and, at the same time, diverging) genes and thus prevent chaos in the control of cellular processes. Rapid evolution and functional divergence have indeed been observed at the level of the transcription of duplicated genes (9, 10), which is usually explained by point mutations in short transcription factor binding motifs. However, because the effectors of gene action are proteins, adaptation might also occur at the posttranslational level of regulation. Because the amino acid sequence motifs for posttranslational modification (PTM), and especially phosphorylation, are short (11) and occur within rapidly evolving unstructured regions (12), we reasoned that changes in PTM sites might present a ready means of rapidly effecting the necessary rewiring. Furthermore, we wanted to explore whether the rapid evolution of these sites might be linked to gene retention. Therefore, we exploited the wealth of proteomic and genomic data available for the baker’s yeast Saccharomyces cerevisiae and examined the relationship between protein phosphorylation, gene retention, and functional divergence following the WGD that occurred in the hemiascomycete yeasts ∼100 mya (13, 14).
Results and Discussion
Retained Duplicates Are Highly Phosphorylated.
There have been several large-scale in vivo studies (15 –20) of the phosphoproteome of S. cerevisiae, using highly reproducible techniques of mass spectrometric analysis. Because the identified phosphopeptides could match one or more open reading frames (ORFs), we generated two phosphorylation datasets. One contains phosphopeptides that have a unique and exact match (designated 6_exp_U; 6 refers to the six datasets used), and the other contains phosphopeptides that exactly match more than one ORF (designated 6_exp_NU) [supporting information (SI) S1.1 and SI_file2]. All subsequent analyses were performed on both these datasets (whenever applicable), and the same conclusions were obtained from both. Tests 1–20 in S1.14 summarize the results of these tests and their statistical significance (through Wilcoxon’s tests). The datasets compiled from these six studies indicate that 8,500–11,300 phosphorylation sites (p-sites) are distributed over 2,200–2,400 proteins in S. cerevisiae. GO_slim analysis confirmed that these datasets were enriched for proteins localized in the nucleus and involved in signal transduction and transcription regulation (S1.3).
Previous analysis of the S. cerevisiae genome (21) identified >500 gene pairs that were produced by the WGD (which we designate WGD genes, or ohnologs, and their products, WGD proteins) and ∼4,000 genes that were duplicated in the WGD but later returned to single-copy status (RSS genes, RSS proteins) (S1.4). Of note, these RSS genes constitute most of the remainder of the S. cerevisiae genome after the WGD-produced duplicate pairs are excluded. We found that a higher fraction of WGD genes encoded phosphoproteins compared to the RSS genes (48–58% vs. 42–43%), a statistically significant difference (P < 1e −3, χ2 test). Furthermore, phosphoproteins of the WGD group have, on average, significantly more p-sites than those of the RSS group (4.6 and 3.5 sites per protein, on average, for 6_exp_U in test 1; P < 1.64e −7, Wilcoxon’s test). This observation is supported not only from experimental data, but also from in silico-predicted data (S1.5 and test 1; P < 3.4e −14, Wilcoxon’s test). Furthermore, this observation is robust with respect to the selection, quality, and evolution of the various experimental datasets (S1.6). A jackknife analysis considering all six experimental datasets showed that, no matter which dataset is excluded from the analysis, WGD phosphoproteins have on average more p-sites (S1.6 and test 2; P < 0.0013, Wilcoxon’s test).
It is possible that this observation is very significant, but is not general, being confined to only a few gene categories. In fact, GO_slim analysis showed that the enrichment of p-sites in WGD phosphoproteins vs. RSS phosphoproteins is statistically significant (S1.7 and test 3) for one-third (6_exp_U) to two-thirds (6_exp_NU) of the GO_slim categories. Only for one category, namely structural molecule activity, in one dataset, did we observe the inverse trend (Table S9). A jackknife analysis, considering all GO_slim categories, showed that, no matter which category was removed, the WGD group still contained more p-sites than the RSS group (S1.7 and test 4; P < 1e −4, Wilcoxon’s test). Signaling and transcription factor (TF) molecules show higher retention than average after a WGD event and also show higher levels of phosphorylation than average. To ensure that the latter observation is not a trivial consequence of the large number of TFs and signaling molecules in our dataset, we removed all TFs, kinases, phosphatases, and cyclins and still found the WGD phosphoproteins to contain more p-sites than the RSS phosphoproteins (S1.7 and test 5; P < 1.5e −6, Wilcoxon’s test). Furthermore, to account for gene dosage imbalances (22, 23), we also removed (i) all of the ribosomal proteins or (ii) all known protein complexes and found that our conclusions are still robust (S1.7 and test 6, P < 2.2e −9, Wilcoxon’s test; and S1.7 and test 7, P < 9.8 e −7, Wilcoxon’s test, respectively). We also controlled for potential biases arising from (i) differences in protein abundance (24) (S1.8 and test 8; P < 3.3 e −5, Wilcoxon’s test), (ii) coverage (S1.8 and test 9; P = 0.009, Wilcoxon’s test) in the various experiments, (iii) essentiality of genes (25 –29) (S1.8 and test 10; P < 2.3e −6, Wilcoxon’s test), or (iv) protein interaction network centrality (27, 29, 30) (S1.8 and test 11; P < 3e −7, Wilcoxon’s test) and still found that WGD proteins contained more p-sites. The importance of taking into consideration protein function in evolutionary analyses as performed here has been highlighted previously (31).
Inference of Ancestral Phosphorylation Sites in the Pre-WGD Ancestor.
On the basis of known S. cerevisiae p-sites and multiple sequence alignments of orthologs in three species that diverged from S. cerevisiae just before the WGD event (Fig. 1A), namely Ashbya (Eremothecium) gossypii (32), Kluyveromyces lactis (33), and Kluyveromyces waltii (13), we inferred the presence of ancestral p-sites in the proteins of the pre-WGD ancestor (S1.9). To achieve this, we used both the 6_exp_U and the 6_exp_NU datasets and, by applying various levels of stringency on the basis of the variation of amino acids surrounding the p-site (S1.9), we generated eight different sets of ancestral p-sites (Materials and Methods). In all eight sets, we again observed that, on average, the ancestral proteins of the WGD group contained significantly more p-sites than ancestral RSS proteins (S1.9 and test 12; P < 3.4e −9, Wilcoxon’s test). Because we used the ORFs of both retained duplicates to infer the ancestral p-sites of WGD proteins and only one RSS-ORF to infer the ancestral p-sites of RSS proteins, it is possible that the observed difference reflects this bias. Therefore, we repeated the analysis using only one of the two ohnologs (the one with most p-sites) to infer the ancestral p-sites. Although we consider this already stringent, because we underestimated the p-sites of the ancestral molecule that later gave rise to subfunctionalized copies (see further), we still observed that ancestral proteins of the WGD group contained significantly more p-sites (see S1.9 and test 13; P < 8.4e −5, Wilcoxon’s test). Recently, concerns have been raised about the possibility that many p-sites are not functional (34, 35). However, because our evolutionary analysis is based on p-sites that have been conserved for >100 million years, we believe that there is little chance that our conclusions are a trivial consequence of an accumulation of nonfunctional p-sites in WGD duplicates.
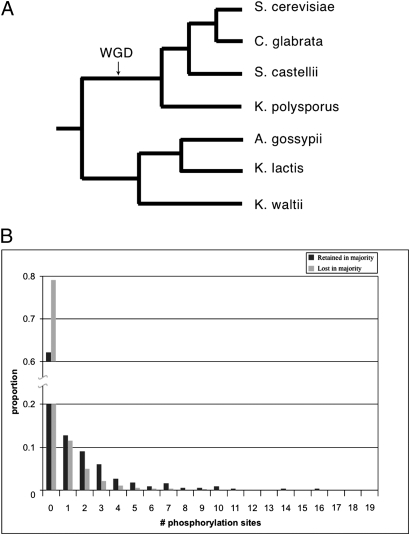
Fig. 1.
The number of protein phosphorylation sites in the pre-WGD ancestor affects the probability of gene retention in the four post-WGD yeast lineages. (A) Species tree showing the evolutionary relationships of the yeast species discussed. (B) The number of ancestral p-sites affects the fate of duplicate retention in four post-WGD lineages (S. cerevisiae, C. glabrata, S. castellii, and K. polysporus). The graph shows that the ancestors of the “retained-in-majority” bins had more p-sites than the ancestors of the “lost-in-majority” bins (S1.9) (see text for details).
To see whether the link between the number of ancestral p-sites and gene duplicate retention is a general phenomenon, and not just confined to S. cerevisiae, we examined the gene retention patterns in three more post-WGD species [Candida glabrata (33), Saccharomyces castelli (33), and Kluyveromyces polysporus (4)]. We are aware of the fact that the evolutionary process is not entirely independent in all these species, because they share a common ancestor. However, they diverged from each other shortly after the WGD event (4). Products of genes that survived as duplicates in the genomes of at least three of the four post-WGD yeast species are designated “retained-in-majority,” whereas products of genes that have returned to single-copy status in the genomes of at least three of those four post-WGD species are designated “lost-in-majority.” For all eight ancestral p-site datasets (Materials and Methods), the pre-WGD ancestors of the retained-in-majority category had, on average, more p-sites than the pre-WGD ancestors of the lost-in-majority category, meaning that proteins with more p-sites have repeatedly been retained in duplicate, compared to proteins with fewer p-sites (S1.9 and test 14; P < 2.2e −8, Wilcoxon’s test) (Fig. 1B).
Sub- and Neofunctionalization of Phosphorylation Sites.
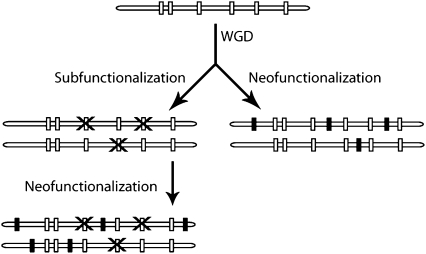
How might protein phosphorylation affect the retention of duplicates? Previous analyses have suggested that the partitioning of functions (subfunctionalization) between the two copies or the emergence of new functions (neofunctionalization) for one or both copies of a duplicated gene favors their retention (Fig. 2). To measure the effect of phosphorylation-related subfunctionalization, we assumed that such a partition had occurred if duplicates lost a complementary set of p-sites. We used all of the pre-WGD genes (ancestors of both WGD and RSS genes) whose proteins had at least two ancestral p-sites and measured how many of these ancestral genes duplicated to give pairs with signs of subfunctionalization (S1.9). We found that between 2.5 and 7% (depending on dataset) of those ancestral genes gave rise to subfunctionalized copies in S. cerevisiae. For example, 5–12 (depending on stringency of criteria to infer ancestry) ancestral p-sites underwent subfunctionalization in the BOI1/YBL085W-BOI2/YER114C WGD pair; these proteins are involved in bud emergence and polar growth. In addition, we observed that ancestral proteins whose ohnologs subfunctionalized had, on average, more p-sites than all other ancestral proteins (S1.9 and test 15; P < 1.4e −4, Wilcoxon’s test) and also had more p-sites than those retained in duplicate without subfunctionalization (S1.9 and test 16; P < 0.011, Wilcoxon’s test). This finding supports the idea of a stochastic process of reciprocal loss of functional p-sites. The more p-sites are in the ancestral protein, the greater the chance is of reciprocal loss in future duplicates, thus leading to subfunctionalization and retention, a pattern in accordance with a model proposed for regulatory sequences (36). It should be noted that the current phosphorylation dataset is incomplete and, as more phosphorylation data are generated, the number of subfunctionalization cases is likely to increase.
Fig. 2.
Protein phosphorylation and the retention of duplicated genes. Subfunctionalization is the partitioning of p-sites among the duplicates via point mutations and stochastic reciprocal loss. A parallel or (more likely) subsequent event is neofunctionalization, the emergence of new p-sites, again via point mutations. Open boxes refer to ancestral p-sites, and solid boxes refer to recently emerged p-sites.
We also identified potential cases of neofunctionalization by looking for p-sites not present in the pre-WGD ancestor (S1.9). We refer to such sites as neo-p-sites and assume that some new regulatory interaction may have evolved. Without mutation data, we do not know whether these neo-p-sites are truly functional (35). Furthermore, without equally extensive phosphorylation data from other yeasts we cannot take into account those p-sites that have undergone evolutionary turnover (37), where a functional p-site is lost, but a new neighboring p-site emerges and rescues its function. Thus, we might overestimate the significance of neofunctionalization in yeast ohnologs; on the other hand, the current phosphorylation dataset is incomplete. Nevertheless, 29–40% of ohnologs seemed to have acquired one or more novel p-sites; moreover, 73–94% of ohnologs that undergo subfunctionalization simultaneously seem to undergo neofunctionalization, which is in agreement with a complex model of neosubfunctionalization (38). The high incidence of novel p-sites is in accordance with previous reports on the importance of neofunctionalization in TF regulatory motifs (39). Over 80% of p-sites are found within unstructured and fast-evolving loops that comprise ∼55% of the protein length (S1.10) and these regions are linked to tight regulation (40). We observed a significant correlation (Pearson coefficient, 0.44–0.45) between the absolute length of the unstructured loops and their number of p-sites. The retained duplicates encode phosphoproteins with loops that are, on average, 14% longer than those of RSS proteins. To see whether the higher incidence of p-sites, and therefore retention, was affected by WGD-protein loops being longer, we normalized our phosphorylation data for loop length and confirmed again that WGD proteins have more p-sites than RSS proteins (test 17; P < 0.0083, Wilcoxon’s test). We repeated the same analysis for intrinsic disorder and again noted that WGD proteins have more p-sites than RSS proteins (test 17; P < 8.5e −5, Wilcoxon’s test).
Posttranslational Modifications in General and Not Only Phosphorylation Likely Affect the Retention of Duplicated Genes.
As we have shown here, WGD proteins are subject to more phosphorylation than RSS proteins. Whereas the only extensive in vivo data on PTM concern protein phosphorylation, there are indications that other PTMs are linked to increased levels of retention following duplication (S1.11). A higher fraction of WGD proteins than RSS proteins are ubiquitinated (23.5% vs. 19.5%; P < 0.004, χ2 test); furthermore WGD proteins seem to have shorter half-lives than RSS proteins (S1.11 and test 18; P < 7e −4, Wilcoxon’s test). All these results are congruent with our hypothesis that changes in posttranslational modification represent rapid and facile routes to the sub- and neofunctionalization of duplicated genes following WGD and thereby promote the retention of duplicate pairs, although they do not explain all cases of duplicate retention (i.e., selection for higher dosage for genes encoding ribosomal proteins).
The impact of phosphorylation on gene retention is probably not confined to WGD, but to small-scale gene duplication (SSD) as well. Several studies have shown that the mode of duplication (WGD vs. SSD) has different effects on the evolution of the genome (7, 41, 42). Because SSDs occur continuously and at various times, it is not possible to repeat the evolutionary analysis that was possible for the WGD. Nevertheless, when we compared properties of SSD vs. singleton proteins (S1.12), we observed that (i) a higher fraction of SSD proteins are phosphorylated (42–43% vs. 33–34%; P < 4e −9, χ2 test), (ii) a higher fraction of SSD proteins are ubiquitinated (20% vs. 14%; P < 2.8e −9, χ2 test), (iii) SSD phosphoproteins have, on average, more p-sites than singleton phosphoproteins (test 19; P < 3.2e −4, Wilcoxon’s test), and (iv) SSD proteins have shorter half-lives (test 20; P < 0.0455, Wilcoxon’s test). Experimental data (43) from Schizosaccharomyces pombe, a very distant relative of S. cerevisiae that did not undergo a WGD, also show a higher fraction of SSD proteins being phosphorylated compared to singletons (22.5% vs.. 14%; P < 2e −14, χ2 tests), although the paucity of functional data for this species limits the analysis (S1.13).
The higher level of phosphorylation observed, not only for WGDs but also for SSDs, seems to imply that the retention of highly phosphorylated proteins in the yeast lineages cannot directly be attributed to stoichiometric constraints. According to the dosage balance hypothesis (2, 23, 44), it is conceivable that proteins involved in phosphorylation might need to maintain relative stoichiometry with their kinases, thus promoting coretention. Therefore, retention of these highly phosphorylated proteins, if due to stoichiometric balances, should be favored only after a WGD event and not after SSD events. Further research is necessary to see why this does not seem to be the case.
Conclusions
It is clear from this study that proteins retained in duplicate are subject to more posttranslational control, and particularly to more phosphorylation, than RSS proteins. Posttranslational regulation repeatedly affected the future of gene duplicates in the various post-WGD yeast lineages, suggesting that gene retention is, to some extent, predetermined. The evolutionary analyses performed are congruent with our hypothesis that changes in posttranslational modification represent rapid and facile routes to the sub- and neofunctionalization of duplicated genes and thereby promote the retention of duplicate pairs. Our observation is also in accordance with previous observations that “complex” genes, where complexity is defined by the number of protein domains encoded and cis-regulatory elements, tend to be retained more frequently (45). An alternative explanation (that does not exclude the previous one) is that tighter regulatory control can buffer the slightly deleterious mutations of duplicated copies that are under relaxed selection and thus provide them with more time to explore the fitness landscape. It may be, after all, that the cell does not favor the survival (for a long time) of a degenerate gene copy that can act like a “loose cannon.”
Materials and Methods
Phosphorylation Data.
For S. cerevisiae, we used six publicly available experimental data sets (15 –20), and for every one of them the filter proposed in that specific study for identifying the exact location of phosphorylation sites was applied. All those experiments rely on affinity-based methods (IMAC) for phosphopeptide isolation and their results are estimated to be up to 93% reproducible (46). The identified phosphopeptides could match (exactly) either one or more than one ORF. Accordingly, we generated two phosphorylation datasets: one that contains phosphopeptides that have a unique match (designated as 6_exp_U) and one that contains phosphopeptides that may exactly match one or more ORFs (designated as 6_exp_NU) (SI_file2). The reason is that this data treatment may have an effect on the analyses of gene duplication. Transposable elements were removed from the two datasets. For S. pombe, we used one publicly available phosphoproteomics experiment (43).
The NetPhosYeast software (47) was used to predict protein phosphorylation sites for the entire S. cerevisiae proteome. The NetPhosYeast software is specifically tailored to S. cerevisiae and has been shown to outperform all other predictors. We ran the predictions with two different stringent cutoffs, 0.75 and 0.85.
Statistics and Gene Ontology Analyses.
The R programming language was used for statistical analyses. For Gene Ontology-related analyses, we used the GO-slim annotation of yeast (48).
Duplication Datasets for S. cerevisiae.
We analyzed genes of S. cerevisiae, a species that underwent a WGD ∼100 mya (21). Genes were separated into those 1,096 that were retained in duplicate following the WGD until today (designated as WGD genes) and those 4,002 that underwent the duplication but later lost the duplicate (designated as RSS genes) (SI_file2). The assignment of orthologs and ohnologs (gene duplicates resulting from a WGD) was based on syntenic information (21), using the S. cerevisiae genome (49) as well as genomes that did not undergo the WGD. For the small-scale gene duplication analysis, we first removed the 1,096 WGD genes from the original list of the 5,795 protein-coding genes. Next, we defined 2,439 S. cerevisiae genes as singletons, on the basis of the fact that their proteins had no Blast-p hit against any other S. cerevisiae proteins, at a cutoff level of 1e-3. As SSDs, we identified 2,260 genes that belong neither to the WGD group nor to the singletons group (SI_file2).
Inferring the Phosphorylation Sites of the Pre-WGD Ancestor.
On the basis of orthology–paralogy relationships identified previously (21), we aligned each S. cerevisiae protein (and ohnolog, whenever appropriate) with its orthologs from the three pre-WGD species (13, 32, 33) [A. (Eremothecium) gossypii, K. lactis, and K. waltii], using the t-coffee software (with default parameters) (50).
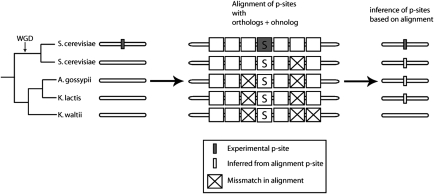
If the exact site and its neighboring amino acids were conserved in any of the orthologs from the three pre-WGD species, or in the other ohnolog (for WGD proteins only), then we inferred that this site was also present in their last common ancestor that was living just before the WGD, ∼100 mya (Fig. 3). It has been shown that the neighboring three amino acids to the left and the three to the right of the exact phosphorylation site are more conserved than the average (11). Therefore, taking into account this information, we generated eight different datasets (we call them ancestral) of ancestral p-sites, by arbitrarily allowing a variation of two, three, four, and six amino acids in the designated vicinity of the p-site, for both 6_exp_NU and 6_exp_U, as long as the p-site was not mutated (SI_file2).
Fig. 3.
Inference of p-sites in the pre-WGD orthologous ancestral protein. The inference is based on alignment with pre-WGD orthologs (and ohnolog, whenever appropriate). Here, the threshold for inference is no more than two amino acid mismatches in the window of six amino acids, surrounding the p-site.
By using the Yeast Genome Order Browser, we also identified the orthologs of S. cerevisiae WGD and RSS genes in each of the available three genomes of species (C. glabrata, S. castelli, and K. polysporus) that diverged after the WGD event. For each of the ancestral (pre-WGD) genes, we identified in how many of the four post-WGD species the duplicates were retained. Products of genes that survived as duplicates in the genomes of at least three of the four post-WGD yeast species are designated retained-in-majority, whereas products of genes that have returned to single-copy status in the genomes of at least three of those four post-WGD species are designated lost-in-majority. Thus, we compared the ancestral pre-WGD orthologs of the retained-in-majority bin vs. the lost-in-majority bin, for each of the eight ancestral p-site datasets.
Supplementary Material
Acknowledgments
We thank Tom Michoel, Anagha Joshi, and Stephane Rombauts for helpful technical discussions and Balázs Papp for his critical reading of the manuscript. S.G.O. acknowledges support from the Biotechnology and Biological Sciences Research Council (Grant BBC5051401). Y.V.d.P. acknowledges support from the Institute for the Promotion of Innovation by Science and Technology IWT (SBO-BioFrame) and the Inter-University Network for Fundamental Research (P6/25) (BioMaGNet). G.D.A. acknowledges support from the European Molecular Biology Organization (ALTF-930-2007).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911603107/DCSupplemental.
This article is a PNAS Direct Submission.
References
- 1.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 4.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 6.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis JC, Petrov DA. Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet. 2005;21:548–551. doi: 10.1016/j.tig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Gnad F, et al. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 14.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 15.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodenmiller B, et al. PhosphoPep—A database of protein phosphorylation sites in model organisms. Nat Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi A, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruhler A, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Li X, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 20.Reinders J, et al. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mintseris J, Weng Z. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci USA. 2005;102:10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 24.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 25.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 26.Pache RA, Babu MM, Aloy P. Exploiting gene deletion fitness effects in yeast to understand the modular architecture of protein complexes under different growth conditions. BMC Syst Biol. 2009;3:74. doi: 10.1186/1752-0509-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira-Leal JB, Audit B, Peregrin-Alvarez JM, Ouzounis CA. An exponential core in the heart of the yeast protein interaction network. Mol Biol Evol. 2005;22:421–425. doi: 10.1093/molbev/msi024. [DOI] [PubMed] [Google Scholar]
- 28.Steinmetz LM, et al. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 29.Zotenko E, Mestre J, O’Leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: Reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batada NN, et al. Stratus not altocumulus: A new view of the yeast protein interaction network. PLoS Biol. 2006;4:e317. doi: 10.1371/journal.pbio.0040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunin V, Pereira-Leal JB, Ouzounis CA. Functional evolution of the yeast protein interaction network. Mol Biol Evol. 2004;21:1171–1176. doi: 10.1093/molbev/msh085. [DOI] [PubMed] [Google Scholar]
- 32.Dietrich FS, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 33.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 34.Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25:193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Lienhard GE. Non-functional phosphorylations? Trends Biochem Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL. All duplicates are not equal: The difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson-Grady JT, Villén J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 44.Birchler JA, Veitia RA. The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He X, Zhang J. Gene complexity and gene duplicability. Curr Biol. 2005;15:1016–1021. doi: 10.1016/j.cub.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 46.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 47.Ingrell CR, Miller ML, Jensen ON, Blom N. NetPhosYeast: Prediction of protein phosphorylation sites in yeast. Bioinformatics. 2007;23:895–897. doi: 10.1093/bioinformatics/btm020. [DOI] [PubMed] [Google Scholar]
- 48.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goffeau A, et al. Life with 6000 genes. Science. 1996;274(546):563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 50.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.