Abstract
The 10th type III domain of human fibronectin (Fn3) has been validated as an effective scaffold for molecular recognition. In the current work, it was desired to improve the robustness of selection of stable, high-affinity Fn3 domains. A yeast surface display library of Fn3 was created in which three solvent-exposed loops were diversified in terms of amino acid composition and loop length. The library was screened by fluorescence-activated cell sorting to isolate binders to lysozyme. An affinity maturation scheme was developed to rapidly and broadly diversify populations of clones by random mutagenesis as well as homologous recombination-driven shuffling of mutagenized loops. The novel library and affinity maturation scheme combined to yield stable, monomeric Fn3 domains with 3 pM affinity for lysozyme. A secondary affinity maturation identified a stable 1.1 pM binder, the highest affinity yet reported for an Fn3 domain. In addition to extension of the affinity limit for this scaffold, the results demonstrate the ability to achieve high-affinity binding while preserving stability and the monomeric state. This library design and affinity maturation scheme is highly efficient, utilizing an initial diversity of 2×107 clones and screening only 1×108 mutants (totaled over all affinity maturation libraries). Analysis of intermediate populations revealed that loop length diversity, loop shuffling, and recursive mutagenesis of diverse populations are all critical components.
Keywords: fibronectin type III domain, Fn3, protein engineering, loop shuffling, affinity maturation
Introduction
The 10th type III domain of human fibronectin (Fn3) has been demonstrated as an effective scaffold for molecular recognition.1–9 It is a small (10 kDa), stable (Tm=84 °C)7 cysteine-free beta-sandwich protein that maintains its native fold in reducing environments and can be produced at up to 50 mg/L in Escherichia coli.9 The protein fold resembles an immunoglobulin variable domain and contains three solvent-exposed loops, termed BC, DE, and FG, for the β strands they connect, which are structurally analogous to antibody complementarity-determining regions (CDRs). Modification of the amino acid sequence in these loops can impart novel binding capacity on the scaffold. Koide and colleagues have diversified the BC and FG loops and screened libraries by phage display to yield a 5 µM binder to ubiquitin4 and a 250 nM binder to Src SH3 domain2; FG loop libraries yielded improved integrin binders8 and novel estrogen receptor binders,3 the latter of which was identified by a yeast two-hybrid screen. Phage display and yeast surface display were used to screen a three-loop library with tyrosine/serine diversity to isolate binders of 5, 7, and 30 nM to yeast small ubiquitin-like modifier, human small ubiquitin-like modifier 4, and maltose-binding protein, respectively.5 mRNA display was used to screen three-loop libraries to identify a 20 pM binder to tumor necrosis factor α9 and a 340 pM binder to vascular endothelial growth factor receptor 2.7 Lipovsek et al. used yeast surface display to engineer a 350 pM binder to lysozyme from a library with diversified BC and FG loops.6 Engineered binders have been used as intracellular3 and extracellular inhibitors8 as well as labeling reagents in flow cytometry8 and Western blots2 and also have been immobilized for affinity purification1 and for use in protein arrays.9
Although wild-type Fn3 is highly stable and monomeric, previously reported high-affinity Fn3 clones are oligomeric or unstable. Two clones with 300 pM and 1 nM affinity for tumor necrosis factor α exhibited midpoints of proteolysis susceptibility at approximately 30 and 42 °C, respectively.9 A clone with 340 pM affinity for vascular endothelial growth factor receptor 2 exhibited Tm values of 32–52 °C; a different clone was more stable but only 40–50% monomeric and was of moderate affinity (13 nM).7 Stability engineering yielded a stable, monomeric 2 nM binder. The ability to engineer Fn3 domains with high affinity and stability represents a critical need in scaffold development, since both properties are required for clinical and biotechnological applications.
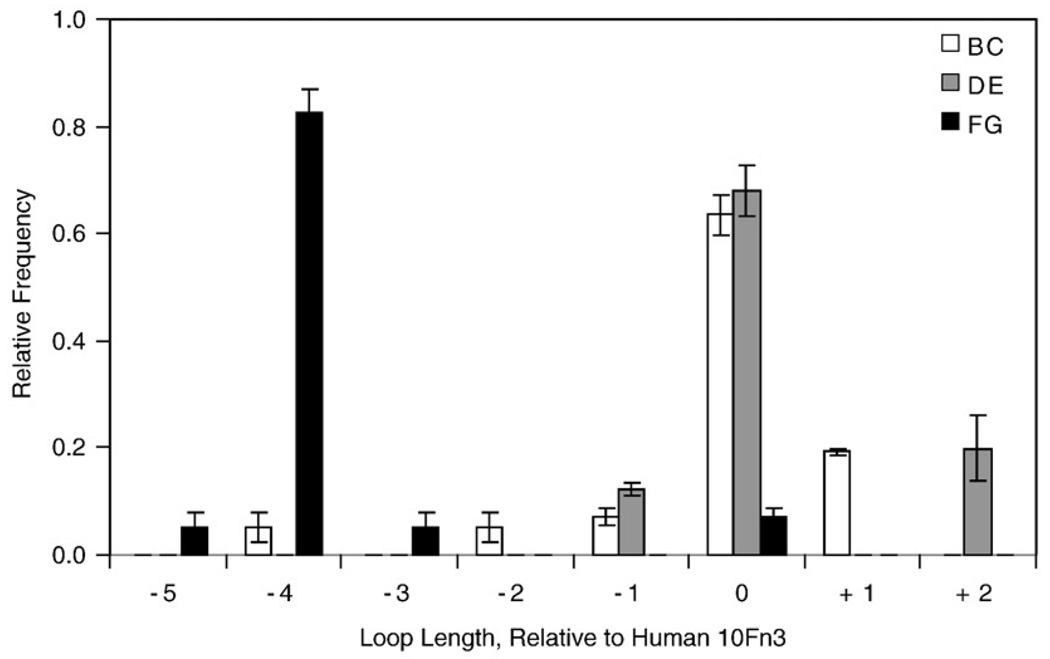
Synthesis errors during our previous library creation resulted in rare clones with non-wild-type loop lengths.6 These loop length variants were preferentially selected during binder isolation, and binding titrations revealed their critical importance to binding. Moreover, sequence analysis reveals non-wild-type lengths in tumor necrosis factor α binders9 and carcinoembryonic antigen binders (B.J.H., unpublished data). The rarity of these length variants in the initial library resulted in very low coverage of theoretical length variant sequence space. These results suggest that more extensive sampling of length variant sequences could improve the binding capabilities of the Fn3 scaffold. This hypothesis is supported by the fact that CDR length diversity is already an acknowledged component of antibody engineering,10,11 and it has been demonstrated that antibody affinity maturation is improved with the inclusion of length diversity.12,13 Koide and colleagues incorporated loop length diversity in a recent Fn3 library and successfully obtained binders with length variation.5 From a structural perspective, Batori et al. demonstrated that the BC, DE, and FG loops can tolerate insertions of four glycine residues while maintaining a native fold.14 In addition, phylogenetic analysis reveals significant length diversity in each of the three loops across different fibronectin type III domains in multiple species (Fig. 1). Thus, we hypothesize that loop length diversity will be stably tolerated and improve the functional binding capabilities of the Fn3 scaffold and perhaps could improve the stability–affinity relationship.
Fig. 1.
Loop length variability of fibronectin type III domains. The relative frequency of loop lengths of the BC (white), DE (gray), and FG (black) loops of sixteen type III domains of fibronectin, relative to the 10th type III domain (10Fn3) of human fibronectin, are shown. Values and error bars represent the average and standard deviation for five species: human, cow, rat, mouse, and chicken.
Yet, loop length diversity increases theoretical sequence space, which is already immense, increasing the demand for efficient protein engineering methods in terms of both clonal selection and sequence diversification. Yeast surface display enables quantitative selections using fluorescence-activated cell sorting (FACS) and correspondingly fine affinity discrimination.15 The eukaryotic protein processing of yeast also improves the functional stringency of selections.16 One potential limitation of yeast surface display is that cellular transformation efficiency holds library size to 107–109 without excessive effort. In vitro display methods enable larger theoretical library size that correlates with selection of improved binders17,18; however, as has been shown for phage display, nominal library size does not necessarily equate with functional diversity.16 The intrinsic mutagenesis from the requisite PCR step in mRNA and ribosome display is a key contributor to the success of these display technologies.19 We hypothesized that an increase in the frequency of mutagenesis during directed evolution will improve the efficiency of cellular display methods by increasing the breadth of the sequence space search in the vicinity of many lead clones, rather than only a select few.
Another important engineering component is the manner in which selected sequences are diversified throughout directed evolution. Successful techniques include DNA shuffling,20 CDR shuffling,21,22 CDR walking,23 and error-prone PCR mutagenesis24 either toward the entire gene or toward the suspected paratopes. In the current work, we combine error-prone PCR and an analog to CDR shuffling to yield both mild and significant changes in sequence space both directed at the expected paratope and throughout the Fn3 domain.
Here we demonstrate that loop length diversity and a novel affinity maturation scheme enable robust and efficient selection of stable, high-affinity binders to lysozyme. The binders are characterized in terms of binding, stability, and structure including detailed analysis of the molecular basis of binding of a picomolar affinity clone.
Results
Fn3 library construction
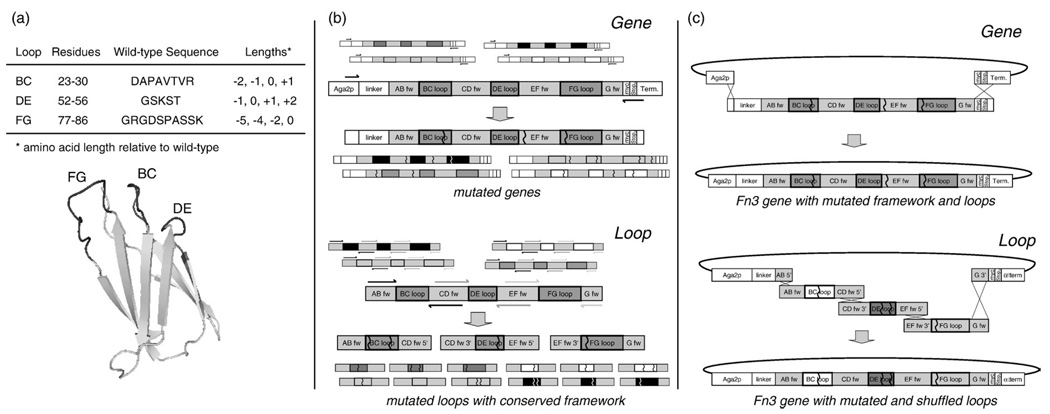
A library was created in which 8, 5, and 10 amino acids of the BC, DE, and FG loops, respectively, were diversified both in amino acid length (Fig. 1) and in composition (Fig. 2a). Amino acid composition was randomized using NNB degenerate codons to yield all 20 amino acids with reduced stop codon frequency. Four different loop lengths were chosen for each loop based partially on the loop lengths observed in fibronectin type III domains in multiple species (Fig. 1). The library of Fn3 genes was incorporated into a yeast surface display system by homologous recombination with a vector incorporating an N-terminal Aga2 protein for display on the yeast surface and a C-terminal c-myc epitope for detection of full-length Fn3.26 Library transformation yielded 6.5×107 yeast transformants. Sixteen (62%) of 26 clones sequenced matched library design. Nine (35%) contained frameshifts and 1 (4%) was annealed improperly or underwent unintentional homologous recombination in yeast. NNB diversification of the loops yields stop codons in approximately 44% of clones. Thus, 34% [16/26×(1–0.44)] of transformed cells should display full-length Fn3. This percentage was verified by flow cytometry analysis (data not shown). The library contains approximately 2.3×107 (6.5×107×0.34) full-length Fn3 clones.
Fig. 2.
Library design and affinity maturation scheme. (a) The naive library is randomized in the BC, DE, and FG loops (structure schematic derived from Main et al25) at the indicated positions to four possible loop lengths each. (b) Error-prone PCR is used to introduce random mutations either into the entire Fn3 gene or into the three loops (separate PCRs). Arrowed lines indicate PCR primers; matching shades correspond to primers in a single PCR reaction. The framework (fw) and loop regions of the gene are indicated. Zigzag lines represent nucleotide mutations. Smaller unlabeled images are used to emphasize that PCR templates and products are part of a repertoire based on many gene variants. (c) Two yeast sublibraries are created by transformation via electroporation. During transformation, the full plasmid is created by homologous recombination of the linearized vector with either the single mutated gene insert or the three mutated loop inserts.
Population mutagenesis design
A negative side effect of loop length diversity is a further increase in the vastness of possible protein sequence space to 1034 possible amino acid sequences for the three loops. Thus, the 2.3×107 clones in the original library grossly undersample sequence space, so an extensive diversification method was desired to broadly search sequence space during affinity maturation. Error-prone PCR directed solely at the solvent-exposed loops was used to focus diversity on the likely paratope. Additionally, to make substantial changes in sequence, a loop shuffling approach was developed. Fn3 genes were constructed with conserved wild-type framework sequence and randomly shuffled mutated loops from the pool of selected clones. Yet, as effective clones are selected, such substantial sequence modification is not desired. Thus, error-prone PCR without shuffling was also employed. This mutagenesis targeted the entire gene to also enable selection of beneficial framework mutations. Both mutagenesis strategies were used in parallel at each mutagenesis step throughout affinity maturation (Supplementary Fig. 1). Analysis of selected clones after the fact indicates that both loop shuffling and whole-gene error-prone PCR contributed to improved phenotypes.
The Fn3 library was screened using yeast surface display and FACS. Plasmids from thousands of lead clones in a partially screened yeast population were collected by yeast zymoprep. Quantitative real-time PCR revealed that full population diversity was recovered (data not shown). Both error-prone PCR of the full gene and shuffling of mutagenized loops were successfully employed to diversify the population of lead clones (Fig. 2b). Gene mutagenesis was performed by error-prone PCR with nucleotide analogs.27 Independent error-prone PCRs were conducted for each of the three loops using primers that overlap either the adjacent loop primer or the plasmid vector. This overlap enabled shuffled gene reconstruction via homologous recombination during yeast transformation (Fig. 2c). The number of diversified transformants during affinity maturation ranged from 2 to 20 million (mean, 8.1 million) for full gene mutagenesis and 0.1 to 6.5 million (mean, 2.5 million) for loop mutagenesis. Sequence analysis exhibited mutagenesis that matched the desired 1 to 5 amino acid mutations per gene (data not shown), which was achieved with a single combination of nucleotide analog concentration and number of PCR cycles determined by mathematical modeling.
High-affinity binder engineering
The efficacy of the new library and affinity maturation scheme was tested by engineering binders to lysozyme, a model protein that we have previously used as an Fn3 target with different library and maturation approaches.6
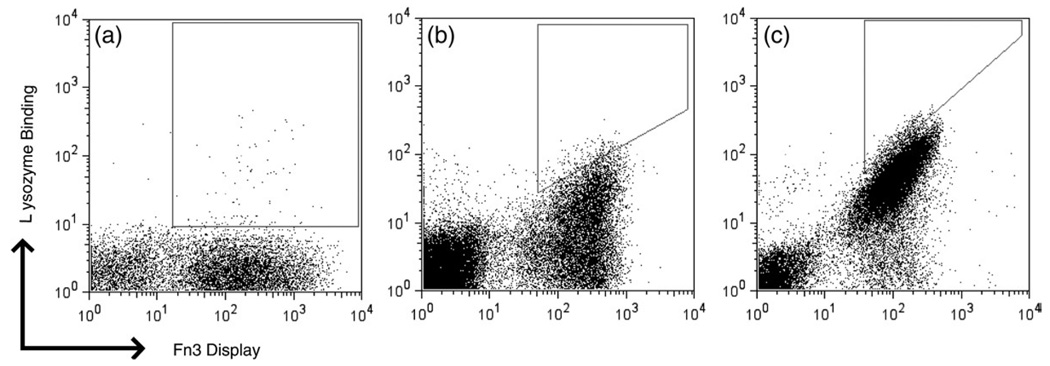
The naive library was sorted three times by FACS using multivalent biotinylated lysozyme preloaded on streptavidin–fluorophore conjugates. The resultant population was diversified by both gene and loop mutagenesis. Transformed mutants as well as the original clones were sorted twice by FACS, yielding enrichment of evident lysozyme-binding clones (Fig. 3). Four additional rounds of isolation and maturation, each consisting of two to three FACS selections followed by mutagenesis (Supplementary Fig. 1), were conducted using monovalent lysozyme, ranging from 1 µM to 20 pM lysozyme. Labeling at low picomolar concentrations while maintaining stoichiometric excess of target to Fn3 would require impractically large volumes; thus, kinetic competition sorting was used for the final three rounds of isolation and maturation. The final two sublibraries were sorted an additional three times and the collected populations were sequenced to identify the highest affinity clones.
Fig. 3.
Binder isolation and affinity maturation. Yeast libraries displaying Fn3 were labeled with mouse anti-c-myc antibody followed by goat anti-mouse fluorophore as well as biotinylated lysozyme and streptavidin–fluorophore and analyzed by flow cytometry. (a) Yeast population during second round of isolation and maturation labeled with 50 nM multivalent lysozyme preloaded in a 3:1 stoichiometry on streptavidin–R-phycoerythrin. (b) Yeast population during sixth round of isolation and maturation labeled with 0.5 nM monovalent lysozyme followed by streptavidin–AlexaFluor488. (c) Yeast population during eighth round of isolation and maturation labeled with 2 nM monovalent lysozyme for 15 min followed by 35 h of dissociation and labeling with streptavidin–R-phycoerythrin. Polygonal regions represent sort gates for cell selection.
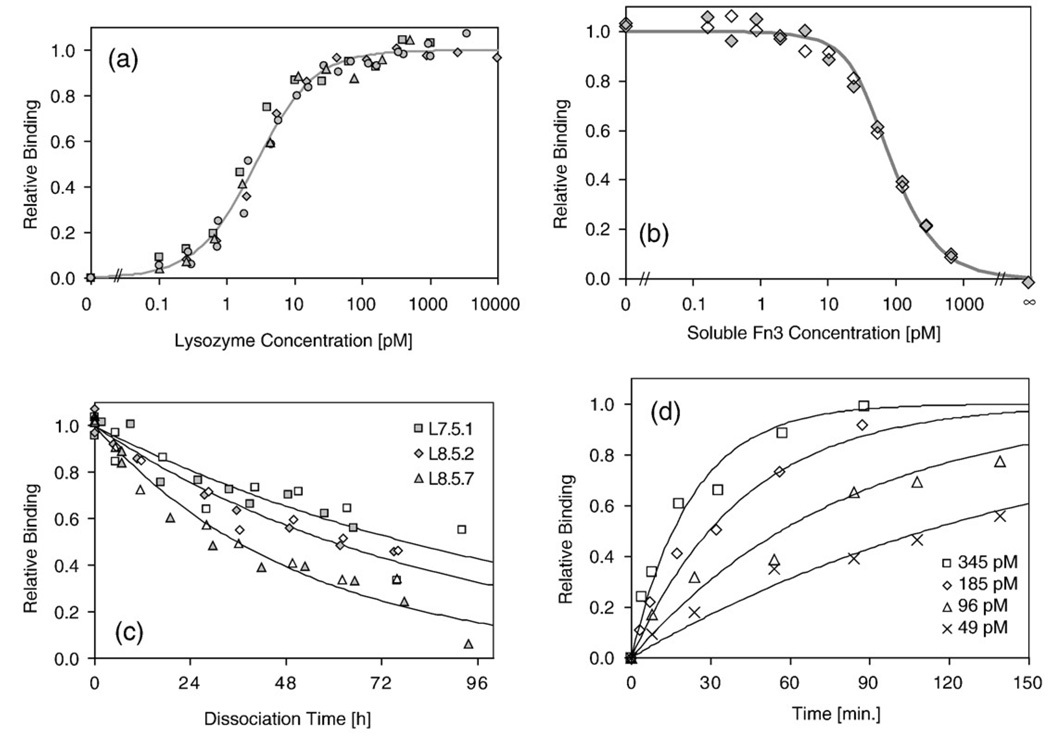
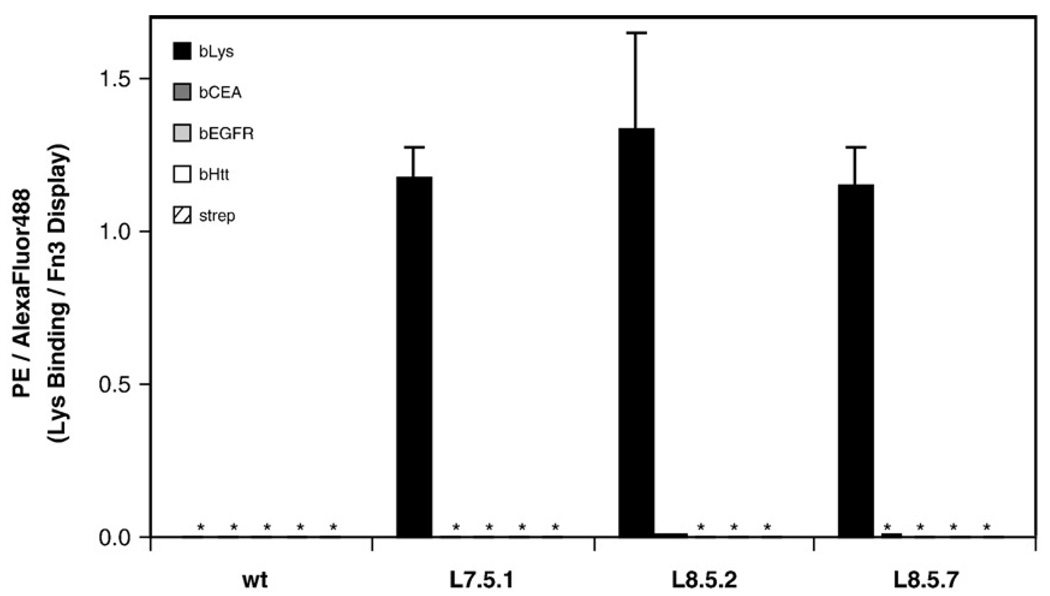
Three dominant clones were identified by sequence analysis (Table 1). Affinity titrations indicate that all three clones have comparable equilibrium dissociation constants (Kd) for binding to biotinylated lysozyme: 2.6±0.6 pM (clone L7.5.1), 2.9±1.4 pM (L8.5.2), and 2.8±0.5 pM(L8.5.7) (Fig. 4a and Table 1). The high-affinity binding was validated using purified Fn3 domains in an equilibrium competition titration, which indicated an affinity of 6.9±0.3 pM for L7.5.1 (Fig. 4b). Dissociation rates for the three clones range from (2.5–5.4)×10−6 s−1, which correspond to dissociation half times of 36–78 h (Fig. 4c). The association rate of L7.5.1 was measured experimentally as (2.0±0.5)×106 M−1 s−1 (Fig. 4d). The calculated value of koff/kon is 1.2±0.3 pM, which is reasonably consistent with the experimentally measured Kd of 2.6±0.6 pM. In addition to high affinity, the clones exhibit target specificity, as they do not show appreciable binding to an array of other molecules (Fig. 5).
Table 1.
Characterization of wild-type, L7.5.1, L8.5.2, and L8.5.7
| Clone | Amino acid sequence |
Kd (pM) |
Koff (10−5 s−1) |
Monomer (%) |
Tm (°C) |
T1/2 (°C) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| BC loop | DE loop | FG loop | Framework | DSC | CD | |||||
| WT | DAPAVTR | GSKST | GRGDSPASSK | >107 | n/d | 99 | 85.7 | 84.2 | n/d | |
| L7.5.1 | RGYPWAT | GVTN | RVGRTFDTPG | P15S, R33G, T35I, P44L, V50M |
2.6±0.6 | 0.25±0.02 | 99 | 58.1 | 58.8±1.6 | 55.7±0.1 |
| L8.5.2 | RGCPWAI | GVTN | RVGRMLCAPG | R33G, I34V, N42S, P44L, V45A, V50M, K63E, K98R |
2.9±1.4 | 0.32±0.01 | 80 | n/d | 52.5±0.2 | 50.6±0.5 |
| L8.5.7 | RDRPWAI | GVTN | RLSIVPYA | D3G, L18I, R33G, N42S, P44L, V50M, Y73H, N91T, S100P |
2.8±0.5 | 0.54±0.06 | 93 | 54.5 | n/d | 53.0±0.4 |
Kd, equilibrium dissociation constant at 25 °C; koff, dissociation rate constant at 25 °C; Monomer, percent of purified protein present as monomer in analytical size-exclusion chromatography; Tm, midpoint of thermal denaturation curve as determined by DSC or CD; T1/2, midpoint of thermal denaturation curve as determined by yeast surface display residual activity assay.
Fig. 4.
Determination of binding parameters for high-affinity clones. L7.5.1, L8.5.2, or L8.5.7 was displayed on the yeast surface and assayed for binding to biotinylated lysozyme at 25 °C. Different symbols indicate replicate experiments. Continuous lines indicate theoretical values. (a) Fraction of displayed L7.5.1 binding to biotinylated lysozyme at equilibrium. Only L7.5.1 is shown for simplicity. (b) Relative binding of 20 pM biotinylated lysozyme to displayed L7.5.1 at equilibrium in the presence of indicated amount of soluble L7.5.1 competitor. ∞ indicates samples without biotinylated lysozyme. (c) Fraction of displayed L7.5.1 (squares), L8.5.2 (diamonds), or L8.5.7 (triangles) binding to biotinylated lysozyme after dissociation at 25 °C for the indicated time. (d) Fraction of displayed L7.5.1 binding to biotinylated lysozyme after association at 25 °C; biotinylated lysozyme was present at 49 pM (crosses), 96 M (triangles), 185 pM (diamonds), or 345 pM (squares). Results are presented from a single experiment, which is representative of triplicate experiments performed at different sets of concentrations.
Fig. 5.
Binding specificity. Wild-type Fn3 (wt), L7.5.1, L8.5.2, or L8.5.7 was displayed on the yeast surface, washed, and incubated with 100 pM biotinylated lysozyme (black) or 100 nM nontarget molecules: biotinylated carcinoembryonic antigen (bCEA, dark gray), biotinylated epidermal growth factor receptor domain IV (bEGFR, light gray), biotinylated huntingtin peptide (bHtt, white), or streptavidin–R-phycoerythrin (strep, striped). Binding was detected with streptavidin–R-phycoerythrin secondary labeling and flow cytometry. The data are presented as the mean fluorescence units from binding (PE signal) normalized by the amount of Fn3 on the yeast surface (AlexaFluor488 signal). Values and error bars represent mean±standard deviation of duplicate measurements. Asterisk (*) indicates a value less than 0.005.
Since previously reported high-affinity clones were unstable or oligomeric, it was desired to examine the behavior of the current Fn3 domains. The clones were produced in bacterial culture and purified for biophysical characterization. Analytical size-exclusion chromatography indicated that all three clones are predominantly monomeric with only L8.5.2 present in a significant oligomeric state (80% monomeric; Table 1 and Supplementary Fig. 2a). It should be noted that L8.5.2 contains two cysteine residues and intermolecular disulfide bonding contributes to oligomerization as indicated by nonreducing SDS-PAGE (data not shown). The thermal stabilities of the proteins were analyzed by differential scanning calorimetry (DSC) and circular dichroism of purified protein as well as thermal denaturation of protein displayed on the yeast surface (Table 1 and Supplementary Fig. 3). L7.5.1 is the most stable clone with midpoints of thermal denaturation ranging from 55.7 to 58.8 °C for the three methods. It should be noted that denaturation of L7.5.1 is not reversible because of aggregation at high temperatures. Both other clones are also stable with Tm values over 50 °C. In addition, far-UV circular dichroism analysis reveals no significant differences in secondary structure between wild-type Fn3 and L7.5.1, indicating that despite loop mutation and length variation the structural integrity of the domain is maintained (Supplementary Fig. 2b).
Analysis of intermediate populations
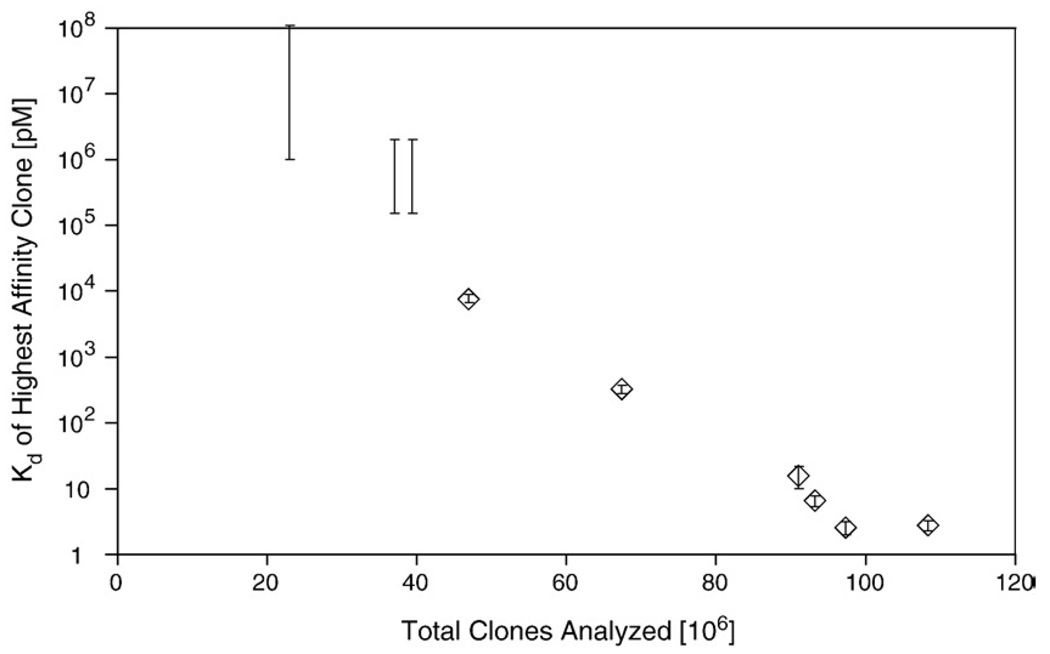
Intermediate populations were investigated to elucidate the progress of affinity maturation. Each sublibrary (i.e., library of mutagenized clones created during affinity maturation) was sorted by FACS without mutagenesis to identify the highest affinity clone at each stage of affinity maturation. The highest affinity clone was identified as the dominant clone from sequencing several clones from the extensively sorted sublibraries. Sequences, equilibrium dissociation constants and dissociation rate constants were determined (Table 2). The highest affinity binder in the original library exhibits apparent midmicromolar affinity. However, affinity maturation progressively improves binding performance to yield nanomolar and then picomolar binders (Fig. 6). It is noteworthy that affinity maturation exhibits a relatively consistent correlation to the number of clones analyzed throughout the course of affinity maturation until the final round of directed evolution. A steady increase characteristic of a consistently progressing affinity maturation scheme is observed rather than one single exceptional increase in affinity, indicative of a fortuitous mutation or recombination.
Table 2.
Highest affinity Fn3 domains in each sublibrary
| Amino acid sequence |
Kd (pM) |
koff (10−5 s−1) |
t1/2 (h) | ||||
|---|---|---|---|---|---|---|---|
| Round | BC loop | DE loop | FG loop | Framework | |||
| 0 | RDCPWAT | WTPVCF | SSQRGCM | none | ≫100,000 | n/d | n/d |
| 1 | SLDNQAN | GQSD | RCEPSRNSAV | none | >100,000 | n/d | n/d |
| 2 | same clone as round 1 | ||||||
| 3 | SLDNQAN | GVTN | RVGRMLDTPG | P44S, V50M | 7600±1100 | 460 | 0.04 |
| 4 | SLDNQAK | GATN | RCKPFRNSAV | P44S, V50M, T97I | 330±50 | n/d | n/d |
| 5 | RDCPWAI | GVTN | RVGRMSCTSG | V1A, S1P, T14A, R33G, P44L, V50M |
16±6 | 4.5±0.3 | 4.2±0.3 |
| 6 | RGCPWAI | GVTN | RVGRMLCTPG | P15S, R33G, N42S, P44L, V50M, K63E |
6.6±1.3 | 0.72±0.07 | 27±3 |
| 7 | RGYPWAT | GVTN | RVGRTFDTPG | P15S, R33G, T35I, P44L, V50M | 2.6±0.6 | 0.25±0.02 | 78±1 |
| 8a | RGCPWAI | GVTN | RVGRMLCAPG | R33G, I34V, N42S, P44L, V45A, V50M, K63E |
2.9±1.4 | 0.32±0.01 | 60±1 |
| RDRPWAI | GVTN | RLSIVPYA | D3G, L18I, R33G, N42S, P44L, V50M, Y73H, N91T |
2.8±0.5 | 0.54±0.06 | 36±4 | |
Round, number of mutagenesis cycles (round 0 indicates naive library); Kd, equilibrium dissociation constant at 25 °C; Koff, dissociation rate constant at 25 °C; t1/2, time for 50% dissociation of the Fn3–lysozyme complex at 25 °C; n/d, not determined.
Two clones with similar affinities were identified from round 8.
Fig. 6.
Affinity maturation progress. The highest affinity clone in each affinity maturation sublibrary was identified by FACS. The equilibrium dissociation constant of the first three clones could not be determined accurately by yeast surface display titration because of the relatively low affinity. Thus, an estimated range of possible affinities consistent with equilibrium labeling at high nanomolar concentrations is indicated. The equilibrium dissociation constant of the latter six clones was determined by titration and is represented as the mean±one standard deviation of replicate measurements. The number of total clones analyzed is the cumulative total of the number of full-length clones in the naive library and total yeast transformants in all affinity maturation sublibraries.
The impact of loop shuffling is evident in multiple cases. The BC loop from the round 1 clone is recombined with new DE and FG loops in round 3 to yield an 8 nM binder. A mutated version of the FG loop present in round 1 is then recombined with mutated BC and DE loops from round 3 to achieve picomolar binding. The highest affinity clones in rounds 5–8 result from apparent shuffling of the BC loop observed in round 0 and the DE and FG loops observed in round 3 as well as multiple framework mutations. The appearance of point mutations is also apparent, both within the loops and in the framework region. The impact of these framework mutations was investigated in more detail in the context of L7.5.1.
L7.5.1 analysis
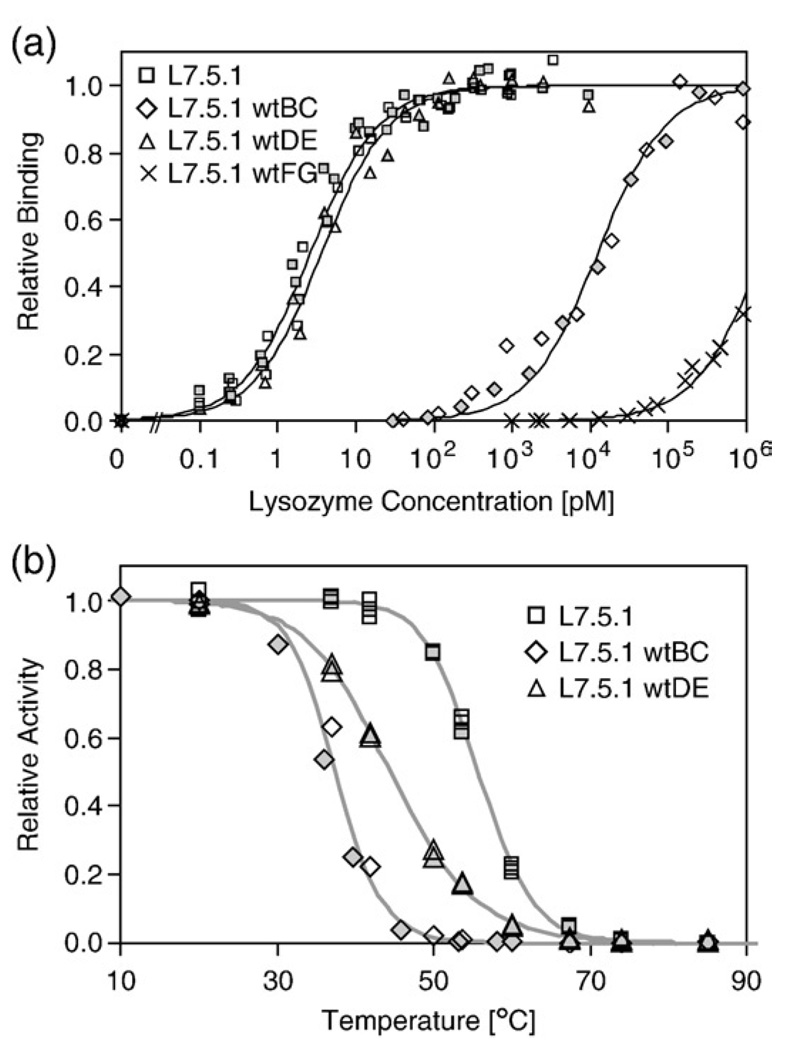
L7.5.1 was selected for more thorough analysis because it has the fewest framework mutations and is the most stable of the high-affinity clones. The engineered elements of the clone were analyzed for their impact on affinity and stability. Each randomized loop and framework mutation was independently restored to wild-type sequence and both affinity and stability were measured using yeast surface display. Reversion of the FG loop had the strongest effect on binding, as wild-type restoration decreased affinity to the micromolar level; conversely, DE loop reversion had a nearly negligible effect on binding, and the BC loop reversion had an intermediate effect with a 4700-fold reduction (Fig. 7a). Both BC and DE loop reversion significantly destabilize the domain (Fig. 7b). Although these loop sequences confer high thermal stability in the wild-type context, modification of adjacent loops apparently adjusts the intramolecular contacts. Thus, during multiloop diversification, selected sequences must provide not only proper intermolecular contacts for high-affinity binding but also proper intramolecular contacts for structural integrity and stability. The stability of the FG loop reversion could not be accurately determined by this method because of its weak binding.
Fig. 7.
L7.5.1 loop analysis. Each engineered loop of L7.5.1 was independently restored to wild-type sequence and the resultant clones were displayed on the yeast surface. Filled and empty symbols indicate replicate experiments. (a) Binding to biotinylated lysozymeat 25°C was quantified by flow cytometry. Clone and equilibrium dissociation constant: L7.5.1 (squares), 2.6±0.6 pM; L7.5.1wtBC (diamonds), 12.4 ± 0.7 nM; L7.5.1wtDE (triangles), 3.7 ± 1.4 pM; L7.5.1wtFG (crosses), 1.6±0.2 µM. (b) Yeast displaying Fn3 were incubated at the indicated temperature for 30 min followed by a flow cytometric assay for biotinylated lysozyme binding. Clone and midpoint of thermal denaturation: L7.5.1 (squares), 55.7±0.1 °C; L7.5.1wtBC (diamonds), 37.4±1.5 °C; L7.5.1wtDE (triangles), 44.5±0.1 °C.
Three of the five framework mutations in the selected clone (T35I, P44L, and V50M) are relatively conservative mutations and were beneficial to both affinity and stability (Table 3). Conversely, R33G replaces a large, positively charged side chain with a single hydrogen; this mutation provides 28-fold stronger binding without a substantial effect on stability. Perhaps the arginine side chain was not accommodated sterically and/or electrostatically at the Fn3–lysozyme interface or the glycine enables a beneficial backbone conformation. The P15S mutation had only a very minor affinity improvement but substantially destabilized the domain. The lack of impact on binding is reasonable given its distance from the expected paratope, and the substantial destabilization is not surprising given the removal of the backbone-constraining proline in the turn between the C and D strands. Selection of this mutation was likely coincident with another beneficialmutation, as it does not impart significant benefit; it should be noted that while the most beneficial mutations, R33G, P44L, and V50M, are consistently observed throughout maturation (Table 2), P15S and T35I are rare. Overall, these data suggest the paratope is focused on the BC and FG loops as well as potential key contacts on the C and D strands of the β sheet.
Table 3.
Affinity and stability of framework mutation reversions of L7.5.1
| AA | WT | L7.5.1 | Kd (pM) | T1/2 (°C) | Location |
|---|---|---|---|---|---|
| L7.5.1-parent clone | 2.6±0.6 | 55.7±0.1 | |||
| 15 | Pro | Ser | 3.4±0.1 | 59.5±0.7 | AB loop |
| 33 | Arg | Gly | 74±23 | 56.2±0.6 | C strand |
| 35 | Thr | lle | 5.8±1.6 | 54.5±0.5 | C strand |
| 44 | Pro | Leu | 14.3±4.0 | 50.3±1.2 | CD loop |
| 50 | Val | Met | 25±16 | 53.7±1.8 | D strand |
The five framework mutations of L7.5.1 were individually reverted to wild-type amino acids. Binding affinity and thermal stability were determined by yeast surface display. AA, amino acid position; WT, wild-type side chain; L7.5.1 clone, 7.5.1 side chain; Kd, equilibrium dissociation constant at 25 °C; T1/2, midpoint of thermal denaturation as determined by yeast surface display.
Focused mutagenesis
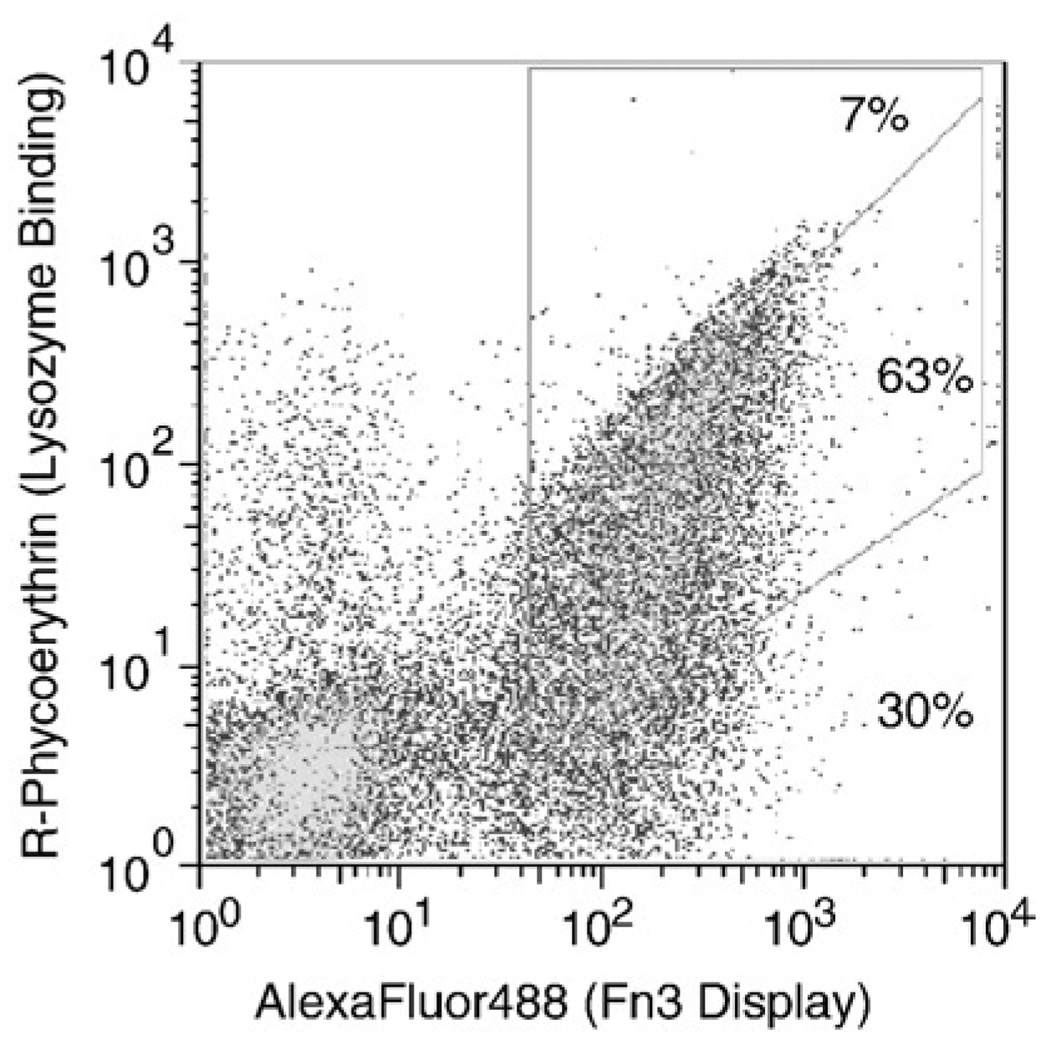
The conservation of the DE loop sequence throughout much of the affinity maturation (Table 2) despite its relatively low impact on affinity relative to wild type prompted further study of this loop. The functionally tolerable diversity and the potential for improved binding were explored through re-randomization of the loop sequence. Within the context of L7.5.1 S15P, the loop was randomized using NNB nucleotide diversity and the same four loop length options as the original library. Labeling the library with 30 pM lysozyme, which yields 90% binding to the parent clone L7.5.1 S15P, yields many clones that provide effective binding, indicating that the loop can tolerate significant diversity and maintain binding. Specifically, 7% of clones exhibit binding at 75% of maximum, which is characteristic of a 10 pM Kd, or within threefold of the parent clone (Fig. 8). Yet, 30% of the full-length clones bind at less than 1% of maximum, indicating that many loop sequences can greatly hinder binding either through direct interaction with the binding partner or through structural modification of the Fn3 paratope. In addition, the library was sorted for binding to biotinylated lysozyme using FACS. Sequence analysis after four selections identified a single clone, DE0.4.1, with GDLSHR replacing GVTN in the DE loop. Binding and stability analyses indicate a 3.1× improvement in binding to 1.1 pM at the expense of an 8.5 °C decrease in the T1/2 (Table 4). The maintenance of glycine at position 52 from wild type to L7.5.1 to DE0.4.1 is noteworthy for possible future library design especially considering the adjacent amino acid is proline; the proline–glycine pair provides an effective turn motif to begin the DE loop.
Fig. 8.
L7.5.1 S15P DE loop randomization library binding analysis. The DE loop of L7.5.1 S15P was randomized using NNB codon diversity with four loop lengths. The yeast surface display library was labeled with 30 pM biotinylated lysozyme and mouse anti-c-myc antibody followed by streptavidin–R-phycoerythrin and goat anti-mouse antibody conjugated to AlexaFluor488. Percentages indicate the relative number of c-myc positive clones with bLys binding: c-myc display signals with 75% of maximum (7%), 1–75% of maximum (63%), or <1% of maximum (30%).
Table 4.
Affinity and stability of clones from focused mutagenesis
| Amino acid sequence |
Kd (pM) |
T1/2 (°C) |
||||
|---|---|---|---|---|---|---|
| Clone | BC loop | DE loop | FG loop | Framework | ||
| L7.5.1 S15P | RGYPWAT | GVTN | RVGRTFDTPG | R33G, T35I, PAAL, V50M | 3.4±0.1 | 59.5±0.7 |
| Cons 0.4.1 | REDPWAK | GVTN | RVGWASYTLG | R33G, T35I, P44L, V50M | 1.1±0.6 | 59±3 |
| DE 0.4.1 | RGYPWAT | GDLSHR | RVGRTFDTPG | R33G, T35I, P44L, V50M | 1.1±0.5 | 51±3 |
Kd, equilibrium dissociation at 25 °C; T1/2, midpoint of thermal denaturation curve as determined by yeast surface display residual activity assay
Lack of improvement from round 7 to round 8 in conjunction with sequence similarities during affinity maturation prompted investigation into a complementary method of affinity maturation. Since only a fraction of protein sequence space is accessible through single nucleotide mutations, a library was constructed in which the DNA encoding nonconserved amino acids in the BC and FG loops was randomized by degenerate oligonucleotides. Nonconserved amino acids were identified as the positions of L7.5.1-homologous clones at which sequence diversity was observed in any sequence during affinity maturation or sublibrary analysis. The nonconserved amino acids were randomized in two modes: either to all 20 amino acids using NNB codons or to amino acids observed during sequence analysis using tailored codons at each position. The library was sorted for binding to biotinylated lysozyme using FACS. Sequence analysis after four selections yielded a single clone, Cons0.4.1. The affinity of Cons0.4.1 was measured as 1.1±0.6 pM by yeast surface display titration and 0.33±0.15 pM by equilibrium competition with purified Fn3 domain. The midpoint of thermal denaturation was measured as 52.5±2.1 °C by circular dichroism analysis and 58.8±3.4 °C by yeast surface display thermal resistance assay. Far-UV circular dichroism indicates that Cons0.4.1 maintains a secondary structure similar to that of the wild-type Fn3 domain (Supplementary Fig. 2b). Two of the eight codon changes were not possible with a single nucleotide mutation making this clone unlikely to be reached by error-prone PCR. Thus, while error-prone PCR provides a highly effective means of diversification, an improvement was observed through more thorough searching of a focused region of sequence space once a consistent binding motif was identified.
Discussion
In this work we explore the impact of multiple components of engineering a high-affinity binding site in the Fn3 scaffold. The combination of loop length diversity and recursive mutagenesis including mutated loop shuffling enabled selection of the highest affinity Fn3 domains yet reported despite a relatively modest initial library size of 2.3×107 full-length Fn3 clones. The results extend the affinity attainable by this single domain scaffold, further validating its use as a molecular recognition scaffold. In addition, insights were gained for both Fn3 design and engineering and protein engineering in general.
Analysis of the highest affinity binders in each sublibrary (Table 2) demonstrates deviation from wild-type loop lengths in all three loops implicating the importance of this diversity element in Fn3 library design. Interestingly, the observed BC loops are all one amino acid shorter than wild type, which is observed both in other fibronectin type III domains (Fig. 1) and in a previously engineered binder.6 DE loops are observed with either one less or one more amino acid than wild type. Clone DE0.4.1 and the highest affinity clone from the naive library (Table 2) justify inclusion of a six-amino acid DE loop despite its lack of existence in native fibronectin type III domains. FG loops of wild-type length as well as both two- and three-amino acid reductions were observed. The observed length diversity, as well as the success of length diversity in a restricted diversity Fn3 library published during preparation of this manuscript,5 support the inclusion of length diversity in all future Fn3 engineering. Moreover, the combination of phylogenetic analysis and sequence analysis of engineered binders should continue to elucidate the relative preference of each length to further improve library design.
Loop length diversity increases sequence space to 1034 possible amino acid sequences for the three loops, although only 2.3×107 clones are present in the yeast library. The two modes of diversity introduced during recursive mutagenesis effectively search this large sequence space despite the sparse sampling. Sequence and binding analyses indicate that each of the elements of the mutagenesis method were beneficial. The loop mutagenesis path of the diversification was effective, as both loop shuffling and loop-focused point mutations are evident (Table 2). In addition, the gene mutagenesis path was advantageous, as an effective loop combination was maintained in rounds 5–8 and beneficial framework mutations were introduced throughout (Table 2 and Table 3). The importance of diversifying many clones during each round of maturation is evident, since homologs of the eventual DE and FG loops were not present in 30 sequences from the enriched populations from the naive library. As a result of these combined components, affinity maturation rapidly and efficiently progressed to yield the clones with 3 pM affinity without rational intervention. The affinity maturation procedure is straightforward and simple; plasmid recovery, mutagenesis, amplification, and yeast transformation can be achieved in 1–2 days. The increased frequency of mutagenesis is obviously applicable to any protein engineering method and is strongly recommended to improve both the speed of binder isolation and the overall efficacy. Shuffling can be implemented in any analogous protein scaffold provided the regions of interest are relatively proximal at the DNA level to enable overlap for homologous recombination. Yeast surface display provides an effective system for shuffling because DNA fragments can be recombined with high fidelity during cellular transformation simplifying the method.
The sequence diversity of the highest-affinity clones is striking. L8.5.2 has two cysteines at locations consistent with formation of an interloop disulfide bond, as was found previously from a different Fn3 library screened by yeast display.6 However, unlike in that case, the disulfide is dispensable for high-affinity binding, since L7.5.1 has highly related loop sequences but lacks both cysteines. A third clone with similar affinity (L8.5.7, 2.8 pM) was found with a substantially different FG loop, but conserved DE and BC loops.
It is noteworthy that the focused mutagenesis results indicate the potential for mild improvement of affinity maturation through targeted randomization at multiple residues identified as variable through sequence analysis. It is not yet clear if the success of this avenue of affinity maturation resulted from the relatively high number of mutations from the parent clone (eight amino acid changes in the two loops) or the ability to reach amino acids that would require more than one nucleotide mutation. Regardless, the fact that a secondary, semirational approach was only able to yield 3.1× enhancement in affinity is indicative of a relatively effective search of sequence space by the initial affinity maturation.
Diversification of the DE loop is an important aspect of Fn3 engineering. Because it is the shortest and least flexible wild-type loop, it is unclear if the binding potential gained by diversification offsets the possible structural destabilization. Thus, analysis of the contribution of the DE loop to binding and stability are valuable to clone maturation and future Fn3 library design. The originally selected DE loop, GVTN, stabilizes clone L7.5.1 relative to wild type but does not significantly aid binding. Although loop reversion analysis suggests that the binding paratope is dominated by the FG and BC loops, the DE loop can be engineered for improved binding (DE0.4.1) albeit at the expense of stability. Since selections were explicit for affinity, the lack of binding performance from the L7.5.1 DE loop likely resulted from a lack of DE loop diversity after a few rounds of directed evolution (Table 2). Thus, a future improvement on loop shuffling will be to incorporate a small percentage of naive loops with the engineered loops. A broader uncertainty regarding the DE loop is the extent to which it should be diversified in future Fn3 libraries. Previous binders have been engineered with wild-type DE loops with 350 pM affinity for lysozyme,6 30 nM for maltose-binding protein,5 250 nM for Src SH3 domain,2 and approximately 5µM for ubiquitin.4 The DE reversion of L7.5.1 represents a 3.7 pM binder with a wild-type DE loop. Collectively, it is clear that binders can be engineered without DE loop diversity. Conversely, engineered DE loops in the L7.5.1 context can either improve the affinity to 1.1 pM or stabilize the molecule by an 11 °C increase in T1/2, the latter of which is not surprising given its extensive contact with the BC loop residues as well as the shortened BC loop length. Moreover, engineered vascular endothelial growth factor receptor 2 binders require their engineered DE loop for binding, although it destabilizes the molecule by 2.0 kcal/mol or ~30 °C decrease in Tm.7 Consequently, as expected, DE loop impact is context dependent. One possible general approach would be to introduce mild DE diversity in the original library to allow for binding constraint removal and structural complementation of selected BC loops as well as the possibility of beneficial binding contacts. After binder selection, affinity or stability maturation could be employed with more diverse DE loop shuffling in the context of effective BC and FG loops.
Unlike several examples of previous high-affinity Fn3 binder engineering, the selected high-affinity clones are relatively stable and monomeric. L7.5.1 is a 2.6 pM binder and >99% monomeric with a midpoint of thermal denaturation of 56–59 °C. Moreover, reversion of serine to proline at position 15 improves the T1/2 to 60 °C. In addition, Cons0.4.1 is a 1.1 pM binder with Tm of 53 °C, and L8.5.7 is a 2.8pMbinder, is 93% monomeric, and has a Tm of 53–55 °C. Although stability was not an explicit element of selection, the eukaryotic secretion machinery of yeast provides some level of quality control against misfolded proteins.28,29 In general, Fn3 clones of higher stability are displayed at high densities on the yeast cell surface (data not shown); thus, unstable clones are slightly selected against based on the two-color sort regions. It remains to be seen if stable clones will generally result from selections by yeast surface display or if this was a fortuitous result; regardless, yeast surface display provides a means for stability engineering either during or after binder selection.
Overall, the method can be further improved through refinement of naive loop length distribution, an increase in magnitude of the initial library, inclusion of naive loops during loop shuffling, and perhaps more expansive mutation than attainable by error-prone PCR with nucleotide analogs. Nevertheless, the combination of yeast surface display, loop length diversity, and dual-mode affinity maturation rapidly yielded stable, high-affinity binders. The method should be valuable toward development of Fn3 binders to additional targets as well as transferable to engineering of other proteins for any screenable functionality.
Materials and Methods
Fn3 library construction
Oligonucleotides were purchased from MWG Biotech (High Point, NC) and Integrated DNA Technologies (Coralville, IA) (see Supplementary Data online for sequences). The Fn3 library was constructed to produce wild-type sequence in the framework regions and to randomize the BC,DE, and FG loops of Fn3. The DNA encoding for amino acids 23–30 (DAPAVTVR) was replaced by (NNB)x where x=6, 7, 8, or 9 to yield a loop length that is −2, −1, 0, or +1 amino acids relative to wild type. Similarly, the DNA for amino acids 52–56 (GSKST) was replaced by (NNB)y where y=4, 5, 6, or 7, and the DNA for amino acids 77–86 (GRGDSPASSK) was replaced by (NNB)z where z=5, 6, 8, or 10.
The library was constructed by sequential annealing and extension of eight overlapping oligonucleotides.6 The following components were combined in a 50-µL reaction: two oligonucleotides (0.2 µM a2, b3nmix, c6t, or d7nmix+ 0.4 µMa1, b4n, c5nmix, or d8n, respectively), 1× polymerase buffer, 0.2mMdeoxynucleotide triphosphate (dNTP),1mM MgSO4, 1 U KOD Hot Start DNA Polymerase (Novagen, Madison, WI), 1 M betaine, and 3% dimethyl sulfoxide. The mixture was denatured at 95 °C for 2 min followed by 10 cycles of 94 °C for 30 s, 58 °C for 30 s, and 68° for 1 min and a final extension of 68 °C for 10 min. Forty microliters of the products (a1+a2, b3nmix+b4n, c5nmix+c6t, or d7nmix+d8n) were combined and thermally cycled at identical conditions. Two microliters of this product was combined with 0.4 µM primer (a1–b4n amplified with p1; c5nmix–d8n amplified with p8) in a new 100-µL reaction and thermally cycled under identical conditions to amplify the appropriate strand. The products were combined and thermally cycled at identical conditions. The final products were concentrated with PelletPaint (Novagen). The plasmid acceptor vector pCTf1f4 (Ref. 6) was digested with NcoI, NdeI, and SmaI (New England Biolabs, Ipswich, MA). Multiple aliquots of ~10 µg of Fn3 gene and 3 µg plasmid vector were combined with 50–100 µL of electrocompetent EBY100 and electroporated at 0.54 kV and 25µF. Homologous recombination of the linearized vector and degenerate insert yielded intact plasmid. Cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) for 1 h at 30 °C, 250 rpm. The number of total transformants was 6.5×107 cells as determined by serial dilutions plated on SD-CAA plates (0.1 M sodium phosphate, pH 6.0, 182 g/L sorbitol, 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 20 g/L glucose). The library was propagated by selective growth in SD-CAA, pH 5.3 (0.07 M sodium citrate, pH 5.3, 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 20 g/L glucose, 0.1 g/L kanamycin, 100 kU/L penicillin, and 0.1 g/L streptomycin) at 30 °C, 250 rpm.
Fluorescence-activated cell sorting
Yeast were grown in SD-CAA, pH 5.3, at 30 °C, 250 rpm to logarithmic phase, pelleted, and resuspended to 1×107 cells/mL in SG-CAA, pH 6.0 (0.1M sodium phosphate, pH 6.0, 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 19 g/L dextrose, 1 g/L glucose, 0.1 g/L kanamycin, 100 kU/L penicillin, and 0.1 g/L streptomycin) to induce protein expression. Induced cells were grown at 30 °C, 250 rpm for 12–24h.
Round 0 (three FACS selections) and round 1 (two FACS selections) were conducted with multivalent lysozyme prepared by incubating streptavidin–fluorophore (R-phycoerythrin, AlexaFluor488, or AlexaFluor633; Invitrogen, Carlsbad, CA) with biotinylated lysozyme (Sigma, St. Louis, MO) in a 1:3 ratio in phosphate-buffered saline with bovine serum albumin (PBSA). Yeast were pelleted, washed in 1 mL PBSA (0.01 M sodium phosphate, pH 7.4, 0.137 M sodium chloride, 1 g/L bovine serum albumin), resuspended in PBSA with 10–40 mg/L mouse anti-c-myc antibody (clone 9E10, Covance), and incubated on ice. Cells were washed with 1 mL PBSA and resuspended in PBSA with multivalent lysozyme (0.5 µM for the first four sorts and 50 nM for the fifth sort) and goat antimouse antibody conjugated to R-phycoerythrin, Alexa-Fluor488, or AlexaFluor633.
Intermediate FACS selections were conducted with near-equilibrium labeling with monovalent lysozyme. Three, two, two, and three selections were performed in rounds 2–5, respectively. Yeast were pelleted, washed in 1 mL PBSA, resuspended in PBSA with biotinylated lysozyme (ranging from 1 µM to 20 pM) and mouse anti-c-myc antibody, and incubated on ice. Cells were then washed with 1 mL PBSA and resuspended in PBSA with streptavidin–fluorophore and fluorophore-conjugated goat anti-mouse antibody.
FACS selections of very high affinity populations were conducted with kinetic competition. Two, three, and two selections were performed in rounds 6–8. Yeast were washed and incubated briefly with 1–2 nM biotinylated lysozyme. Yeast were then washed and resuspended with PBSA with 140 nM unbiotinylated lysozyme (to prevent further association of labeled target) and incubated at room temperature for 2 h to 7 days to enable dissociation of biotinylated lysozyme. Cells were washed in PBSA, resuspended in PBSA with mouse anti-c-myc antibody, and incubated on ice. Cells were washed and labeled with secondary reagents as in equilibrium labeling.
In all cases, labeled cells were washed with 1 mL PBSA, resuspended in 0.5–2.0 mL PBSA and analyzed by flow cytometry using either a MoFlo (Cytomation) or Aria (Becton Dickinson) cytometer. c-myc + cells with the top 0.2–3% of lysozyme binding/c-myc display ratio were selected. Collected cells were grown in SD-CAA, pH 5.3, at 30 °C, 250 rpm and either induced in SG-CAA, pH 6.0, for further selection or used for plasmid recovery.
Fn3 mutagenesis
Plasmid DNA from 1×108 cells was isolated using two columns of Zymoprep kit II (Zymo Research, Orange, CA) according to the manufacturer’s instructions except for additional centrifugation of neutralized precipitate. The zymoprep elution was cleaned using the Qiagen PCR Purification kit (Qiagen, Valencia, CA), and eluted in 40 µL of elution buffer. Error-prone PCR of the entire Fn3 gene was performed in a 50-µL reaction containing 1×Taq buffer, 2 mM MgCl2, 0.5 µM each of primers W5 and W3, 0.2 mM (each) dNTPs, 5 µL of zymoprepped DNA template, 2 mM 8-oxo-deoxyguanosine triphosphate (TriLink, San Diego, CA), 2 mM 2′-deoxy-p-nucleoside-5′-triphosphate (Tri-Link), and 2.5 U of Taq DNA polymerase (Invitrogen). In parallel, error-prone PCR of the loop regions was performed via three separate 50 µL reactions with 20 mM 8-oxo-deoxyguanosine triphosphate and 20 mM 2′-deoxy-p-nucleoside-5′-triphosphate and primers BC5 and BC3 for the BC loop, DE5 and DE3 for the DE loop, and FG5 and FG3 for the FG loop. The reaction mixtures were denatured at 94 °C for 3 min, cycled 15 times at 94 °C for 45 s, 60 °C for 30 s, and 72 °C for 90 s, and finally extended at 72 °C for 10 min. Multiple preliminary mutagenesis reactions of the wild-type plasmid were conducted at different nucleotide analog concentrations. Sequence analysis and comparison to a theoretical framework30 indicated the aforementioned conditions yield one to five amino acid mutations per gene. The PCR products were purified by agarose gel electrophoresis and each amplified in four 100-µL PCR reactions containing 1× Taq buffer, 2 mMMgCl2, 1 µM of each primer, 0.2 mM (each) dNTPs, 4 µL of error-prone PCR product (of 40 µL from gel extraction), and 2.5U of Taq DNA polymerase. The reactions were thermally cycled at the same conditions except that 35 cycles were used. Reaction products were concentrated with PelletPaint (Novagen) and resuspended in 1 µL of water.
Plasmid pCT-Fn3 was digested with PstI, BtgI, and BamHI to create linearized vector pCT-Fn3-Gene with the entire Fn3 gene removed. Plasmid pCT-Fn3 was digested with BclI, BtgI, and PasI to create linearized vector pCT-Fn3-Loop with the wild-type gene removed from the BC loop through the FG loop.
Electrocompetent EBY100 (100 µL) was combined with 0.5–2.0 µg of pCT-Fn3-Gene and the gene-based PCR product and electroporated at 0.54 kV and 25 µF in a 2 mM electroporation cuvette. Similarly, 100 µL of electrocompetent EBY100 was combined with 0.5–2.0 µg of pCT-Fn3-Loop and the three loop-based PCR products and electroporated. Homologous recombination of the linearized vector and mutagenized insert(s) yielded intact plasmid. Cells were grown in YPD for 1 h at 30 °C, 250 rpm. The medium was switched to SD-CAA to enable selective propagation of successful transformants via growth at 30 °C, 250 rpm, for 24–48 h.
DNA sequencing
Plasmid DNA was isolated using the Zymoprep kit II, cleaned using the Qiagen PCR Purification kit, and transformed into DH5α (Invitrogen) or XL1-Blue E. coli (Stratagene, La Jolla, CA). Individual clones were grown, miniprepped, and sequenced using BigDye chemistry on an Applied Biosystems 3730.
Measurement of Kd, kon, and koff
The equilibrium dissociation constant for a clone was determined essentially as described.26 Briefly, yeast containing the plasmid for an Fn3 clone was grown and induced as for FACS selection. Cells were washed in 1 mL PBSA and resuspended in PBSA containing biotinylated lysozyme in concentrations generally spanning 4 orders of magnitude surrounding the equilibrium dissociation constant. The numbers of cells and sample volumes were selected to ensure excess lysozyme relative to Fn3. For clones of low picomolar affinity, this criterion necessitates very low cell density, which makes cell collection by centrifugation procedurally difficult. To obviate this difficulty, uninduced cells are added to the sample to enable effective cell pelleting during centrifugation with no effect on lysozyme binding of the Fn3-displaying induced cells. Cells were incubated at 25 °C for sufficient time to ensure that the approach to equilibrium was at least 98% complete. Cells were then pelleted, washed with 1 mL PBSA, and incubated in PBSA with 10 mg/L streptavidin–R-phycoerythrin for 10–30 min. Cells were washed and resuspended with PBSA and analyzed with an Epics XL flow cytometer. The minimum and maximum fluorescence and the Kd value were determined by minimizing the sum of squared errors.
For determination of the dissociation constant, koff, clonal cell cultures were grown, induced, and washed as above. Cells were incubated in PBSA with a saturating concentration of biotinylated lysozyme at 25 °C. At various times, an aliquot of cells was washed with PBSA with excess unbiotinylated lysozyme, resuspended in PBSA with excess unbiotinylated lysozyme, and incubated at 25 °C. Simultaneously, all samples of differing dissociation times were washed with PBSA and incubated in 10 mg/L streptavidin–R-phycoerythrin for 10–30 min. Cells were washed and resuspended with PBSA and analyzed with an Epics XL flow cytometer. The minimum and maximum fluorescence and the koff value were determined by minimizing the sum of squared errors.
For determination of the association constants, kon, clonal cell cultures were grown, induced, and washed as above. At various times, an aliquot of cells was resuspended in biotinylated lysozyme and incubated at 25 °C. Simultaneously, all samples of differing association times were washed with PBSA with excess unbiotinylated lysozyme and incubated in PBSA with 10 mg/L streptavidin–R-phycoerythrin for 10–30 min. Cells were washed and resuspended with PBSA and analyzed with an Epics XL flow cytometer. The maximum fluorescence and kon were determined by minimizing the sum of squared errors assuming a 1:1 binding model. The experimentally determined value of koff was used to determine the effective association rate, kon[lysozyme]+koff.
The equilibrium dissociation constant was also determined for the soluble forms of L7.5.1 and Cons0.4.1 by equilibrium competition titration. Varying concentration of purified Fn3 domains were incubated with 20 pM biotiny-lated lysozyme in 50 mL of PBSA. Yeast displaying L7.5.1 were added and incubated for 7 days to near equilibrium. Cells were then pelleted, washed with 1 mL PBSA, and incubated in PBSA with 10 mg/L streptavidin–R-phycoerythrin for 15 min. Cells were washed and resuspended with PBSA and analyzed with an Epics XL flow cytometer. A two-state binding model was assumed and the minimum and maximum fluorescence and equilibrium dissociation constant were determined by minimizing the sum of squared errors.
Fn3 production and biophysical characterization
Fn3 clones were produced as previously described.6 Briefly, BL21(DE3)pLysS E. coli (Invitrogen) containing the pET-24b-based Fn3 plasmid were grown in Luria–Bertani medium with 50 µg/mL kanamycin and 34 µg/mL chloramphenicol at 37 °C, 250 rpm, to an A600 of 0.1–0.2 and induced with 0.5 mM IPTG for 18–24 h at 30 °C, 250 rpm. Cells were lysed by sonication and the insoluble fraction was removed by centrifugation at 19,000g for 40 min. His6-tagged Fn3 was purified from the soluble fraction with TALON Superflow Metal Affinity Resin (Clontech), dialyzed against PBS, and concentrated to 0.5 mL with an Amicon Ultra centrifugal filter (Millipore).
The oligomeric state was analyzed by size-exclusion chromatography on a Superdex 75 HR10/300 column (Amersham Pharmacia Biotech, Piscataway, NJ). Monomer was isolated for biophysical analysis. PBS standards or monomeric protein in PBS was thermally denatured from 25 to 95 °C at a rate of 1 °C/min in a differential scanning calorimeter (VP-DSC, MicroCal). Irreversible aggregation of L7.5.1 occurs at high temperatures. The midpoint of thermal denaturation for this clone is identified as the temperature of maximum heat capacity. Samples were dialyzed in 10 mM sodium phosphate buffer, pH 7.0, and diluted to 8–10 µM for far-UV circular dichroism analysis. Ellipticity was measured from 250 to 190 nm on an Aviv 202 spectrometer (Aviv Biomedical, Lakewood, NJ) with a quartz cuvette with a 1-mm path length (New Era, Vineland, NJ). Thermal denaturation was conducted by measuring ellipticity at 216 nm from 25 to 95 °C and calculating Tm from a standard two-state unfolding curve.
Thermal stabilities were also determined using a yeast surface display thermal denaturation assay derived from Orr et al.31 Fn3 was displayed on the yeast surface as for measurement of kinetic and equilibrium binding constants. Cells were washed and resuspended with PBSA, incubated at 20–85 °C for 30 min, and incubated on ice for 5 min. Biotinylated lysozyme was added at a saturating concentration (e.g., 20 nM for L7.5.1) and mouse anti-c-myc antibody was added at 40 mg/L and incubated on ice for 20 min. Cells were washed and incubated in PBSA with 10 mg/L streptavidin–R-phycoerythrin and 25 mg/L AlexaFluor488 conjugated goat anti-mouse antibody. Cells were washed and resuspended in PBSA and analyzed on an Epics XL flow cytometer. The minimum and maximum fluorescence (Fmin and Fmax, respectively), the T1/2, and the enthalpy of unfolding at T1/2 (ΔHm) were determined by minimizing the sum of squared errors between experimental data and theoretical values according to a two-state unfolding equation:
L7.5.1 reversion clone construction
Reversion of engineered loops of L7.5.1 to wild-type sequence was accomplished by annealing and extending two PCR products created with a gene-terminal primer and a primer that annealed adjacent to the loop of interest but was extended to include wild-type sequence. Specifically, one PCR reaction contained a 5′ gene terminal primer and a primer that annealed to the 25 nucleotides immediately 5′ of the loop of interest but included a nonannealing ‘tail’ encoding for the wild-type loop sequence. In parallel, PCR was performed with a 3′ gene terminal primer and a primer annealing to the 25 nucleotides immediately 3′ of the loop and including a nonannealing tail. The first PCR product encodes from the start of the gene to the loop of interest and the second PCR product encodes from the loop of interest to the end of the gene. These two products are annealed and extended to yield the full Fn3 gene containing the wild-type sequence in the loop of interest. Framework reversions were introduced by standard site-directed mutagenesis using the QuikChange Mutagenesis Kit (Stratagene) according to the manufacturer′s instructions. Clone construction was verified by DNA sequencing.
Focused library construction
The DE randomization library was created in a manner similar to that of the L7.5.1 loop reversion clones. One PCR amplified the L7.5.1 S15P gene fragment 5′ of the DNA encoding for the DE loop. A second PCR amplified the gene 3′ of the DNA encoding the DE loop using a primer that included a degenerate (NNB) DE loop sequence and 20 nucleotides of overlap with the other PCR product. The PCR products were annealed, extended to produce the full gene, and amplified. This process was conducted independently with four oligonucleotides encoding the four different DE loop lengths. The gene fragments were electroporated into electrocompetent EBY100 along with pCT-Fn3-Loop vector. The resulting library encoded for L7.5.1 S15P with a fully random DE loop of length 4, 5, 6, or 7 amino acids.
A library randomizing the unconserved residues of clones similar to L7.5.1 was constructed by PCR of L7.5.1 S15P The 5′ primer contained 17 nucleotides 5′ of the BC loop, 21 nucleotides to encode the BC loop, and 22 nucleotides to anneal 3′ of the BC loop. The 3′ primer contained 19 nucleotides 3′ of the FG loop, 30 nucleotides to encode for the FG loop, and 10 nucleotides 5′ of the FG loop (note that the nucleotides encoding the first three conserved amino acids of the FG loop also enable annealing during PCR). The PCR products were amplified with extended primers to increase the length of the conserved sequence flanking the loops to improve homologous recombination. The gene fragments were electroporated into electrocompetent EBY100 along with pCT-Fn3-Loop vector. Two versions of the BC and FG loops were included. One oligonucleotide completely randomized the unconserved residues using NNB degeneracy (BC, RXXPWAX; FG, RVGRXXXXXG). The other oligonucleotide restricted diversity to amino acids observed during affinity maturation [BC, R(D/G)(C/H/R/ Y)PWA(I/T); FG, RVG(R/W)(A/M/T/V)(F/L/P/S)(C/D/ G/Y)(A/T)(L/P/S)(G/S)].
Supplementary Material
Acknowledgements
The research was funded by CA96504, CA101830, a National Defense Science and Engineering Graduate Fellowship and a National Science Foundation Graduate Fellowship. Assistance from the MIT Flow Cytometry Core Facility is greatly appreciated. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and NIH GM68762) is gratefully acknowledged.
Abbreviations used
- CDR
complementarity-determining region
- DSC
differential scanning calorimetry
- FACS
fluorescence-activated cell sorting
- Fn3
10th type III domain of human fibronectin
- PBSA
phosphate-buffered saline with bovine serum albumin
Footnotes
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2008.06.051
References
- 1.Huang J, Koide A, Nettle KW, Greene GL, Koide S. Conformation-specific affinity purification of proteins using engineered binding proteins: application to the estrogen receptor. Protein Expression Purif. 2006;47:348–354. doi: 10.1016/j.pep.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Karatan E, Merguerian M, Han ZH, Scholle MD, Koide S, Kay BK. Molecular recognition properties of FN3 monobodies that bind the Src SH3 domain. Chem. Biol. 2004;11:835–844. doi: 10.1016/j.chembiol.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc. Natl Acad. Sci. USA. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koide A, Bailey CW, Huang XL, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 5.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl Acad. Sci. USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipovsek D, Lippow SM, Hackel BJ, Gregson MW, Cheng P, Kapila A, Wittrup KD. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J. Mol. Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Parker MH, Chen Y, Danehy F, Dufu K, Ekstrom J, Getmanova E, et al. Antibody mimics based on human fibronectin type three domain engineered for thermostability and high-affinity binding to vascular endothelial growth factor receptor two. Protein Eng. Des. Sel. 2005;18:435–444. doi: 10.1093/protein/gzi050. [DOI] [PubMed] [Google Scholar]
- 8.Richards J, Miller M, Abend J, Koide A, Koide S, Dewhurst S. Engineered fibronectin type III domain with a RGDWXE sequence binds with enhanced affinity and specificity to human alpha v beta 3 integrin. J. Mol. Biol. 2003;326:1475–1488. doi: 10.1016/s0022-2836(03)00082-2. [DOI] [PubMed] [Google Scholar]
- 9.Xu LH, Aha P, Gu K, Kuimelis RG, Kurz M, Lam T, et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 10.Rock E, Sibbald P, Davis M, Chien Y. CDR3 length in antigen-specific immune receptors. J. Exp. Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlin M, Borrebaeck CAK. Characteristics of human antibody repertoires following active immune responses in vivo. Mol. Immunol. 1996;33:583–592. doi: 10.1016/0161-5890(96)00018-1. [DOI] [PubMed] [Google Scholar]
- 12.Lamminmaki U, Pauperio S, Westerlund-Karlsson A, Karvinen J, Virtanen PL, Lovgren T, Saviranta P. Expanding the conformational diversity by random insertions to CDRH2 results in improved anti-estradiol antibodies. J. Mol. Biol. 1999;291:589–602. doi: 10.1006/jmbi.1999.2981. [DOI] [PubMed] [Google Scholar]
- 13.Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High-affinity human antibodies from phage-displayed synthetic fab libraries with a single framework scaffold. J. Mol. Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 14.Batori V, Koide A, Koide S. Exploring the potential of the monobody scaffold: effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 2002;15:1015–1020. doi: 10.1093/protein/15.12.1015. [DOI] [PubMed] [Google Scholar]
- 15.VanAntwerp JJ, Wittrup KD. Fine affinity discrimination by yeast surface display and flow cytometry. Biotechnol. Prog. 2000;16:31–37. doi: 10.1021/bp990133s. [DOI] [PubMed] [Google Scholar]
- 16.Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng. Des. Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- 17.Ling MM. Large antibody display libraries for isolation of high-affinity antibodies. Comb. Chem. High Throughput Screening. 2003;6:421–432. doi: 10.2174/138620703106298608. [DOI] [PubMed] [Google Scholar]
- 18.Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindqvist E, Schier R, et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc. Natl Acad. Sci. USA. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanes J, Schaffitzel C, Knappik A, Pluckthun A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat. Biotechnol. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 20.Stemmer WPC. Rapid evolution of a protein in-vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 21.Jirholt P, Ohlin M, Borrebaeck CAK, Soderlind E. Exploiting sequence space: shuffling in vivo formed complementarity determining regions into a master framework. Gene. 1998;215:471–476. doi: 10.1016/s0378-1119(98)00317-5. [DOI] [PubMed] [Google Scholar]
- 22.Marks JD, Griffiths AD, Malmqvist M, Clackson TP, Bye JM, Winter G. Bypassing immunization—building high-affinity human-antibodies by chain shuffling. Bio/Technology. 1992;10:779–783. doi: 10.1038/nbt0792-779. [DOI] [PubMed] [Google Scholar]
- 23.Barbas CF, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry RM, et al. In-vitro evolution of a neutralizing human-antibody to human-immunodeficiency-virus type-1 to enhance affinity and broaden strain cross-reactivity. Proc. Natl Acad. Sci. USA. 1994;91:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 25.Main AL, Harvey TS, Baron M, Boyd J, Campbell ID. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 26.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat. Protocols. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 27.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 28.Kowalski JM, Parekh RN, Mao J, Wittrup KD. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J. Biol. Chem. 1998;273:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- 29.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 30.Moore GL, Maranas CD. Modeling DNA mutation and recombination for directed evolution experiments. J. Theor. Biol. 2000;205:483–503. doi: 10.1006/jtbi.2000.2082. [DOI] [PubMed] [Google Scholar]
- 31.Orr BA, Carr LM, Wittrup KD, Roy EJ, Kranz DM. Rapid method for measuring ScFv thermal stability by yeast surface display. Biotechnol. Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.