Abstract
The mechanisms by which microRNA dysfunction contributes to the pathogenesis of diffuse large B cell lymphoma (DLBCL) are not well established. The identification of the genes and pathways directly targeted by these small regulatory RNAs is a critical step to advance this field. Using unbiased genome-wide approaches in DLBCL, we discovered that the oncogenic microRNA-155 (miR-155) directly targets the bone morphogenetic protein (BMP)-responsive transcriptional factor SMAD5. Surprisingly, we found that in DLBCL a noncanonical signaling module linking TGF-β1 signals to SMAD5 is also active. In agreement with these data, miR-155 overexpression rendered DLBCLs resistant to the growth-inhibitory effects of both TGF-β1 and BMPs, via defective induction of p21 and impaired cell cycle arrest. In confirmatory experiments, RNAi-based SMAD5 knockdown recapitulated in vitro and in vivo the effects miR-155 overexpression. Furthermore, in primary DLBCLs, miR-155 overexpression inhibited SMAD5 expression and disrupted its activity, as defined by individual and global analyses of its transcriptional targets. Together, our data helped explain miR-155 function, highlighted a hitherto unappreciated role of SMAD5 in lymphoma biology, and defined a unique mechanism used by cancer cells to escape TGF-β’s growth-inhibitory effects.
Keywords: lymphoma, miR-155, transforming growth factor β, B lymphocytes, bone morphogenetic protein
Significant progress has been made recently in elucidating the molecular basis of diffuse large B cell lymphoma (DLBCL) (1). These studies were primarily centered on classic mRNA genes, whereas the role of microRNAs (miRNA) in lymphomagenesis remains to be fully appreciated. MiRNAs are non–protein-coding RNAs that function by regulating the expression of target transcripts (2). Thus, to capture the biologic impact of these regulatory molecules it is necessary to identify the genes that they inhibit. MicroRNA-155 (miR-155) is overexpressed in aggressive DLBCLs (3, 4), and its aberrant expression in a transgenic, Eμ-miR-155, mouse model was associated with the development of lymphoblastic leukemia/high-grade lymphoma (5). However, although several bona fide miR-155 targets have been identified (6–10), it is unclear how their excessive down-regulation in DLBCLs that overexpress miR-155 contribute to lymphomagenesis. A direct way to address this problem is to propose that miR-155 physiologically targets B cell growth-suppressing pathways and that its abnormal expression in DLBCL curtails these inhibitory signals, contributing to an advantageous prosurvival behavior.
Herein, we report that miR-155 binds to the 3′ UTR of the SMAD5 gene. Detailed characterization of this interaction showed that genetic modulation of miR-155 expression in DLBCL cell lines concomitantly changed SMAD5 levels. Although SMAD5 activity is classically associated with signals transduced by the BMP (bone morphogenetic protein) family of cytokines (11), we found that in DLBCL TGF-β1 also activated SMAD5. Thus, DLBCL cell lines engineered to express miR-155 became resistant to the cytostatic effects derived from both BMPs and TGF-β1, via a defective induction of p21 and impaired cell cycle arrest. Further, we found that stable shRNA-based SMAD5 knockdown recapitulated in vitro and in vivo the effects miR-155 overexpression in DLBCL. Finally, we confirmed the functional repercussions of these findings by showing that miR-155 influenced SMAD5 expression and activity in primary DLBCLs.
Results
SMAD5 Is a Direct Target of miR-155.
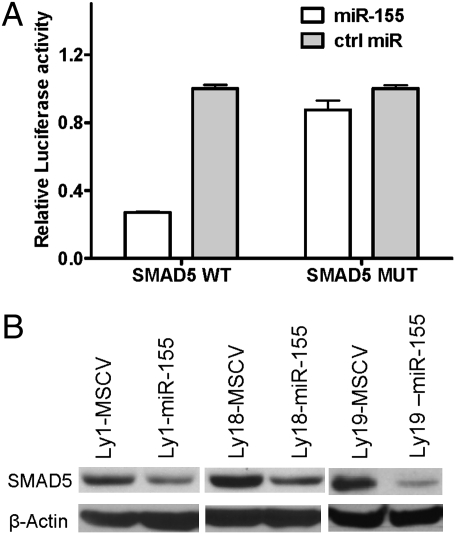
We previously identified an inverse correlation between the expression of SMAD3 and primary-miR-155 (3), suggesting that a blockade in the tumor-suppressing TGF-β signals could be involved in miR-155 oncogenesis. Thus, we searched for miR-155 binding sites in all SMAD genes. Putative binding sites were found in the 3′ UTRs of SMAD1, SMAD3, and SMAD5 (Fig. S1A). We tested the functionality of these interactions with reporter assays and found that miR-155 decreased the luciferase activity of WT SMAD5 but had no major effect on the seed sequence mutant constructs (P < 0.01, Student’s t test) (Fig. 1A). MiR-155 did not influence SMAD3 reporter activity and had a more modest effect on SMAD1 (Fig. S1B). To validate these findings, we stably expressed miR-155 (Fig. S2A) in DLBCL cell lines (3). In agreement with the reporter assays, ectopic expression of miR-155 down-regulated SMAD5 but not SMAD1 or SMAD3 (Fig. 1B and Fig. S2B). In addition, inhibiting miR-155 expression in relevant DLBCL cell lines elevated SMAD5 levels (Fig. S2C). These data indicated that SMAD5 is a direct target of miR-155. Finally, our findings suggest that mechanisms other than miR-155 activity account for the inverse correlation between the expression of this miRNA and SMAD3 noted earlier in DLBCLs (3).
Fig. 1.
SMAD5 is a direct target of miR-155. (A) Luciferase reporter constructs containing the 3′ UTR of the SMAD5 gene [WT or with point mutations in both miR-155 binding sites (MUT)] were cotransfected with pre-miR-155 or control oligos. Pre-miR-155 inhibited luciferase activity in the SMAD5-WT but had no effect in the mutant (MUT) construct (P < 0.05, Student's t test). Data shown are mean ± SD of the ratio of luciferase activity in pre-miR-155 and control oligo transfections. (B) Western blots depict SMAD5 inhibition in DLBCL cell lines stably expressing miR-155.
In DLBCL, Both BMP2/4 and TGF-β1 Activate SMAD5.
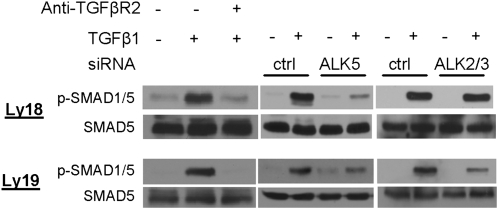
Classically, TGF-β1 signals are transduced via the TGFRB2 and TGFRB1 (ALK5) receptors to activate SMAD2 and SMAD3 (11). Conversely, signals derived from the BMP family of cytokines use BMPR2 and a host of type I receptors (ALK1, -2, -3, and -6) to activate SMAD1/5/8. However, noncanonical signals linking TGF-β1 to SMAD1/5 have been recently described in endothelial and epithelial tissues (12–15). Because miR-155 specifically targets SMAD5, we investigated whether this alternative route was active in malignant B lymphocytes. In DLBCL cell lines, BMP2/4 induced phosphorylation of SMAD1/5 and, expectedly, had no effect on SMAD2/3 (Fig. S3A). Surprisingly, exposure to TGF-β1 resulted in phosphorylation of SMAD2/3 but also of SMAD1/5 (Fig. 2 and Fig. S3A). Interestingly, in murine or human normal mature B cells we could not detect these noncanonical signals, albeit TGF-β promptly phosphorylated SMAD2 (Fig. S3A). However, because the canonical BMP-mediated phosphorylation of the SMAD1/5 was also not present, we cannot exclude the possibility that in normal B cells the levels of SMAD1/5 expression/phosphorylation are below the sensitivity of available antibodies and Western blot measurements. In addition, to separate the effects of TGF-β1 on SMAD1 and SMAD5, we used immunoprecipitation assays and confirmed that SMAD5 is phosphorylated after TGF-β1 treatment (Fig. S3B). Finally, we showed that the phosphorylation of SMAD2/3 is not modified by miR-155 (Fig. S3C).
Fig. 2.
Noncanonical TGF-β signals in DLBCL. Western blot analyses of phospho-SMAD1/5 in DLBCL cell lines exposed to TGF-β1, with or without pretreatment with a blocking anti-TGFBR2 antibody (Left), or with siRNA-mediated knockdown of ALK5 or ALK2/3 (Center and Right). The noncanonical phosphorylation of SMAD1/5 was abolished by blocking the TGFBR2 receptor and inhibiting ALK5 expression in both Ly18 (Top) and Ly19 (Bottom) cells, whereas ALK2/3 knockdown impaired TGF-β1 signals exclusively in the Ly19 cell line.
The noncanonical TGF-β1 signaling in endothelial and epithelial cells requires both BMP and TGF-β type I receptors (12–15). To study this issue in DLBCL, we first measured the expression of ALK1-7, TGFRB2, and BMPR2. We found that ALK2, -3, -5, TGFRB2, and BMPR2 were consistently expressed in DLBCL (Fig. S3D). To define the contribution of the distinct classes of type I and II receptors in the noncanonical TGF-β1 signal transduction in our model, we used siRNA oligos toward ALK2, ALK3, and ALK5, a blocking antibody anti-TGFBR2, and kinase inhibitors specific to BMP (dorsomorphin) or TGF-β receptors (SB-431542) (13, 15). In the cell lines examined, TGF-β1–mediated phosphorylation of SMAD1/5 was markedly abolished by pretreatment with the blocking TGFBR2 antibody and ALK5 siRNA, indicating a significant contribution of the TGF-β receptors to this process (Fig. 2). Conversely, the role of the BMP receptors, investigated with ALK2/3 siRNAs, albeit clear, was limited to the Ly19 cell line (Fig. 2 and Fig. S4 A and B). Importantly, identical results were obtained with the kinase inhibitors SB-431542 and dorsomorphin (Fig. S4C).
DLBCLs Overexpressing miR-155 Are Resistant to the Growth-Inhibitory Effects of TGF-β1 and BMPs and Display Impaired G0/G1 Arrest and p21 Induction.
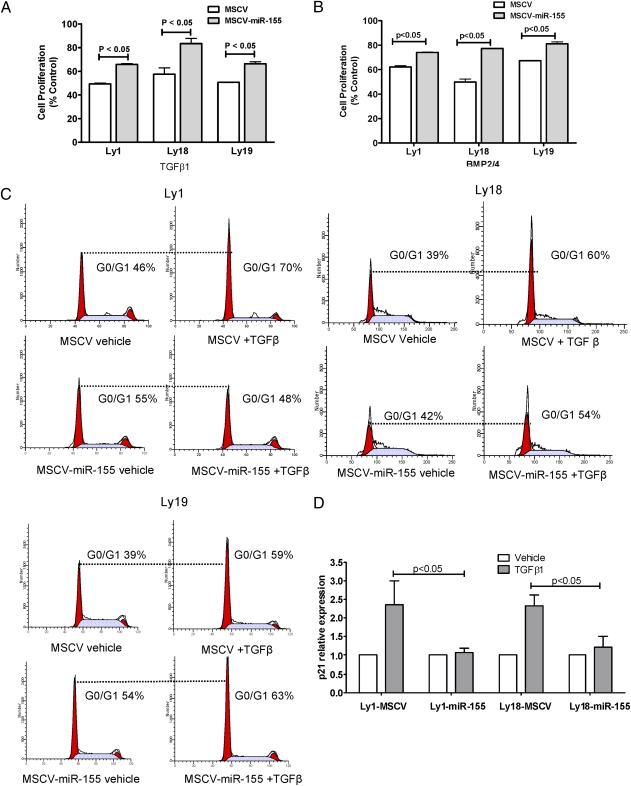
We tested whether expression of miR-155 in DLBCL, via targeting of SMAD5, ablated the growth-inhibitory effects of TGF-β1 and BMP2/4 toward B lymphocytes (16). Indeed, ectopic expression of miR-155 significantly limited the cytostatic activities of these cytokines (P < 0.05, Student’s t test) in all cell line models analyzed (Fig. 3 A and B) and at multiple doses (Fig. S5A). Next, we determined the influence of miR-155 expression on TGF-β1–mediated cell cycle arrest (17) and found that DLBCL cell lines expressing miR-155 became either refractory or had a diminished response to TGF-β1–mediated G0/G1 arrest (Fig. 3C), which was accompanied by an impaired TGF-β1–mediated induction of p21 (P < 0.05, Student’s t test) (Fig. 3D).
Fig. 3.
MiR-155 limits the cytostatic effects of TGF-β1 and BMP2/4 in DLBCL. DLBCL cell lines ectopically expressing miR-155 were significantly more resistant (P < 0.05, Student's t test) to the cytostatic effects of TGF-β1 (A) and BMP2/4 (B) than their isogenic controls. TGF-β1 doses were 1 ng/mL (Ly1 and Ly18) and 2 ng/mL (Ly19); see Fig. S4A for the complete dose range. Data shown are mean ± SEM of the percentage inhibition of cells exposed to TGF-β1 or BMP2/4, normalized by vehicle-treated cells. (C) Cell cycle analyses show G0/G1 arrest after TGF-β1 exposure in DLBCL cell lines low/null for miR-155 (MSCV), and an absent or more limited effect in MSCV-miR-155 cells. (D) Real-time RT-PCR quantification of p21 induction by TGF-β1 (2.5 ng/mL). MiR-155 expression significantly blocked TGF-β1–mediated induction of p21 in DLBCL (P < 0.05, Student's t test). Data shown are mean ± SEM of cells exposed to TGF-β1 normalized by vehicle-treated cells. TGF-β1 consistently did not induce p21 expression in the Ly19 cell line.
MiR-155 Expression Enhances Tumor Aggressiveness in a Xenograft Model of Human DLBCL.
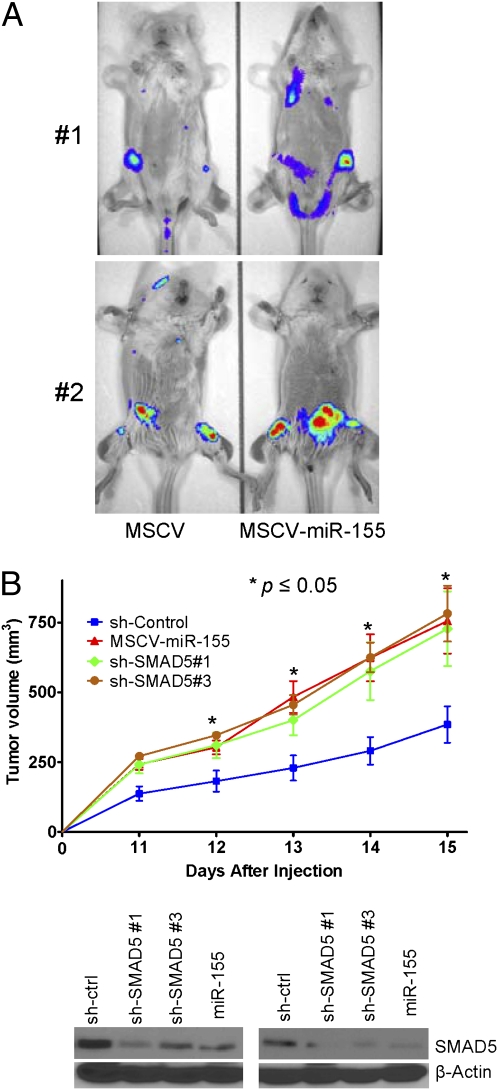
In Eμ-miR-155 transgenic mice, an early pre-B cell proliferation leads to the development of B cell tumors (5), suggesting that miR-155 may facilitate the acquisition of mutations needed for the growth of a monoclonal neoplasm. However, the oncogenic contribution of miR-155 to fully established mature B cell tumors has not been defined. Thus, we created DLBCL cell lines constitutively coexpressing miR-155 (or vector alone) and the luciferase gene. These cells were injected by the tail vein in sublethally irradiated SCID/NOD (nonobese diabetic) mice and live bioluminescent imaging performed (Fig. 4A). In parallel experiments, carried out in an additional mouse cohort, the same DLBCL cell lines were injected s.c. and tumor volume measured daily. In both models, miR-155 expression led the development of larger and more widespread tumors (P < 0.05 tumor volume, P < 0.05 photon flux quantification, Student’s t test; Fig. S5B).
Fig. 4.
MiR-155 overexpression and SMAD5 knockdown enhance DLBCL aggressiveness in vivo. (A) Bioluminescent imaging of mice injected with the Ly18 lymphoma cells ectopically expressing miR-155 (MSCV-miR-155) or their isogenic controls (MSCV). MiR-155 expression yielded larger and more widespread tumors, as is visually evident, and was confirmed by photon flux quantification (P < 0.05, Student’s t test; Fig. S5B). Data shown are for four independent mice imaged at day 14 (group 1) and day 24 (group 2) after cells injection. (B) Tumor volume in mice inoculated s.c. with Ly18 cells stably expressing SMAD5 shRNAs, shRNA vector control, or miR-155. SMAD5 knockdown and miR-155 expression gave raise to tumors of similar size, which were consistently larger than tumor originated from the isogenic sh-control cells (P < 0.05, Student’s t test). The data shown are mean ± SEM for five mice per group (n = 20, two independent experiments) in each day. Western blots (Bottom) of eight independent xenografts confirm the lower levels of SMAD5 in tumors expressing miR-155 or SMAD5-shRNAs.
SMAD5 Knockdown Recapitulates the in Vitro and in Vivo Effects of miR-155 Overexpression in DLBCL.
Using two independent targeting sequences, in transient and stable RNAi strategies, we created DLBCL cells with specific down-regulation of SMAD5 (Fig. S6 A and B). Supporting the relevance of SMAD5 targeting by miR-155, DLBCLs with stable SMAD5 knockdown became resistant to the cytostatic effects of BMP2/4 and TGF-β1 and showed a defective induction of p21 (Fig. S6 A and B) (P < 0.05, Student’s t test). In miR-155-overexpressing and SMAD5 knockdown DLBCLs, the disruption of p21 induction was independent of the inhibitory effects of TGF-β1 toward v-myc myelocytomatosis viral oncogene homolog (MYC) (18). Importantly, the tumor-suppressor properties of SMAD5 were confirmed in vivo: we found that DLBCLs stably expressing SMAD5 shRNAs (or overexpressing miR-155) developed into larger and more aggressive tumors than their isogenic counterparts (P < 0.05, Student’s t test) (Fig. 4B and Fig. S6C), thus substantiating the functional relevance of our in vitro assays (Fig. 3, Fig. S5A, and Fig. S6 A and B). Notably, numerous attempts to reconstitute SMAD5 expression (lacking a miR-155 binding site) in our miR-155 overexpression DLBCL models were unsuccessful, supporting the tumor-suppressive activities of SMAD5 in these tumors.
MiR-155 Expression Impacts on SMAD5 Function in Primary DLBCL.
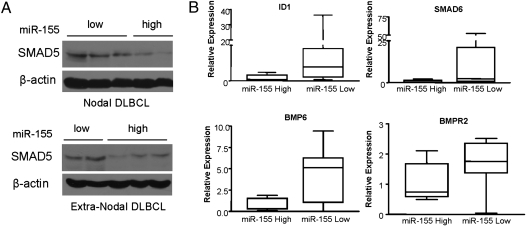
To confirm the relevance of our findings beyond the cell line models, we quantified the expression of mature miR-155 in a collection of 20 primary DLBCLs (Table S1). We were able to isolate intact protein from a subset of these tumors (n = 10), and using Western blotting we found an inverse correlation between miR-155 and SMAD5 expression (Fig. 5A).
Fig. 5.
MiR-155 modulates SMAD5 expression and activity in primary DLBCLs. (A) Immunoblot detection of SMAD5 in primary DLBCL. Nodal and extranodal DLBCLs with high levels of miR-155 have significantly lower expression of SMAD5 expression [R = −0.82 (nodal) and R = −0.93 (extranodal), Pearson’s correlation]. (B) Quantitative real-time RT-PCR measurement of SMAD5 transcriptional targets in primary DLBCL with high (n = 7) and low miR-155 (n = 8) expression. Levels of ID1, BMP6, BMPR2, and SMAD6 were significantly lower in DLBCL with high miR-155 levels (P < 0.05, Mann-Whitney test).
We wished to test whether the down-regulation of SMAD5 had physiologic consequences. A well-validated list of SMAD5 transcriptional targets in mature B cells is not available. However, mining expression datasets derived from other tissues (19–21), we defined a small catalog of genes that were commonly up-regulated by SMAD5 (or SMAD1/5 but not SMAD2/3) (SI Materials and Methods). To confirm the relationship between SMAD5 and these putative transcriptional targets in B lymphocytes, we used the DLBCL cell lines with SMAD5 knockdown and found that TGF-β1–induced expression of ID1, ID2, BMP6, BMPR2, SMAD6, and SMAD7 was significantly impaired (Fig. S7A). Importantly, further supporting the functional link between miR-155 and SMAD5 in DLBCL, the expression of these transcriptional targets was also defective in a DLBCL cell line ectopically expressing miR-155 (Fig. S7B).
Next we sought to confirm these findings in primary DLBCLs. We hypothesized that genes transcriptionally induced by SMAD5 would be underexpressed in tumors with high levels of miR-155. Indeed, comparing the primary DLBCLs with the highest (n = 7) or lowest (n = 8) miR-155 expression (Table S1), we found that ID1, BMP6, BMPR2, and SMAD6 expression was significantly lower in primary DLBCLs with high miR-155 levels than in those with low levels of this miRNA (P < 0.05, Mann-Whitney test) (Fig. 5B); the expression of SMAD7 and ID2 despite being lower in miR-155-overexpressing tumors did not reach statistical significance (Fig. S7C). Interestingly, the expression of p21, which we identified as the TGF-β–induced cycle regulator that is modulated by miR-155 and SMAD5 expression, was also significantly lower in primary DLBCLs overexpressing this miRNA (Fig. S7C). Finally, to expand these findings to a larger and independent tumor cohort, we mined a dataset of DLBCL (22) for which we had previously defined the expression of the primary miR-155 transcript (BIC gene) (3). In this investigation, using unsupervised hierarchical clustering and the gene set enrichment analysis (GSEA) computational method (SI Materials and Methods), we found that the expression of approximately two thirds (13 of 19) of the putative SMAD5 transcriptional targets was significantly lower in DLBCLs with high miR-155 levels than in lymphomas expressing low levels of this miRNA [P < 0.05, t statistic in clusters comparison; false discovery rate (FDR) = 0.02, enrichment analysis] (Fig. S8). The lack of correlation between six of the SMAD5 target genes and miR-155 expression may indicate that the SMAD5 effect on these genes is tissue specific and not present in B cells or that additional regulatory pathways that control their expression are disrupted in DLBCL. Future studies in larger tumor cohorts and precise measurements using real-time RT-PCR should clarify this issue.
Discussion
MiR-155 overexpression is associated with aggressive forms of DLBCL and contributes to the development of lymphoid and myeloid malignancies (3, 5, 6). Herein, we described SMAD5 as a direct target of miR-155, establishing a link between this miRNA, the TGF-β pathway, and lymphomagenesis.
Although SMAD5 primarily relays signals initiated by the BMP family of cytokines, we found that in DLBCL SMAD5 is also phosphorylated by TGF-β1. Our investigations suggest that in DLBCLs, the type I and II TGF-β receptors are essential for this noncanonical activation of SMAD5. However, similarly to the findings in epithelial tissues (13, 15), we identified heterogeneity in this response: inhibition of type I BMP receptors also partially blocked the TGF-β1–mediated phosphorylation of SMAD5. These data suggest that transduction of these noncanonical signals may occur via heteromultimeric complexes that include TGFBR2, ALK5, ALK2, and/or ALK3, as proposed (13). In our models, the expression levels and phosphorylation of SMAD1, SMAD2, and SMAD3 were not modified by miR-155 overexpression or SMAD5 knockdown. Yet various downstream effects of TGF-β1 signaling were disrupted by the exclusive down-modulation of SMAD5, indicating the existence of a previously unappreciated layer of specificity in the effector functions of SMAD proteins.
A tumor-suppressive role for SMAD5 in lymphoid neoplasms has not been previously reported. However, recent data showed that conditional deletion of SMAD1/5 induces granulosa cell and testicular tumors in mice (20), thus suggesting a broad effect of the “BMP-SMADs” in tumor control. The mechanism by which loss of SMAD5 function contributes to DLBCL pathogenesis remains to be elucidated, although the defective induction of p21 found in miR-155 overexpressing and SMAD5 knockdown lymphomas suggests a participation of this cell cycle regulator in this process. Loss of SMAD5 disrupted the expression of numerous putative transcriptional targets, indicating that the tumor-suppressive activities of SMAD may reflect the contributions multiple downstream pathways. These findings lend support to the recent suggestion that the activation of SMAD1/5 by TGF-β1 results in the formation of mixed SMAD1/5–SMAD2/3 complexes (13). Thus, it is possible that the targeting of SMAD5 by miR-155 disrupts two independent signaling modules: the traditional, BMP-induced, that leads to the formation of the putative SMAD5-SMAD4 complex, and a unique, TGF-β1–mediated, signaling branch that results in mixed complexes composed of SMAD5–SMAD2/3.
A few miR-155 targets have been well validated. PU.1, AID, and SOCS1 were shown to mediate specific miR-155 effects on B cell homeostasis and T regulatory cell function (7, 8, 10). Recently, SHIP1 was shown to be a critical mediator of miR-155–induced myeloid neoplasia (9). These studies suggest that the pleiotropic effects of miR-155 may be achieved by the modulation of unique target genes, within specific cell types and physiologic contexts. Thus, our data showing that the exclusive, RNAi-based, SMAD5 down-regulation recapitulates in vitro and in vivo the effects of miR-155, suggest that SMAD5 is one of the relevant targets associated with miR-155–mediated lymphomagenesis. These findings also concur with the emerging concept that a very limited number of target genes can account for the malignant phenotype found in association with miRNA dysfunction (9, 23, 24). Finally, our data also raise the possibility that the reduced number of germinal-center B cells and defective germinal-center reaction described in miR-155 null mice (25) could relate, at least in part, to inappropriate SMAD5 regulation and excessive inhibitory TGF-β signaling. Future studies, that test in a combinatorial fashion SMAD5 and additional miR-155 targets, should more specifically quantify and qualify their individual contribution to the benign and malignant phenotypes associated with miR-155 dysfunction.
In summary, our work helped explain miR-155 function, highlighted the activity of the noncanonical TGF-β signaling branch in DLBCL, exposed a hitherto unappreciated role of SMAD5 in lymphoma biology, and defined a unique, miRNA-mediated mechanism used by cancer cells to escape TGF-β’s growth-inhibitory effects.
Materials and Methods
Additional methodologic details are provided in SI Materials and Methods.
Cell Lines and Primary DLBCL.
The cell lines Ly1, Ly3, Ly10, Ly18, Ly19, and HEK-293 were cultured as previously described (26). In all experiments that included activation with TGF-β1 or BMP, the cell lines were grown in 2% FBS. Twenty primary DLBCLs were obtained from our tumor bank. The detailed features of this collection are described in Table S1. These studies were approved by the institutional review board of the University of Texas Health Science Center at San Antonio (UTHSCSA).
Reporter assays.
The 3′ UTR of SMAD1, SMAD3, and SMAD5 genes were cloned into the pMIR luciferase reporter vector (Ambion). These constructs were cotransfected in HEK-293 cells with the pCMVβ plasmid (Clontech) and 20 nM of pre-miR-155 or control RNA oligos (Ambion), using Fugene (Roche). Cells were harvest 24 h after transfection. All experiments were performed in triplicate and repeated three times.
Modulation of miR-155 Levels in DLBCL Cell Lines.
A fragment of the BIC gene containing the miR-155 precursor sequence was cloned into the murine stem cell virus (MSCV)-eGFP vector. Virus production and cell line transduction were performed as previously described (27). MiR-155 expression was determined with stem-loop real-time RT-PCR (Applied Biosystems), as previously described (26). Knockdown of miR-155 expression in DLBCL cell lines was performed by transiently transfecting anti-miR-155 oligos (Ambion) by electroporation (SI Materials and Methods).
TGF-β1 and BMP Stimulation of DLBCL Cell Lines.
DLBCL cells were exposed to recombinant TGF-β1 (1 ng/mL for 1 h) or BMP2/4 (50 ng/mL for 1 h) (R&D Systems), with or without pretreatment with SB-431542 (Sigma-Aldrich; 10 μM for 30 min), dorsomorphin (Sigma-Aldrich; 10 μM for 1 h), an anti-TGFBR2 blocking antibody (R&D Systems, 50 μg/mL for 1 h), or after transient transfection of siRNA oligos toward ALK2, -3, or -5 (Dharmacon). Next, protein was isolated and total or phospho levels of the SMAD proteins defined by immunoblotting with anti SMAD1/5, SMAD2, SMAD3, and SMAD5 antibodies (R&D Systems and Cell Signaling).
Cell Proliferation, Cell Cycle Analysis, and CDKN1A (p21) Expression.
Proliferation rate of relevant DLBCL cell lines was measured with CellTiter Proliferation Assay (Promega) after exposure to TGF-β1 or BMP2/4. Cell cycle analyses were performed by propidium iodide staining. Expression of the CDKN1A was determined by quantitative real-time RT-PCR (SI Materials and Methods).
Transient and Stable Knockdown of SMAD5 in DLBCL Cell Lines.
Two SMAD5 siRNA duplexes (Dharmacon) were transiently transfected in the Ly18 cell line and SMAD5 down-regulation confirmed by Western blot. These sequences were also cloned into the pLTR-H1-eGPP retrovirus and DLBCL cell lines stably expressing SMAD5 shRNAs established as previously described (27).
Xenograft Model of Human DLBCL with miR-155 Overexpression or SMAD5-Specific shRNA-Mediated Knockdown.
The Ly18 MSCV-miR-155 or MSCV-only cells were cotransduced with a luciferase expressing retrovirus and stable populations established with neomycin selection. SCID-NOD mice were irradiated (300 rads) and injected by tail vein (n = 4) or s.c. (n = 8) with 5 × 106 and 10 × 106 cells, respectively. Development of hematogenously disseminated tumors was monitored weekly by in vivo imaging (Xenogen IVIS 200 imaging system). Subcutaneous tumors were measured daily using an electronic caliper. In additional cohorts, Ly18 cells stably expressing shRNA-control, SMAD5-shRNAs, or MSCV-miR-155 were injected s.c. (5 × 106 cells) in nude mice (n = 20) after irradiation (400 rads) and tumor volume measured as above. These studies were approved by the Institutional Animal Care and Use Committee of the UTHSCSA.
Expression of TGF-β/BMP Receptors and SMAD5 Target Genes in Primary DLBCL and DLBCL Cell Lines.
Expression of TGFRB2, BMPR2, and all type I receptors (ALK1-7) was determined by semiquantitative RT-PCR (SI Materials and Methods). Expression of the putative SMAD5 target genes ID1, ID2, SMAD6, SMAD7, BMP6, and BMPR2 was measured by quantitative real-time RT-PCR in primary DLBCL (n = 15) and relevant DLBCL cell lines.
Statistics.
The Mann-Whitney test was used to determine the difference in the expression of SMAD5 target genes in primary DLBCL. For all other in vitro and in vivo assays, the statistical analyses were performed with two-tailed Student’s t test. P < 0.05 was considered significant. Data analyses were performed with Prism software (version 5.0; GraphPad) and Excel software (Microsoft).
Supplementary Material
Acknowledgments
Supported by grants from the Elsa Pardee Foundation and Concern Foundation. R.A. is a Scholar of the American Society of Hematology and the recipient of a Voelcker Fund Young Investigator Award. P.D. is a Kimmel Scholar.
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910667107/DCSupplemental.
References
- 1.Abramson JS, Shipp MA. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106:1164–1174. doi: 10.1182/blood-2005-02-0687. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rai D, Karanti S, Jung I, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 12.Goumans MJ, et al. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFbeta/ALK5 signaling. Mol Cell. 2003;12:817–828. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 13.Daly AC, Randall RA, Hill CS. Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol. 2008;28:6889–6902. doi: 10.1128/MCB.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu IM, et al. TGFbeta-stimulated Smad1/5 phosphorylation requires the ALK5 L45 loop and mediates the pro-migratory TGFbeta switch. EMBO J. 2009;28:88–98. doi: 10.1038/emboj.2008.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrighton KH, Lin X, Yu PB, Feng XH. Transforming growth factor beta can stimulate Smad1 phosphorylation independently of bone morphogenic protein receptors. J Biol Chem. 2009;284:9755–9763. doi: 10.1074/jbc.M809223200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 17.Massagué J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Chen CR, Kang Y, Siegel PM, Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 19.Ota T, et al. Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-beta in human umbilical vein endothelial cells. J Cell Physiol. 2002;193:299–318. doi: 10.1002/jcp.10170. [DOI] [PubMed] [Google Scholar]
- 20.Pangas SA, et al. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol. 2008;28:248–257. doi: 10.1128/MCB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McReynolds LJ, Gupta S, Figueroa ME, Mullins MC, Evans T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood. 2007;110:3881–3890. doi: 10.1182/blood-2007-04-085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monti S, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 23.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 26.Li C, et al. Copy number abnormalities, MYC activity, and the genetic fingerprint of normal B cells mechanistically define the microRNA profile of diffuse large B-cell lymphoma. Blood. 2009;113:6681–6690. doi: 10.1182/blood-2009-01-202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith PG, et al. The phosphodiesterase PDE4B limits cAMP-associated PI3K/AKT-dependent apoptosis in diffuse large B-cell lymphoma. Blood. 2005;105:308–316. doi: 10.1182/blood-2004-01-0240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.