Abstract
Keap1 is a substrate adaptor of a Cullin 3-based E3 ubiquitin ligase complex that recognizes Nrf2, and also acts as a cellular sensor for xenobiotics and oxidative stresses. Nrf2 is a transcriptional factor regulating the expression of cytoprotective enzyme genes in response to such stresses. Under unstressed conditions Keap1 binds Nrf2 and results in rapid degradation of Nrf2 through the proteasome pathway. In contrast, upon exposure to oxidative and electrophilic stress, reactive cysteine residues in intervening region (IVR) and Broad complex, Tramtrack, and Bric-à-Brac domains of Keap1 are modified by electrophiles. This modification prevents Nrf2 from rapid degradation and induces Nrf2 activity by repression of Keap1. Here we report the structure of mouse Keap1 homodimer by single particle electron microscopy. Three-dimensional reconstruction at 24-Å resolution revealed two large spheres attached by short linker arms to the sides of a small forked-stem structure, resembling a cherry-bob. Each sphere has a tunnel corresponding to the central hole of the β-propeller domain, as determined by x-ray crystallography. The IVR domain appears to surround the core of the β-propeller domain. The unexpected proximity of IVR to the β-propeller domain suggests that any distortions generated during modification of reactive cysteine residues in the IVR domain may send a derepression signal to the β-propeller domain and thereby stabilize Nrf2. This study thus provides a structural basis for the two-site binding and hinge-latch model of stress sensing by the Nrf2-Keap1 system.
Keywords: Nrf2, oxidative stress, single particle analysis, transmission electron microscopy, three-dimensional reconstruction
Keap1 (Kelch-like ECH-associated protein 1) is a multifunctional protein that represses activity of the transcriptional factor Nrf2 (NF-E2-related Factor 2). Nrf2 promotes expression of various cytoprotective genes in response to xenobiotic and oxidative insults (1–3). Whereas Keap1-null mutant mice are juvenile-lethal due to hyperkeratosis of the esophagus (4), hepatocyte-specific knockout of the Keap1 gene robustly elevates accumulation of Nrf2 in the nucleus and protects hepatocytes against acute drug toxicity and inflammatory liver injury (5, 6). In contrast, the risk of oxidant-induced acute lung injury is significantly increased upon dysfunction of the Nrf2 gene (7). Similarly, Nrf2-null mutant mice are susceptible to environmental toxicants and various pathologies, including hepatotoxicity, pneumotoxicity, neurotoxicity, carcinogenicity, and inflammation (8–10). The induction of Phase 2 enzymes through augmentation of the Nrf2 activity has been widely accepted as a promising approach for cancer chemoprevention, for protection against oxidative stress, and for chemoprophylaxis against stress-related disorders (11–14). Indeed, a number of Keap1 loss-of-function somatic mutations have been identified in human lung cancers and cancer-derived cell lines in which Nrf2 is constitutively active (15–17). The latter finding suggests that Keap1 may sensitize cancer cells to chemotherapy.
Keap1 is a member of newly described BTB and C-terminal Kelch (BACK) domain-containing protein family (18). Keap1 is a substrate adaptor component of a Cul3-dependent E3 ubiquitin ligase complex that contributes to the rapid degradation of Nrf2 (19–22). Oxidative and electrophilic stresses covalently modify cysteine thiol residues of Keap1 and abrogate the Keap1-E3 ubiquitin ligase activity, which leads to the stabilization and activation of Nrf2 (23). Thus, in response to such stress, Nrf2 is released from Keap1 repression, accumulates in the nucleus, and activates transcription of cytoprotective enzyme genes.
Keap1 retains four discrete domains: i.e., BTB (Broad complex, Tramtrack and Bric-à-Brac), IVR (intervening region), DGR (double glycine repeat), and CTR (C-terminal region). The DGR and CTR domains in collaboration form a β-propeller structure and interact with the Neh2 domain of Nrf2 (hereafter we refer to these two domains of Keap1 as the DC domain). The IVR domain contains two critical cysteine residues, Cys272 and Cys288, that are important for repression of Nrf2 activity. This suggests that these residues are important for the electrophilic stress sensing (23, 24). Whereas the BTB domain acts to dimerize Keap1, the Cys151 residue in the BTB domain also appears to be important in stress sensing, even though it is not required for the repression of Nrf2 activity per se.
We describe a unique molecular mechanism employed in the repression of Nrf2 activity. The substrate adaptor Keap1 forms a dimer to accommodate Nrf2 and the stoichiometry of Nrf2: Keap1 is 1∶2. So that the Keap1 dimer recognizes and binds two evolutionarily conserved degrons, DLG and ETGE in Nrf2. These two motifs show two orders of magnitude difference in the association constant to the DC domain of Keap1. The two-site binding enables Keap1 to initiate efficient ubiquitination of Nrf2 (25–27). Importantly, perturbation of the Keap1-Nrf2 binding, especially at the weak DLG site, blocks Nrf2 degradation (a hinge and latch mechanism) and this constitutes the critical electrophilic/oxidative stress sensor function (28, 29). The necessity of both degrons for efficient Nrf2 turnover has been supported through structural, biochemical, and clinical analyses (27, 30).
To understand the molecular mechanisms underlying the interaction of Keap1 with Nrf2, two groups of investigators have solved the crystal structure of the substrate-binding DC domain of mouse and human Keap1 (15, 31). Nonetheless, the molecular mechanism whereby modification of IVR Cys residues affects the binding of Nrf2 degron motifs to the DC domain remains to be clarified. This is partly due to the difficulty in crystallizing the full-length dimeric Keap1 protein. To this end, we examined the three-dimensional imaging/reconstruction analysis of mouse Keap1 homodimer by EM by use of negatively stained proteins.
Results
Purification of Mouse Keap1 Protein.
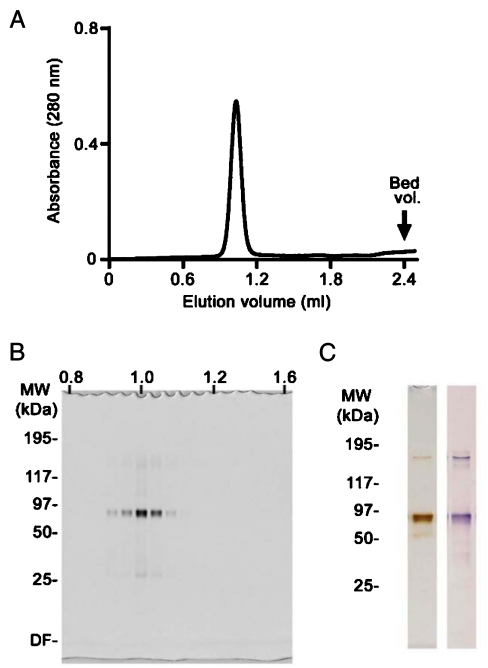
Recombinant mouse Keap1 (M1—R614) protein with a C-terminal six-His tag was purified by Ni-NTA agarose affinity chromatography and twice by Superdex S-200 size exclusion chromatography (SEC). Keap1 was eluted from the second SEC as a sharp peak at 1.03 mL (Fig. 1A), which corresponds to a major protein band with an apparent molecular weight of approximately 80 kDa in SDS-PAGE (Fig. 1B). The major protein band was confirmed to be Keap1 by immunoblotting analysis with anti-Keap1 antibody (Fig. 1C, right). A faint band at 150 kDa corresponds to the size of Keap1 dimer. An aliquot at 1.03 ml elution was used for the EM study.
Fig. 1.
(A) Size exclusion chromatography. Recombinant Keap1 was purified using a Superdex S200 SEC column before EM analysis. Protein was routinely eluted at 1.03 mL as a single sharp peak; this fraction was used for EM image analysis. (B) SDS-PAGE and silver staining. The purity of fractions eluted from the SEC was monitored by SDS-PAGE and silver staining. (C) Purity of Keap1 was verified by silver staining (Left) and immunoblotting analysis using anti-Keap1 antibody (Right).
Molecular Shape of Keap1 by EM.
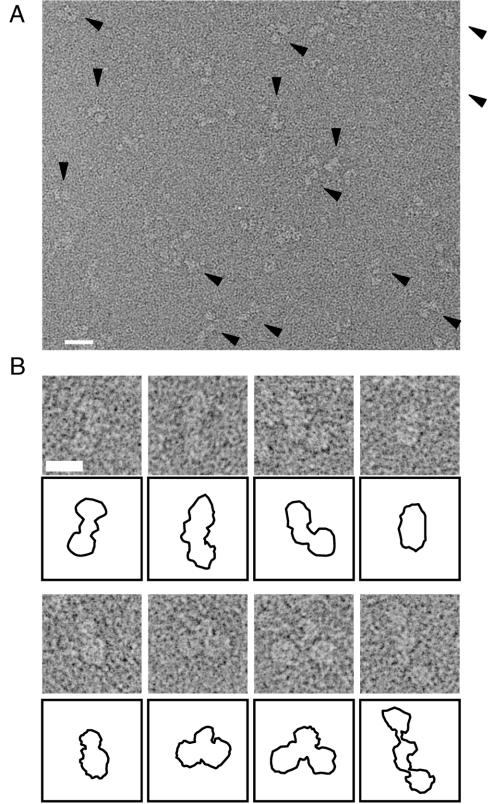
Mouse Keap1 recombinant protein was negatively stained and imaged by EM at x 52,100. Keap1 displayed variously shaped particles of uniform size with dimeric assembly (Fig. 2A). Most particles had two isolated spherical bodies joined via linkers to a forked-stem structure (Fig. 2B). The distance between the two spherical masses varied, reflecting different orientations of the same molecule. Although rarely observed, the image of two spheres joined by a straight linker (Fig. 2B, Upper Left) is postulated to be the top view, while particles with a hinged linker likely represent side views (Fig. 2B, Lower Second from Right). Minor projections with extremely distant spherical bodies were also observed (Fig. 2B, Lower Right).
Fig. 2.
EM image of negatively stained Keap1. (A) Keap1 particles were observed as uniformly sized projections (arrowheads). Protein is shown in bright shades. Scale bar, 200 Å. (B) Examples of the Keap1 projections with schematic diagrams below each panel. An example of a minor population with extremely distant round spheres is also displayed at the rightmost bottom. Scale bar, 100 Å. For statistical analysis, 216 particles were automatically picked up by the auto-accumulation method and utilized as training data for the three-layer neural network (NN) auto-picking system. The trained NN selected 12,651 particles for analysis.
Three-Dimensional Reconstruction of Keap1.
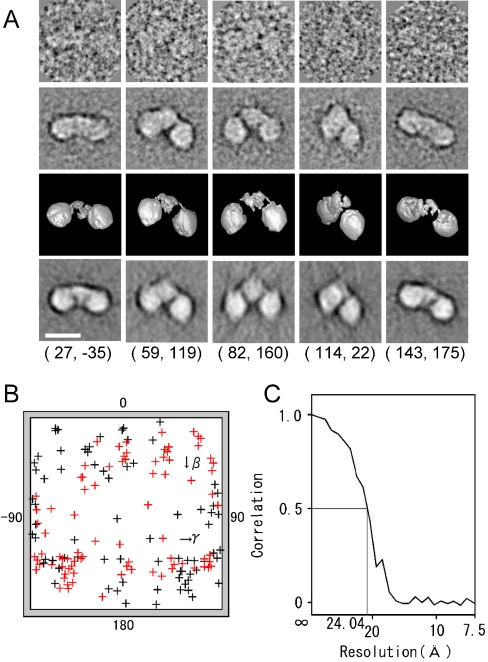
We then performed a three-dimensional reconstruction (32) using our Single Particle Image analysis method using Neural Networks and Simulated annealing programs (33–36) and the IMAGIC V software (37). From the initial 12,651 EM images, 9,827 particles (77.7%) were selected during the image analysis for the final reconstruction. Representative raw images are presented in Fig. 3A (First Row) with their corresponding class averages (Second Row) and with surface representations and projections of the reconstructed three-dimensional structure (Third and Fourth Rows). A high degree of consistency was observed in size, shape, and inner structure among these datasets (Fig. 3A), indicating successful reconstruction from the original particle images.
Fig. 3.
Three-dimensional reconstruction of Keap1. (A) Raw images of Keap1 with different Euler angles (Row 1), compared with the corresponding two-dimensional averages (Row 2), the surface views of the three-dimensional reconstruction (Row 3), and the reprojections of the three-dimensional reconstruction (Row 4) consistent through the reconstruction. The Euler angle (β, γ) is denoted below each column. Protein is displayed in bright shades. Scale bar, 100 Å. (B) Surface projection of Euler angle (β, γ) distribution of 162 adopted class averages. The distribution covers the whole angular range, with a small bias reflecting a somewhat preferred orientation of Keap1 molecules on the carbon surface. (C) Fourier shell correlation function indicates the resolution limit of 24 Å by the FSC > 0.5 criterion.
Euler angle distribution of 162 adopted class averages (Fig. 3B) appears to cover the whole angular range, with a small bias reflecting a somewhat preferred orientation. However, this is not considered to cause distortion in the final structure. The resolution is 24 Å if a Fourier shell correlation function (38) of > 0.5 was used as the resolution criterion (Fig. 3C).
Structural Features of the Keap1 Molecule.
The surface representation shows two spherical bodies joined by thin linkers to a forked-stem structure, roughly resembling a pair of cherries (Fig. 4, Left). From the top, the two round domains form a nearly straight line with the stem-like joint in the center (Fig. 4B, Left). Keap1 measures about 160 Å across the width of the “cherries” and is 60 Å at its greatest depth (Fig. S1A). Molecular height estimated from the side view was 103 Å. Each globular domain was a vertically elongated sphere with dimensions of 60 × 60 × 78 Å; the surface was basically continuous and smooth. In contrast, the surface of the stem region was rough and thin; one linker leading from the stem structure near the point of connection appeared to be partially disengaged from its “cherry.” Two small orifices were also observed on each sphere: A larger orifice approximately 15 × 17 Å is prominent on the top (Fig. S1B, 18–25) and a relatively smaller elliptical orifice of approximately 10 × 12 Å or smaller, appears at the bottom (Fig. S1B, 1–3). Between these orifices is a vertical tunnel (described below). For surface representation, the three-dimensional map is contoured at an isosurface containing a volume corresponding to 155.3 kDa, which is 110.3% of the molecular mass of dimeric Keap1 (140.8 kDa) calculated from the amino acid composition. This additional volume is attributable to minor thermal-vibration-induced structural variations, which cannot be ascertained by current biochemical techniques or software.
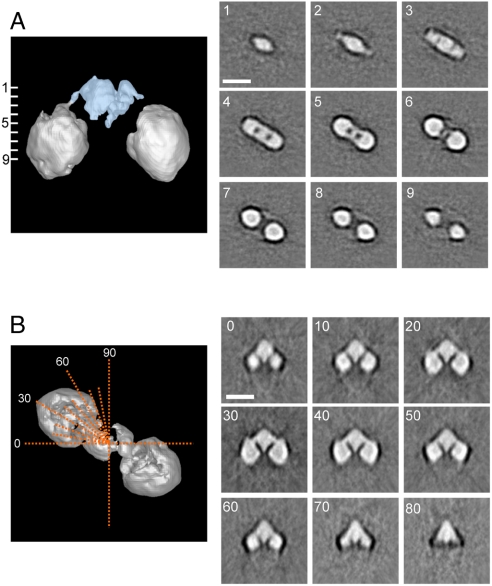
Fig. 4.
Sections through Keap1 molecule. (A) Horizontal slices perpendicular to the pseudo twofold symmetric axis show the characteristic cherry-shaped structure of Keap1 (Right). Positions of the cross sections, at 9.43 Å intervals throughout the molecule, are numbered from one to nine on the side surface view (Left). The stem structure, occupying 13.5% of the total volume (Blue). (B) Axial slices (Orange Dotted Lines; top surface view) every 10 ° from 0 ° to 90 ° (Left). Each sectional image is shown with corresponding angle (Right). Two globular domains are connected to the central stem-like density with slender linkers. The central stem-like density comprises two thin layers connected to one another but separated by a thin gap. The globular domains on either side are dense cylinders with round corners, pierced by an apparent low-density tunnel between the two surface orifices. Protein is displayed in bright shades. Scale bars, 100 Å.
Internal Structure of Keap1.
Sections through the Keap1 molecule are displayed in Fig. 4 (Right). As demonstrated by the density map of the vertical sections, the two globular domains connect indirectly through narrow linkers to a central mass (Fig. 4B, Right 20–40). The stem-shaped central mass roughly comprised two thin, interacting layers separated by a narrow gap. Each globular cherry is a rounded, high-density cylinder with an internal low-density tunnel formed between the two small surface orifices (Fig. 4A, 4–9 and Fig. 4B, 0–60).
Assignment of the DC Domain to the Globular Densities.
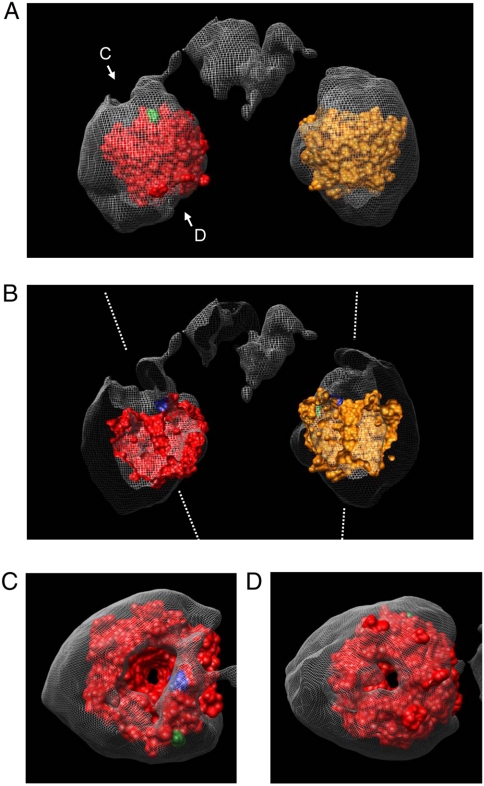
We recently reported on the crystal structure of the DC domain formed by residues 324 to 613 of mouse Keap1 (PDB accession code 1X2J) (15). We searched the low-resolution three-dimensional map for densities that could accommodate the DC structure by visual inspection using Chimera. Only the prominent globular domains showed a good fit (Fig. 5). One of the internal tunnels (mentioned above) is inclined at 20 ° to the pseudosymmetric axis, while the other is almost parallel to the axis (Fig. 5B, White Dotted Lines). Each of the tunnels fits the central channel of the drum-shaped β-propeller DC domain in the atomic model (Fig. 5B), although in the reconstruction an additional bulging density is apparent around the DC domain (Fig. 5C, D).
Fig. 5.
Molecular fitting of the DC crystal structure to the Keap1 volume. (A) Atomic model of the Keap1-DC domain (PDB accession code 1X2J (15)), (Orange or Red), fitted into the EM density map. (B) Front half cutaway view of Keap1 molecule. One tunnel is inclined at 20 ° from the pseudosymmetry axis, while the other is almost parallel to the axis (White Dotted Lines). Tunnels closely coincide with those in the superimposed atomic model. (C) View from direction “C” as depicted in A. (D) View from direction “D” as depicted in A. The small exit of each tunnel closely coincides with that in the x-ray model. Extra density observed in the globular domain is composed of the IVR domain and part of the BTB domain, as described in Fig 6D. Distance between the two Nrf2 binding sites located on the bottom of the DC domains is approximately 80 Å. The N and C termini of the DC domain are denoted by Blue and Green, respectively.
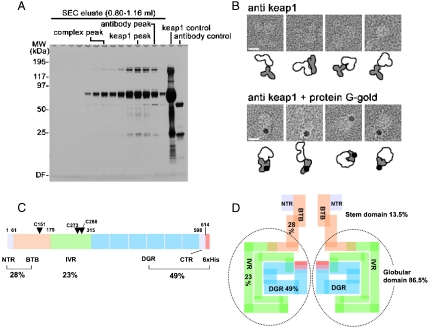
To further confirm the molecular fit of the DC domain, the protein was mixed with either anti-Keap1 or gold-conjugated anti-Keap1 monoclonal antibodies, which recognize epitopes within the DC domain. To remove unbound Keap1 from samples, the Keap1/anti-DC antibody complex was isolated by using SEC (Fig. 6A). Because the Keap1 dimer (140 kDa)/antibody (150 kDa) complex is bigger than the free Keap1 dimer or the antibody alone, the complex was clearly separated as the earliest peak. Notably, free Keap1 molecules, although smaller than IgG in molecular mass, were subsequently eluted before the free antibody. IgG has a maximum dimension of 150Å, and its other dimensions are comparable with those of Keap1. Considering the flexible nature of the linker among the two Fabs and Fc, this elution profile suggests a relatively stable, stiff architecture between Keap1 domains.
Fig. 6.
BTB, IVR, and DC domains of Keap1.(A) Purification of the anti-DC antibody-bound Keap1 complex. SDS-PAGE analysis shows isolation of the antibody/Keap1 complex by SEC. The purified complex was eluted in early fractions at approximately 290-kDa region. (B) The large cherries of the molecule contain the DC domains. Gallery of negatively stained anti-DC/Keap1 complexes (Upper) and protein-G-gold/anti-DC antibody/Keap1 complexes (Bottom). Schematic diagrams of Keap1 (White), antibodies (Gray), and gold particles (Black) are illustrated below each panel. Scale bars,100 Å. (C) Protein architecture of Keap1 showing BTB, IVR, DGR, and CTR domains with corresponding primary sequence boundaries. Reactive cysteines (Cys151, Cys273, and Cys288) important for stress sensing are indicated with single letter codes and residue numbers. Percentages (%) of each functional domain are also shown. (D) Schematic distribution of the four functional domains in the Keap1 homodimer, as predicted by volume ratio. The stem structure occupies only 13.5% of the whole volume, while the globular regions comprise about 86.5%, suggesting that the entire IVR and part of the BTB domain are integrated into the globular cherries.
Indeed, the negatively stained images revealed that the antibody attaches to one of the large spheres of Keap1, confirming the presence of the DC domain in the sphere (Fig. 6B, Upper). The antibody binding is further verified through the binding of protein G-gold conjugate (Fig. 6B, Lower).
Location of BTB, IVR, and DC Domains.
It has been believed that in the Keap1 molecule, the IVR forms a long helical linker that connects the BTB domain and DC domain, or the β-propeller substrate-binding domain (Fig. 6C). To our surprise, however, as shown in molecular fitting with the crystal structure of Keap1-DC, the β-propeller domain only partially occupies the EM density of the large spheres (Fig. 5). While the EM Keap1 three-dimensional model also shows a linker region between the two major structural entities (i.e., BTB and DC domains) on either side, the linkers are quite short and lead into the upper polar ends of the large globular domains.
The stem region occupies only 13.5% of the total molecular volume (Fig. 4A, Blue Colored), whereas the two globular domains occupy 86.5%. Compared with the theoretical mass ratio of the functional domains in Keap1 based on their primary amino acid sequence (BTB, 28%; IVR, 23%; DC, 49%; Fig. 6C), this ratio indicates that the stem region is part of the BTB domain, whereas the globular domains are mostly composed of the IVR and DC domains (Fig. 6D). This is consistent with the bulkiness and extra volume observed around the DC domain in the fitted EM density map as described above (Fig. 5).
Structure of BTB Domain.
To further support the interpretation of the Keap1 structure, we attempted to fit the EM density of the forked-stem domain to the crystal structure of a BTB-zinc finger protein known as LRF (PDB-ID: 2NN2) homologous to the Keap1 BTB domain (39). As illustrated in Fig. S2A, B, each LRF model almost fits into the main body of the stem structure, except helices 2 and 3 (H2 and H3). This suggests that a part of the Keap1 BTB could have protruded from the stem region to form the short arm linkers, which connect to or even intrude into the beginning of the spheres. In fitting the LRF model into the stem domain, Asn103 (Magenta Colored) corresponding to Cys151 of Keap1 is located at the outermost edge, directly connecting to the linkers. This is consistent with the volume ratio interpretation (Fig. 6C, D).
Discussion
The x-ray structure of the Keap1 DC β-propeller domain has been solved both with and without substrate peptides (15, 30, 31, 40). In this study we present an EM reconstruction of the entire mouse Keap1 homodimer structure, including the BTB and IVR domains, at 24-Å resolution. The forked-stem structure in the center attaches to two short arm linkers, which further connect to two large spheres in pseudo-twofold symmetry. We surmise that there are several critical findings in this study. First, the BTB domain forms both the stem-like region and short arm linker. The forked-stem is made of two thin layers, with an observable gap between them. This is consistent with the known dimerization role of the BTB domain (2), and indicates that each of the layers comes from a single Keap1 monomer. Second, the IVR and DC domains constitute the bulk of the sphere, and the C-terminal part of the BTB domain consists of the short linker between the forked-stem structure (BTB dimerization domain) and the sphere (IVR and DC domains). Third, IVR appears to be attached closely to the outer surface of the DC domain, instead of forming a long helical linker. This notion is further supported by the continuous, smooth surface of the spheres. Finally, the narrow tunnel through each sphere is characteristic of Keap1 and coincides well with the central channel in the crystal structure of the DC domain. This helps us to identify accurately the location of the DC domain within the sphere, with the remaining regions attributable mainly to IVR. These lines of structural information provide insight into the molecular basis of Keap1 sensor function.
It has been shown that oxidative and electrophilic stress signals lead to the modification of Cys273 and Cys288 residues within IVR. Our current finding of the structural proximity of IVR to the DC domain further supports the hypothesis that covalent modification of these cysteines may induce conformational change in IVR and affect the structural integrity of neighboring substrate-binding DC domain, which eventually alters the substrate-binding ability of Keap1. Two DC domains in the Keap1 homodimer recognize two distinct degron motifs of Nrf2. Binding of DLG degron is weaker than that of ETGE degron, and the DLG degron appears to dissociate upon modification of the cysteine residues (27). We refer to this mechanism as the hinge and latch model (27).
In addition to the cysteine residues in the IVR, Cys151 in the BTB domain also plays an important role in the stress sensing. In the present reconstruction analysis, the assigned position of Cys151 is remote from the substrate-binding DC domain. Therefore, we suggest that stress modification of Cys151 may not impair recognition of Nrf2, but rather that it induces dysfunction of the E3 ligase by intervening in the other domains of the modular holoenzyme. High-resolution imaging by a cryoEM single particle analysis is essential to determine the precise location and the structure-function relationship of this important cysteine in the oxidative/electrophilic stress response.
In the x-ray atomic model, the DC domain has a drum-shaped β-propeller structure with a narrow channel in the center. The substrate-binding interface is located at the bottom of the molecule and is composed of positively charged residues, which stabilize its interaction with both ETGE and DLG degron motifs. We infer that the asymmetry observed within Keap1 might facilitate its complex formation with asymmetric Nrf2. In this scenario the ETGE motif first binds to one DC domain of the homodimer and this facilitates the DLG motif binding to the other DC domain. The two-site binding stabilizes the Keap1-Nrf2 complex structure and gives rise to the structural basis for the efficient ubiquitination of Nrf2. Perturbations of this structural integrity by cysteine modifications reduce the E3 ubiquitin ligase activity and result in stabilization (or derepression) of Nrf2.
The estimated distance between the two binding pockets of DC domains in the reconstruction is 80 Å. In addition, our particle images of Keap1 produced minor populations with both relatively distant and relatively closer globular domains, suggesting that the stem domain or linker arm region may be flexible. Previous NMR structural studies (27) revealed that 47 amino acid residues (Leu30 to Leu76) reside between the two degrons in Nrf2 and, of these, 33 residues form an α-helix (Phe39 to Phe71), which theoretically accounts for approximately 49 Å. The NMR analysis further revealed that the α-helix is mostly rigid, whereas the region from the N-terminal to the α-helix is rather flexible (27). Given that an extended polypeptide chain is, in theory, 3.5-Å long per amino acid, the rest of the 14 residues should result in a length of approximately 49 Å, if this region were fully stretched. This results in approximately 98 Å between the two degrons. One possibility is that the distance between the two DC domains may be critical for the two-site substrate-binding or recruitment of the Neh2 domain to Keap1. This is one of the important questions for future study.
Methods
Detailed methodology is provided in SI text.
Protein Purification and EM.
Recombinant Keap1 was expressed as C-terminal His-tagged fusion protein and purified using Ni-NTA agarose. The Keap1 dimer was purified twice with Superdex S-200 SEC. Keap1 protein at approximately 50 μg/mL was adsorbed by thin carbon film supported by copper mesh grid, negatively stained, and observed by using a JEOL 100CX transmission EM at a magnification of 52,100 at a 100-kV acceleration voltage. Images were recorded on films and digitized at a pixel size of 1.92 Å at the specimen level. Molecular complexes between Keap1 and antibodies were generated, separated from unbound proteins, negatively stained, and observed by EM (41). Anti-Keap1 antibody/protein G-gold/Keap1 protein complexes were similarly purified, isolated using SEC, negatively stained, and observed by EM.
Automated Particle Selection and Image Analysis.
Image analysis was performed with our SPINNS program (33–36) and IMAGIC V (37). Two-hundred and sixteen Keap1 projections were selected (34) and used to train a neural network (NN) (33, 42). With the trained NN, 12,651 particles were selected, aligned (32,43), and sorted (35). Their class averages were adopted as new references, and this cycle was repeated. Euler angles of the class averages were determined by the echo-correlated three-dimensional reconstruction method with simulated annealing (36). A primary three-dimensional density map was calculated and refined, also in cycles (32, 36). Particle images that correlated poorly with the three-dimensional projections were automatically rejected, so that the final reconstruction includes 9,827 particles, 77.7% of all the selected images.
Supplementary Material
Acknowledgments.
We thank M. Mio, S. Manaka, and M. Yanagihara (National Institute of Advanced Industrial Science and Technology) for technical assistance in biochemistry and image analysis. Molecular fitting of the x-ray model was performed using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081). This work was supported by grants from the Targeted Proteins Research Program of the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.Y., C.S., H.K.), a Grant-in-Aid for Scientific Research on Priority Areas (to M.Y.), Structure of Biological Macromolecular Assemblies (to C.S. and K.M.), the Japan New Energy and Industrial Technology Development Organization (to C.S.) and KAKENHI Grant-in-Aid for Scientific Research B from the Japan Society for the Promotion of Science (to T.O.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The EM reconstruction volume has been deposited in the Electron Microscopy Data Bank, www.emdatabank.org (EMD ID code EMD-1675).
This article contains supporting information online at www.pnas.org/cgi/content/full/0914036107/DCSupplemental.
References
- 1.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 2.Itoh K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 5.Okawa H, et al. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 6.Osburn WO, et al. Genetic or pharmacologic amplification of nrf2 signaling inhibits acute inflammatory liver injury in mice. Toxicol Sci. 2008;104:218–227. doi: 10.1093/toxsci/kfn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marzec JM, et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007;21:2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 8.Copple IM, Goldring CE, Kitteringham NR, Park BK. The Nrf2-Keap1 defense pathway: Role in protection against drug-induced toxicity. Toxicology. 2008;246:24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Osburn WO, Kensler TW. Nrf2 signaling: An adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters DM, Cho HY, Kleeberger SR. Oxidative stress and antioxidants in the pathogenesis of pulmonary fibrosis: A potential role for Nrf2. Antioxid Redox Signal. 2008;10:321–332. doi: 10.1089/ars.2007.1901. [DOI] [PubMed] [Google Scholar]
- 11.Talalay P. Chemoprotection against cancer by induction of phase 2 enzymes. Biofactors. 2000;12:5–11. doi: 10.1002/biof.5520120102. [DOI] [PubMed] [Google Scholar]
- 12.Ramos-Gomez M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iida K, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 14.Innamorato NG, et al. The transcription factor Nrf2 is a therapeutic target against brain inflammation. J Immunol. 2008;181:680–689. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
- 15.Padmanabhan B, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Ohta T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 17.Singh A, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLOS Med. 2006;3:1865–1876. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stogios PJ, Prive GG. The BACK domain in BTB-kelch proteins. Trends Biochem Sci. 2004;29:634–637. doi: 10.1016/j.tibs.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakabayashi N, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh Y, et al. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch Biochem Biophys. 2005;433:342–350. doi: 10.1016/j.abb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 26.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 27.Tong KI, et al. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong KI, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 32.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State. New York: Oxford Univ Press; 2006. pp. 72–276. [Google Scholar]
- 33.Ogura T, Sato C. An automatic particle pickup method using a neural network applicable to low-contrast electron micrographs. J Struct Biol. 2001;136:227–238. doi: 10.1006/jsbi.2002.4442. [DOI] [PubMed] [Google Scholar]
- 34.Ogura T, Sato C. Auto-accumulation method using simulated annealing enables fully automatic particle pickup completely free from a matching template or learning data. J Struct Biol. 2004;146:344–358. doi: 10.1016/j.jsb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Ogura T, Sato C. A fully automatic 3D reconstruction method using simulated annealing enables accurate posterioric angular assignment of protein projections. J Struct Biol. 2006;156:371–386. doi: 10.1016/j.jsb.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 37.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 38.Harauz G, Van Heel M. Exact filters for general geometry 3-dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 39.Stogios PJ, Chen L, Prive GG. Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci. 2007;16:336–342. doi: 10.1110/ps.062660907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato C, Sato M, Iwasaki A, Doi T, Engel A. The sodium channel has four domains surrounding a central pore. J Struct Biol. 1998;121:314–325. doi: 10.1006/jsbi.1998.3990. [DOI] [PubMed] [Google Scholar]
- 42.Ogura T, Sato C. Automatic particle pickup method using a neural network has high accuracy by applying an initial weight derived from eigenimages: A new reference free method for single-particle analysis. J Struct Biol. 2004;145:63–75. doi: 10.1016/s1047-8477(03)00139-4. [DOI] [PubMed] [Google Scholar]
- 43.van Heel M, et al. Single-particle electron cryo-microscopy: Towards atomic resolution. Q Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.