Abstract
We show that translation initiation factor EF-Tu plays a second important role in cell shape maintenance in the bacterium Bacillus subtilis. EF-Tu localizes in a helical pattern underneath the cell membrane and colocalizes with MreB, an actin-like cytoskeletal element setting up rod cell shape. The localization of MreB and of EF-Tu is interdependent, but in contrast to the dynamic MreB filaments, EF-Tu structures are more static and may serve as tracks for MreB filaments. In agreement with this idea, EF-Tu and MreB interact in vivo and in vitro. Lowering of the EF-Tu levels had a minor effect on translation but a strong effect on cell shape and on the localization of MreB, and blocking of the function of EF-Tu in translation did not interfere with the localization of MreB, showing that, directly or indirectly, EF-Tu affects the cytoskeletal MreB structure and thus serves two important functions in a bacterium.
Keywords: Bacillus subtilis, bacterial cytoskeleton, cell shape
Maintenance of cell morphology is an essential trait for most cells. Except for the cell wall-less Mollicutes, bacterial cell shape is dictated by the peptidoglycan (murein) cell wall. However, it is still unclear how the murein sacculus of bacteria obtains its specific shape—be it rod shape, vibroid, spiral, or any other type of morphology. Several membrane proteins of unknown function [MreC, MreD, RodA, RodZ (1–5)] have been implicated in the maintenance of rod shape in several bacteria, as well as MreB (6, 7), an ortholog of actin. MreB has a structure that is very similar to that of actin and forms magnesium- and ATP- or GTP-dependent straight double filaments in vitro (8–10), while actin forms right-handed helical double filaments (11). MreB localizes as helical filaments underneath the cell membrane, and is essential for viability, in many bacteria analyzed so far (12). In Bacillus subtilis and in Caulobacter crescentus, MreB filaments remodel within a time scale of few minutes and appear to extend at the plus end and to retract at the minus end along a helical path (13–15). Depletion of MreB results in the loss of rod shape in many bacteria, showing that it is a genuine cytoskeletal element. MreB interacts with cell wall-synthesizing enzymes (16, 17), and with the MreC membrane protein (18, 19), which in turn interacts with cell wall-synthesizing enzymes (20). According to a model, the helical MreB filaments in the cytosol position the cell wall synthetic machinery within the envelope, setting up cell wall synthesis, which indeed occurs in a helical pattern along the lateral cell wall in several rod-shaped bacteria (21). MreB has also been shown to play an important role in the positioning of proteins to the cell poles (22). However, it is still unclear how MreB obtains its helical localization underneath the cell membrane, which is key to understanding how helical incorporation of new cell wall material is achieved. When expressed in yeast cells, Escherichia coli MreB forms straight filaments within the cytosol (23), suggesting that helicity is not an intrinsic property to MreB. Except for an interaction with RNA polymerase (24), it has been unclear whether MreB interacts with any other cytosolic proteins, and whether there are regulators of MreB filaments in bacteria.
Under rapid growth conditions, the translation elongation factor EF-Tu is the most abundant protein in most bacterial cells, reaching ≈10 times the concentration of ribosomes (i.e., up to 700,000 molecules per cell) (25). The concentration of EF-Tu varies greatly with growth rate, like that of ribosomes (26–28). EF-Tu binds to GTP, which is hydrolyzed upon delivery of the correct amino acyl-tRNA to the A site of the ribosome. GDP is exchanged for GTP through exchange factor EF-Ts, which exists at approximately one-tenth of the concentration of EF-Tu within the cytosol. EF-Tu has been reported to form filamentous structures in vitro (29, 30), and it has been speculated that EF-Tu may be a cytoskeletal element in prokaryotes. The latter idea is based on immuno-gold staining of electron micrographs (31, 32), where EF-Tu was observed in defined structures just under the cytoplasmic membrane in a variety of different bacterial species (e.g., Mycoplasma, E. coli, Thermoaerobacter) (33). However, it has remained unclear whether these structures also exist in live, nonfixed cells.
Here, we present evidence that EF-Tu plays a dual role in the model bacterium Bacillus subtilis, its established role in translation and an additional role in cell shape maintenance. We show that MreB and EF-Tu interact within B. subtilis cells, and also in vitro, and affect each other’s localization underneath the cell membrane. We therefore provide proof that EF-Tu is associated with the bacterial cytoskeleton and plays an important role in the maintenance of the MreB cytoskeletal elements.
Results
MreB and EF-Tu Interact in Vivo and in Vitro.
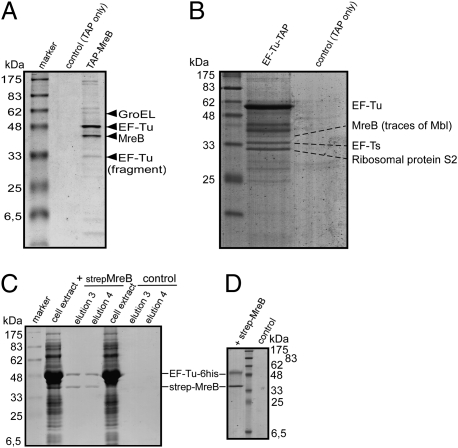
To identify further cytoskeletal elements, which we reasoned might be MreB interacting proteins in B. subtilis, we performed TAP-tag experiments with a functional N-terminal TAP-tag fusion of MreB, which was generated by a Campbell-like integration of a tap-mreB fusion at the original gene locus. After elution from a protein A Sepharose column by proteolytic cleavage of the first part of the TAP fusion, the calcium-binding domain fusion of MreB (CBD-MreB) was retrieved from the second affinity column (Fig. 1A). Several other proteins coeluted with MreB, in contrast to the control (in which only the TAP tag was expressed), which showed little or no signal (Fig. 1A). GroEL and EF-Tu were among the proteins that strongly interact with MreB, as identified by mass spectrometry. We investigated further the MreB/EF-Tu interaction because EF-Tu has been discussed as a cytoskeletal element in bacteria (29, 31–33). MreB was detected as an interacting protein in the inverse EF-Tu-TAP-tag assay in B. subtilis, together with GTP exchange factor EF-Ts, a specific EF-Tu interacting protein (Fig. 1B). Note that there is approximately one-tenth of EF-Ts relative to EF-Tu; i.e., up to 70,000 molecules per cell versus ≈15,000 molecules of MreB, suggesting that the MreB band indicates a considerable interaction between EF-Tu and MreB. To further verify the interaction between MreB and EF-Tu, we either expressed EF-Tu-6his alone (“control” in Fig. 1C) or coexpressed strep-MreB and EF-Tu-6His in E. coli cells, and purified MreB using streptavidin columns. Both MreB and EF-Tu proteins were recovered from the columns in an ≈1:1 stoichiometry after coexpression, but not in the control (Fig. 1C). As further controls for this experiment, we coexpressed EF-Tu-6his with the chromosome segregation protein ScpB or with the second MreB paralog, Mbl. EF-Tu-6his did not coelute with ScpB-strep and little with strep-Mbl (Fig. S1A). To rule out that an unknown factor from E. coli cell extract mediates the interaction, we purified strep-MreB, strep-Mbl, ScpB-strep, and EF-Tu-6his separately, and loaded streptavidin columns with either strep-MreB or the corresponding elution fraction of strep-columns from extract of E. coli cells carrying the empty MreB expression vector (“control” in Fig. 1D). EF-Tu-6his was retrieved quantitatively from the MreB-containing columns, but neither from control columns (Fig. 1D) nor from columns preloaded with strep-Mbl or ScpB-strep (Fig. S1B), showing that EF-Tu and MreB specifically interact in vivo and in vitro. EF-Tu could be purified as fully soluble protein dependent on the presence of GDP, and likewise, MreB only in the presence of ATP (and low salt); therefore, we were not able to determine whether the interaction of MreB and EF-Tu depends on different nucleotide cofactors.
Fig. 1.
Coomassie-stained SDS/PAGE experiments showing the interaction of MreB and EF-Tu. The identities of coeluting bands are indicated by triangles or lines. (A) TAP-tag experiment of cells expressing TAP-MreB (JS82); shown are the elution fractions of the second affinity column. Control, extracts from cells expressing TAP-tag only; TAP-MreB, extracts from cells expressing TAP-MreB. (B) TAP-tag experiments of cells expressing EF-Tu-TAP or TAP-tag only (“control”); shown are second elution fractions. (C) Strep-MreB and EF-Tu-6His coelution from streptavidin columns: overexpression of both proteins (“strep-MreB”) and or just of EF-Tu-6His (“control”) in E. coli cells. Shown are main MreB elution fractions. (D) Coelution of purified EF-Tu-6His from Streptavidin columns preloaded with purified strep-MreB (“strep-MreB”) or with elution fractions from a Streptavidin column from E. coli cell extract lacking any overexpressed protein (“control”).
EF-Tu Colocalizes and Interacts with MreB in Helical Filaments Underneath the Cell Membrane.
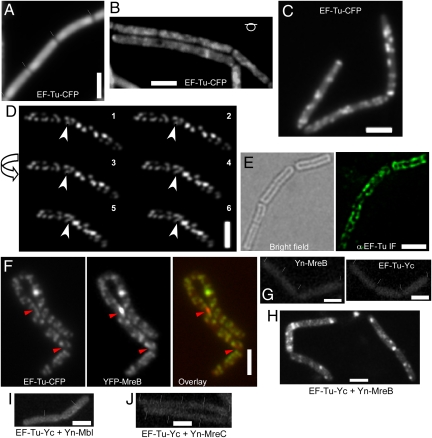
MreB localizes as helical filaments (most likely bundles of protofilaments) underneath the cell membrane in B. subtilis, E. coli, and C. crescentus cells (7). We wanted to investigate whether the MreB/EF-Tu interaction is also reflected by a specific subcellular localization of EF-Tu. Cells expressing EF-Tu-CFP as sole source of the proteins (under control of the native promoter) grew with indistinguishable doubling time as wild-type cells and expressed EF-Tu solely in the form of the CFP fusion (Fig. S2), showing that EF-Tu is fully functional. EF-Tu-CFP was mostly diffuse throughout the cytosol and visibly enriched at the cell poles during the early exponential growth phase (OD 0.1–0.5) (Fig. 2A), in full agreement with its function in translation, because ribosomes are also highly enriched at the cell poles (34). However, starting at mid exponential growth (OD 0.5–1.5), EF-Tu-CFP was observed as many discrete foci or transverse bands underneath the cell membrane, when the focal plane was shifted to the top of the cells (Fig. 2B). These foci became most clearly visible during late exponential growth (OD 1.5–4) (Fig. 2C), and their pattern was somewhat reminiscent of the MreB cytoskeleton (12, 14). Three-dimensional deconvolution of Z stacks taken from cells bearing EF-Tu-CFP revealed an obvious punctuate or band-like distribution of EF-Tu (Fig. 2D and Movies S1 and S2), with many elongated assemblies of EF-Tu along the length of the cells, in what appeared to be a helical arrangement. To confirm this striking localization pattern, we performed immunofluorescence experiments on cells growing at the late exponential phase. As shown in Fig. 2E, B. subtilis EF-Tu signals were observed as punctuate staining along the cell periphery, with transversal bands evocative of helical structures. It should be noted that IF experiments usually overemphasize the brightest signals, such that the diffuse staining of EF-Tu within the cytosol as seen with the CFP fusion is not apparent. These experiments verify the existence of distinct accumulations of EF-Tu along the cell periphery, in addition to the cytosolic localization of EF-Tu. We favor the idea that the accumulations of EF-Tu are also present during early exponential growth but are masked through the diffuse EF-Tu molecules, because a larger fraction of EF-Tu may be involved in translation at this growth period, and more EF-Tu may switch from translation to the peripheral structures as cells grow slower during late exponential growth.
Fig. 2.
Localization of EF-Tu in B. subtilis cells. (A–C) Cells expressing EF-Tu-CFP in B. Subtilis (JS88) at the original locus early exponential phase (A) (septa between cells are indicated by white lines and can usually be seen due to a lack of fluorescence between the cells), mid exponential phase (B), late exponential phase (C). (D) Three-dimensional deconvolution images. Arrow, direction of the turning angle of the cell; white arrowhead, an apparent helix. (E) Immunofluorescence on B. subtilis wild-type cells (late exponential growth) using anti-EF-Tu antiserum. (F) Two-dimensional deconvolution images showing colocalization of YFP-MreB and EF-Tu-CFP (strain JS89, late exponential growth). Red arrowheads, YFP-MreB foci (red in the overlay) that do not colocalize with EF-Tu-CFP foci (green in the overlay image). (G–J) BiFC interaction studies. Yn, N-terminal part of the split YFP; Yc, C-terminal part [note that coexpression of EF-Tu-Yc and just the Yn fragment (JS92) or of Yn-MreB and just the Yc fragment (JS93) did not show any defined signal]. (G) Expression of Yn-MreB (JS71; Left) or of EF-Tu-Yc (JS90; Right). (H) Cells expressing both EF-Tu-Yc and Yn-MreB (JS94). (I) Cells expressing of both EF-Tu-Yc and Yn-Mbl (JS95). (J) Cells expressing both EF-Tu-Yc and Yn-MreC (JS96). Cells were grown in S750 minimal media at room temperature supplemented with necessary antibiotics or inducers (0.5% xylose for YFP-MreB or 1 mM IPTG for Yn-fusions). (Scale bars: 2 μm.)
Based on their interaction (Fig. 1), we assumed that the EF-Tu structures along the cell membrane might colocalize with MreB structures, and therefore analyzed a strain expressing EF-Tu-CFP and YFP-MreB. A minor fraction of cells showed irregular cell shape and aberrant MreB localization (10–20%), suggesting that the strain expressing both fluorescent protein fusions may be somewhat dysfunctional. Longer cells contained more EF-Tu-CFP foci and YFP-MreB foci than shorter cells. In most cells, YFP-MreB foci/filaments colocalized with EF-Tu-CFP foci (Fig. 2F), although nonoverlapping signals were also observed. In 82% of all foci, these were coincident, whereas 18% of the foci were clearly distinct (generally, these were YFP-MreB foci without visible EF-Tu-CFP signal) (150 cells analyzed). To ensure that the MreB and EF-Tu structures indeed interact, we performed BiFC analysis. When EF-Tu or MreB were fused to either part of split-YFP (EF-Tu-Yc was expressed from the original gene locus, Yn-MreB from the ectopic amy site under the control of the IPTG inducible hyper-spank promoter), no fluorescence above background fluorescence was observed (Fig. 2G), like in cells expressing EF-Tu-Yc and the Yn part, or Yn-MreB and just the Yc part. Clear signals underneath the cell membrane were observed when EF-Tu-Yc and Yn-MreB were coexpressed (Fig. 2H). In 55 cells analyzed, fluorescence intensity of BiFC signals was 225 ± 25 AU, whereas fluorescence in control cells was 123 ± 3 AU (n = 60), similar to values in wild-type cells lacking any fluorescence protein fusion. MreB has been shown to interact with the other two MreB-like proteins, Mbl and MreBH, in B. subtilis, using BiFC (18). Mbl, but not MreB, has been shown to interact with the membrane protein MreC (18), so we used Yn-MreC as a control for the specificity of interaction between MreB and EF-Tu. No fluorescence different from that of wild-type cell was observed in the EF-Tu-Yc/Yn-MreC-expressing cells (Fig. 2I). These experiments show that the interaction between MreB and EF-Tu takes place between the cytoskeletal structures formed by the two proteins. We also found Mbl as an interacting partner in EF-Tu-TAP tag experiments, as determined by mass spectrometry. To further investigate this potential interaction, we performed BiFC with EF-Tu and Mbl, which did not reveal any specific signal (Fig. 2J), reinforcing the idea that EF-Tu specifically interacts with MreB, and only indirectly with Mbl, via MreB.
EF-Tu-CFP Structures Are More Static Than YFP-MreB Filaments and Depend on MreB.
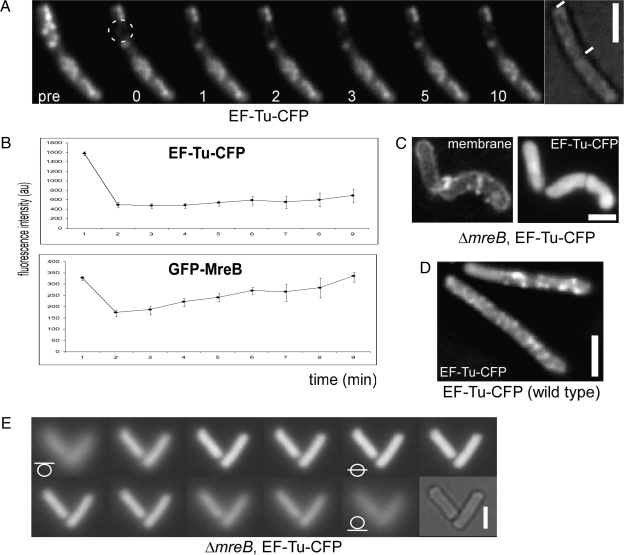
GFP-MreB filaments have been shown to be highly dynamic and need only a few minutes to fully recover after photobleaching (13, 18). We performed time-lapse experiments with EF-Tu-CFP, which revealed that the helical structures formed by EF-Tu are much more static than MreB filaments. EF-Tu-CFP structures did not change in their pattern between 1-min intervals for at least 10 min in growing cells (Movie S3), whereas MreB filaments changed their position within 10-s intervals (Movie S4). FRAP (fluorescence recovering after photobleaching) experiments verified that EF-Tu structures are rather static. EF-Tu-CFP structures did not recover after 10 min (Fig. 3A) following bleaching, in contrast to GFP-MreB filaments that show fast recovery within 1–2 min (18) (Fig. 3B). Thus, EF-Tu and MreB cytoskeletal structures frequently overlap but show different dynamics. These findings could also explain why MreB and EF-Tu signals are nonoverlapping in some cases.
Fig. 3.
Dynamics of EF-Tu-CFP filaments and effect of the loss of MreB. (A) FRAP experiment on EF-Tu-CFP-expressing cells (JS88). pre, before bleaching; dotted circle, area of bleaching; numbers, minutes after bleaching. (B) FRAP profiles of EF-Tu-CFP (JS88) and of GFP-MreB (JS12). (C) Two-dimensional deconvoluted images of the localization of EF-Tu-CFP in mreB null cells (JS97). (D) Two-dimensional deconvoluted image of EF-Tu-CFP in wild-type cells (JS88). (E) Z-stack through mreB mutant cells expressing EF-Tu-CFP (JS97). White lines through circles, position of the focal plane. For FRAP experiments, strains JS12 and JS88 were grown in S750 minimal media at room temperature supplemented with necessary antibiotics or inducers (0.5% xylose for GFP-MreB). Strains JS88 and JS97 were grown in PAB/SMM medium (high magnesium and sucrose concentration). Late exponentially growing cells (OD 1.5–3) were used for the microscopy. (Scale bars, 2 μm.)
To assess whether MreB affects the cytoskeletal EF-Tu structures, we moved the EF-Tu-CFP fusion into a strain carrying an in-frame mreB deletion, which is only possible under special media conditions (high magnesium and sucrose concentrations) (35). Strikingly, only homogeneously localized EF-Tu-CFP could be detected in the absence of MreB (Fig. 3C), in contrast to wild-type cells grown under the same conditions (Fig. 3D). Optical sectioning of mreB mutant cells verified the absence of any defined structures underneath the cell membrane (Fig. 3E), and Western blotting confirmed that the fusion is not cleaved and still expressed at a level comparable to that in wild-type cells (Fig. S2), showing that the formation of the EF-Tu structures depends on the presence of MreB.
Lowering of the Cellular EF-Tu Levels Affects Cell Shape and the Localization of MreB, but Has a Minor Effect on Translation.
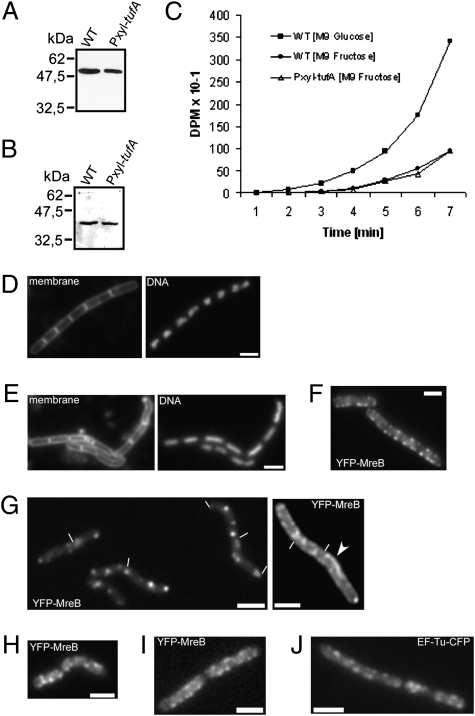
We next addressed the question of whether EF-Tu affects MreB. This is technically difficult because EF-Tu is essential for translation and thus for viability. However, to obtain some further information on the role of EF-Tu in cell shape determination, we investigated the effect of lowering the EF-Tu level in growing cells. EF-Tu is present in an ≈10:1 ratio to the number of ribosomes within bacterial cells (25, 31), and it has been speculated that only a fraction of the EF-Tu pool may be required for efficient translation. We were able to reduce the amount of EF-Tu by placing the EF-Tu gene under the control of the inducible Pxyl promoter. Approximately 40% of EF-Tu protein levels were detectable in the Pxyl strain (grown with the maximum induction of Pxyl with 0.5% xylose) compared with wild-type cells (Fig. 4A). Under these decreased EF-Tu levels, cells showed a long lag phase before exponential growth but were able to grow with a similar doubling time between OD 0.2 and 0.5 as wild-type cells (Fig. S3A). Interestingly, between 30% and 50% of the cells showed a strong defect in cell morphology (400 cells counted), being strongly bent, bulgy, or oval instead of rod-shaped [Fig. 4E (compare with Fig. 4D)]. To determine whether the observed phenotype is due to a decrease in the general translation capacity or the consequence of a more direct effect on cell shape maintenance, we measured the S35-methionine incorporation rate. In these translation efficiency experiments (Fig. 4C), the Pxyl-tufA strain did not show any significant difference compared with wild-type cells growing under the same condition [i.e., mid exponential growth at OD 0.5 in M9 fructose medium (Fig. S3A)]. It appears from these data that lowering the levels of EF-Tu has a strong effect on cell shape, but a small effect on translation. This finding could be further stipulated as the amount of MreB present in the Pxyl-tufA strain during mid exponential growth was similar to that in wild-type cells (Fig. 4B). Based on this, it was possible to ask whether the localization of MreB is affected by lowering the amount of EF-Tu. Instead of regular helical structures (Fig. 4F), irregular and often straight and extended GFP-MreB filaments were observed, which are never observed in wild-type cells, as well as irregularly positioned foci along the lateral cell membrane (Fig. 4G) (at OD 0.5 in M9 medium). Many foci contained a high amount of GFP-MreB, indicating that MreB filaments may bundle or aggregate in an uncontrolled manner. Importantly, MreB filaments were no longer dynamic and did not change their position within 5 min (Movies S5 and S6), showing that, under lowered levels of EF-Tu, MreB forms rather static filaments that lose their helical path underneath the membrane. Because EF-Tu failed to form defined structures in the ΔmreB strain, our analyses suggest that both proteins require each other’s presence in sufficient amounts to properly localize in helical structures. The analyses also indicate that EF-Tu affects the dynamics of MreB filaments in vivo. It is possible, though, that lowering of EF-Tu levels has an indirect effect on MreB localization and filament dynamics, via an effect on a different protein affecting cell morphology.
Fig. 4.
Effect of the depletion of EF-Tu in B. subtilis cells. (A) Western blot using EF-Tu antiserum showing levels of EF-Tu in wild-type cells (WT) and in the pxyl-tufA strain (JS91, OD = 0.5, M9 minimal medium plus fructose and xylose). (B) Western blot using MreB antiserum showing levels of MreB in wild-type (WT) cells and in the pxyl-tufA strain (JS91, OD = 0.5). (C) Translation efficiency (incorporation of radioactive methionine into cellular proteins) in wild-type and pxyl-tufA (JS91) cells. Cells were grown in M9 medium with fructose and xylose until early exponential phase (OD600 = 0.5), where wild-type and JS91 cells showed the same doubling time (Fig. S3). See SI Materials and Methods for details. (D–G) Fluorescence microscopy of early exponential phase (OD600 = 0.5) growing B. subtilis cells in M9 medium with fructose and xylose. (D) Wild-type cells. (E) pxyl-tufA cells (JS91, EF-Tu depletion). (F) Localization of YFP-MreB in wild-type cells (JS36). (G) pxyl-tufA cells (JS98). Arrowhead, irregular nonhelical filament; white lines, ends of cells. (H) Cells of strain JS98 (YFP-MreB, pxyl-tufA) growing in SMM/PAB medium (high magnesium and sucrose). (I–J) Cells 1 h after addition of kirromycin, where growth had ceased, after growth in S750 minimal medium to late exponential phase. (I) Cells expressing YFP-MreB (JS36). (J) Cells expressing EF-Tu-CFP (JS88). (Scale bars, 2 μm.)
The addition of a high concentration of magnesium (in PAB medium) was reported to rescue the lethality and cell morphology defect of mreB mutant cells (35). Addition of magnesium to PAB medium did not restore the cell shape defect in pxyl-tufA cells, but interestingly, YFP-MreB structures became somewhat more regular (Fig. 4H) (yet still not as regular as in wild-type cells) compared with the highly defective assemblies in normal medium (Fig. 4G), suggesting that magnesium may stabilize the defective MreB filaments in the EF-Tu depletion strain.
The Function of EF-Tu in Cell Shape Maintenance Is Not Affected Through the Inhibition of EF-Tu in Translation.
The above experiments suggest that EF-Tu has a dual function, in translation and in cell shape maintenance via MreB modulation. To address the question of whether, possibly, two different populations of EF-Tu confer these different roles, we used kirromycin, which specifically blocks the release of EF-Tu from the ribosome, thereby inhibiting elongation of translation (36, 37). The addition of 100 μg of kirromycin to exponentially growing B. subtilis cells expressing EF-Tu-CFP or YFP-MreB (both from the respective original gene loci) arrested cell growth after 1 h of incubation, showing that the block in translation had become effective. However, in these cells, clear YFP-MreB or EF-Tu-CFP structures underneath the cell membrane similar to those in wild-type cells were present (Fig. 4 I and J and Fig. S4), showing that a block in the function of EF-Tu in translation does not affect the localization of EF-Tu to the helical structures or the localization of MreB filaments. These experiments show that EF-Tu functions in translation and in cell shape maintenance can be separated.
Discussion
Our work provides evidence that the bacterial translation factor EF-Tu serves two distinct vital roles in the bacterium Bacillus subtilis. Besides its function in the elongation cycle of translation, which involves a fraction of EF-Tu at sites close to the cell poles, where most of the ribosomes are accumulated (34, 38), a distinct pool of EF-Tu affects cell morphology, apparently through an effect on the cytoskeletal element MreB. We show that EF-Tu is a major interaction partner of MreB in vivo and interacts in a 1:1 stoichiometry with MreB in vitro. Because MreB and EF-Tu have been suggested to be interaction partners in E. coli (39), it is likely that our findings are also valid for many other bacteria. Intriguingly, eukaryotic elongation factor EF1a has also been shown to interact with actin in eukaryotic cells (40, 41), so the interaction between these two proteins appears to be conserved through the domains of organisms. The interaction of EF-Tu and MreB is reflected in the localization of EF-Tu: a functional EF-Tu-CFP fusion localizes to distinct sites along the lateral cell membrane and extensively colocalizes with MreB. Because MreB forms helical filaments underneath the cell membrane, it follows that EF-Tu structures are also positioned in a helical fashion. BiFC experiments show that EF-Tu and MreB interact at these sites, showing that a fraction of EF-Tu is associated with the MreB cytoskeleton. EF-Tu has been shown to be present at an ≈10-fold excess over ribosomes (25, 31), and it has been speculated that there may be no need for such an excess of the elongation factor. We therefore generated a strain in which transcription of the tufA gene encoding for EF-Tu is under the control of an inducible promoter. Even under maximum induction, cells contained <50% of EF-Tu compared with wild-type cells. In these cells, translation occurred at a rate comparable to that in wild-type cells, but the cells displayed a highly aberrant cell morphology, in that they were twisted and bent. Interestingly, in these cells, MreB was present at wild-type levels but had lost its helical localization; rather, extended straight or curved filaments were observed localizing along the long axis of the cells. These experiments show that a decrease in the concentration of EF-Tu leads to a defect in cell morphology and in MreB localization, while translation is not detectably affected. In addition, MreB filaments did not show the dynamic turnover that characterizes MreB filaments in wild-type cells (13, 14). We have shown that the dynamics of MreB filaments are essential for the proper function of the cytoskeletal element, indicating the need to ensure sufficient filament turnover (18). Based on the finding that EF-Tu structures are rather static and do not turn over within a few minutes like MreB filaments, we speculate that EF-Tu serves as tracks for MreB filaments, which (based on the interaction between the two proteins) could extend along the more static EF-Tu structures (Fig. S5). Interestingly, the defined EF-Tu structures in B. subtilis depended on MreB, showing that the proteins mutually affect each other’s pattern of localization and that, therefore, EF-Tu is a true component of the bacterial cytoskeleton. We speculate that EF-Tu is positioned in its GDP form (in which it interacts with MreB) through MreB filaments and thereby forms the relatively static extended structures, which may not be available for EF-Ts to exchange the GDP cofactor. MreB may assemble on these structures in its ATP bound form, and may disassemble after ATP hydrolysis, thus dynamically remodeling along EF-Ts tracks.
To further analyze the two functions of EF-Tu, we blocked the activity of EF-Tu in translation by the specific antibiotic kirromycin, which resulted in an eventual arrest in cell growth but did not affect the localization of MreB or the formation of the helical EF-Tu structures underneath the membrane. These analyses indicate that two distinct EF-Tu fractions are involved in translation, which mostly occurs at polar zones in B. subtilis (Fig. S5) and as a cytoskeletal element in conjunction with MreB. It will be important to investigate how the recruitment of EF-Tu to the cytoskeletal structure is regulated and how the cell ensures that both essential functions of EF-Tu are granted to occur in a proper fashion.
Materials and Methods
Growth Conditions.
For genetic manipulations, E. coli and B. subtilis strains were grown in LB rich medium. Transformants were selected under antibiotics pressure, as required, with 100 μg/mL ampicillin, 50 μg/mL streptomycin, 5 μg/mL chloramphenicol, 50 μg/mL spectinomycin, and 5 μg/mL kanamycin. For microscopy, B. subtilis cells were grown in S750 or M9 minimal medium, except for mreB mutant cells, which were grown in PAB medium supplemented with 20 mM magnesium and 0.5 M sucrose. Expression of YFP-MreB in the Pxyl-tufA strain was driven by 0.5% xylose in S750 medium containing fructose instead of glucose. The generation of strains is described in SI Materials and Methods.
Strains and Plasmids.
All plasmids and Bacillus strains generated in this work are listed in Tables S1 and S2, respectively.
Immunofluorescence and Microscopy.
EF-Tu antiserum was obtained through immunization of rabbits with an EF-Tu-derived peptide (derived from the last 20 amino acids of EF-Tu). Cells were fixed with glutaraldehyde and were treated with 1:5,000 diluted EF-Tu antiserum, and with secondary Alexa Fluor 488-coupled secondary antibody (Molecular Probes/Invitrogen). For fluorescence microscopy, DNA was stained with 4′,6-diamidino-1-phenylindole (DAPI) (0.2 ng/mL), and membranes were stained with FM4-64 (1 nM). For further details, see SI Materials and Methods. Fluorescence intensities were determined with MetaMorph 6.5 (MDS Analytical Technologies).
Supplementary Material
Acknowledgments
We thank David Rudner (Harvard Medical School) for the gift of BsCFP. This work was supported by Deutsche Forschungsgemeinschaft grant FOR 929.
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911979107/DCSupplemental.
References
- 1.Matsuzawa H, Hayakawa K, Sato T, Imahori K. Characterization and genetic analysis of a mutant of Escherichia coli K-12 with rounded morphology. J Bacteriol. 1973;115:436–442. doi: 10.1128/jb.115.1.436-442.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wachi M, Doi M, Okada Y, Matsuhashi M. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:6511–6516. doi: 10.1128/jb.171.12.6511-6516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendezú FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alyahya SA, et al. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wachi M, et al. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- 8.van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 9.Esue O, Cordero M, Wirtz D, Tseng Y. The assembly of MreB, a prokaryotic homolog of actin. J Biol Chem. 2005;280:2628–2635. doi: 10.1074/jbc.M410298200. [DOI] [PubMed] [Google Scholar]
- 10.Bean GJ, Amann KJ. Polymerization properties of the Thermotoga maritina actin MreB: roles of temperature, nucleotides, and ions. Biochemistry. 2008;47:826–835. doi: 10.1021/bi701538e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 12.Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 13.Carballido-López R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- 14.Graumann PL, Defeu Soufo HJ. An intracellular actin motor in bacteria? Bioessays. 2004;26:1209–1216. doi: 10.1002/bies.20126. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- 17.Vats P, Shih YL, Rothfield L. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol Microbiol. 2009;72:170–182. doi: 10.1111/j.1365-2958.2009.06632.x. [DOI] [PubMed] [Google Scholar]
- 18.Defeu Soufo HJ, Graumann PL. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol Microbiol. 2006;62:1340–1356. doi: 10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- 19.Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 20.Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci USA. 2005;102:18602–18607. doi: 10.1073/pnas.0507937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 22.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan R, Mishra M, Murata-Hori M, Balasubramanian MK. Filament formation of the Escherichia coli actin-related protein, MreB, in fission yeast. Curr Biol. 2007;17:266–272. doi: 10.1016/j.cub.2006.11.069. [DOI] [PubMed] [Google Scholar]
- 24.Kruse T, et al. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 2006;20:113–124. doi: 10.1101/gad.366606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furano AV. Content of elongation factor Tu in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krásný L, et al. Structure and expression of elongation factor Tu from Bacillus stearothermophilus. J Mol Biol. 1998;283:371–381. doi: 10.1006/jmbi.1998.2102. [DOI] [PubMed] [Google Scholar]
- 27.Krásný L, Vacík T, Fucík V, Jonák J. Cloning and characterization of the str operon and elongation factor Tu expression in Bacillus stearothermophilus. J Bacteriol. 2000;182:6114–6122. doi: 10.1128/jb.182.21.6114-6122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young FS, Furano AV. Regulation of the synthesis of E. coli elongation factor Tu. Cell. 1981;24:695–706. doi: 10.1016/0092-8674(81)90096-9. [DOI] [PubMed] [Google Scholar]
- 29.Beck BD, Arscott PG, Jacobson A. Novel properties of bacterial elongation factor Tu. Proc Natl Acad Sci USA. 1978;75:1250–1254. doi: 10.1073/pnas.75.3.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck BD. Polymerization of the bacterial elongation factor for protein synthesis, EF-Tu. Eur J Biochem. 1979;97:495–502. doi: 10.1111/j.1432-1033.1979.tb13137.x. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson GR, Rosenbusch JP. Abundance and membrane association of elongation factor Tu in E. coli. Nature. 1976;261:23–26. doi: 10.1038/261023a0. [DOI] [PubMed] [Google Scholar]
- 32.Cremers AF, Bosch L, Mellema JE. Characterization of regular polymerization products of elongation factor EF-Tu from Escherichia coli by electron microscopy and image processing. J Mol Biol. 1981;153:477–486. doi: 10.1016/0022-2836(81)90293-x. [DOI] [PubMed] [Google Scholar]
- 33.Mayer F. Cytoskeletal elements in bacteria Mycoplasma pneumoniae, Thermoanaerobacterium sp., and Escherichia coli as revealed by electron microscopy. J Mol Microbiol Biotechnol. 2006;11:228–243. doi: 10.1159/000094057. [DOI] [PubMed] [Google Scholar]
- 34.Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 2000;19:710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Formstone A, Errington J. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 36.Wolf H, Chinali G, Parmeggiani A. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc Natl Acad Sci USA. 1974;71:4910–4914. doi: 10.1073/pnas.71.12.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blumenthal T, Douglass J, Smith D. Conformational alteration of protein synthesis elongation factor EF-Tu by EF-Ts and by kirromycin. Proc Natl Acad Sci USA. 1977;74:3264–3267. doi: 10.1073/pnas.74.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascarenhas J, Weber MH, Graumann PL. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2001;2:685–689. doi: 10.1093/embo-reports/kve160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 40.Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 41.Liu G, et al. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.