Abstract
Stannous chloride (SnCl2) and UVA induce DNA lesions through ROS. The aim of this work was to study the toxicity induced by UVA preillumination, followed by SnCl2 treatment. E. coli BER mutants were used to identify genes which could play a role in DNA lesion repair generated by these agents. The survival assays showed (i) The nfo mutant was the most sensitive to SnCl2; (ii) lethal synergistic effect was observed after UVA pre-illumination, plus SnCl2 incubation, the nfo mutant being the most sensitive; (iii) wild type and nfo mutants, transformed with pBW21 plasmid (nfo+) had their survival increased following treatments. The alkaline agarose gel electrophoresis assays pointed that (i) UVA induced DNA breaks and fpg mutant was the most sensitive; (ii) SnCl2-induced DNA strand breaks were higher than those from UVA and nfo mutant had the slowest repair kinetics; (iii) UVA + SnCl2 promoted an increase in DNA breaks than SnCl2 and, again, nfo mutant displayed the slowest repair kinetics. In summary, Nfo protects E. coli cells against damage induced by SnCl2 and UVA + SnCl2.
1. Introduction
Numerous agents, from endogenous and exogenous sources, can induce DNA damage in cell genome. At times, the lesion induction mechanisms of these agents are mediated by reactive oxygen species (ROS) production. These compounds can play a role in the initial events involved in aging, mutagenesis, and cancer [1, 2].
Stannous salts, used in different areas of human daily life, and ultraviolet radiation A (UVA or near-UV light—320–400 nm), a fraction from solar light that reaches the Earth surface, are examples of exogenous agents which produce lesions in DNA by ROS generation. Different forms of stannous salts are used in food industry [3–6], but major concern lies in its inorganic form such as stannous chloride (SnCl2), since this is widely used in nuclear medicine as a reducing agent in the labeling of molecules and cells with technetium-99 m [7, 8]. In this case, SnCl2 is intravenously administered to the patient [8].
Several authors have been demonstrated the cytotoxic, genotoxic, and mutagenic effects of SnCl2 and UVA. As to SnCl2, it was described that this salt is able to produce ROS in a Fenton-like reaction, inducing lethality in Escherichia coli (E. coli) [9], cytotoxicity and genotoxicity in eukaryotic cells [10], and genotoxicity in plasmid DNA [11–13]. Mutagenic properties were also reported to SnCl2, being 8-hydroxyguanine (8-oxodGuo) the predominant lesion, accountable for G:C-T:A transversions [14], as well as mutagenicity in yeast and bacteria [15]. Concerning UVA, ROS production was observed in bacteria [16], calf thymus DNA, and HeLa cells [17]. In 1986, Caldeira de Araujo and Favre [18], demonstrated, by using the SOS chromotest assay, that this radiation was able to induce SOS functions in E. coli, suggesting that this agent had mutagenic properties. Recent works have pointed out that UVA mutagenic effects are associated to 8-oxodGuo production [17, 19, 20]. Another phenomenon observed by Favre and coworkers, in 1985 [21], was that UVA sublethal doses were able to induce growth delay effect in E. coli.
In addition to many papers pointing toward ROS production by SnCl2 and UVA, some researchers have demonstrated that these agents are also able to induce a direct effect on DNA. For instance, De Mattos and collaborators [22] suggested that stannous ions were able to bind to DNA, inducing ROS formation very close to this target; and Courdavault et al. [23], upon studying human fibroblasts and keratinocytes submitted to UVA, described production of thymine cyclobutane dimer (CPD) level higher than both 8-oxodGuo and single strand breaks. Similar results were found in Chinese hamster ovary [24] and in rodent cells [25].
Different authors have pointed to base excision repair (BER) as the most important system capable of repairing oxidative damage from broad oxidative sources [26, 27]. Thus, a number of studies have been pointed to the participation of BER enzymes in lesion repair induced by SnCl2 [14, 28] and UVA [29–32]. In BER pathway, the injured base excision is initiated by the action of a class of DNA repair enzymes called DNA glycosylases, which recognize oxidative lesions and sever these from DNA. The initial enzymatic event during BER produces abasic (AP) sites in DNA. The removal of AP sites is initiated by a second class of BER enzymes called AP endonucleases. AP sites are extremely toxic to cells and can be generated in DNA during normal aerobic metabolism or by exogenous agents, such as ionizing radiation, UV light, and hydrogen peroxide (H2O2) [26].
The XthA protein accounts for 90% of AP endonucleolytic activity found in E. coli, being an important member of the BER mechanism [26]. The E. coli xthA mutants are sensitive to oxidative lesions produced by agents such as near-UV and H2O2 [33]. Most of the residual AP endonucleolytic activity present in xthA mutants is encoded by the nfo gene [34]. The endonuclease IV (Nfo protein), codified by nfo gene, recognizes and cleaves DNA in a similar way to exonuclease III [35], albeit some authors suggest that the Nfo enzyme is able to recognize lesions not recognized by exonuclease III [36]. Moreover, endonuclease IV is the only BER protein which, besides existing constitutively, can have its expression increased by the SoxRS system [37]. It has recently been described that endonuclease IV plays a role in an alternative pathway to classic BER, called nucleotide incision repair (NIR) [38], in which it cleaves DNA generating terminus which constitutes the DNA polymerase target. Thus, this pathway has the advantage of avoiding the genotoxic intermediates generated in BER mechanism [38].
Based on the above, we decided to investigate cytotoxicity and genotoxicity induced by the pretreatment of UVA followed by SnCl2 incubation in E. coli BER mutants. Cytotoxicity was assessed through bacterial survival assays, and genotoxicity was studied by using alkaline gel electrophoresis, described by De Mattos et al. 2008 [39].
2. Material and Methods
2.1. Chemicals
Stannous chloride (SnCl2·2H2O) and sodium dodecyl sulfate (SDS) were acquired from Sigma Chemical Co. (USA). Sodium chloride and ethylene diaminetetracetic acid (EDTA) were obtained from Lafan (Brazil). Sodium hydroxide (NaOH) and Tris (hydroxymethyl) amino-methane were obtained from Nuclear (Brazil). Normal point agarose (NPA), low-melting point agarose (LMPA) and ethidium bromide were acquired from Invitrogen (Brazil).
2.2. Bacterial Strains
The strains and plasmid used and their relevant genetic characteristics for this work are listed in Table 1.
Table 1.
Escherichia coli strains and plasmid with their relevant genetic characteristics for this study.
| Strains | Relevant genetic characteristics | Source |
|---|---|---|
| AB1157 | Wild type | Our laboratory |
| BH20 | fpg (formamidopirimidine-DNA glicosilase) | Our laboratory |
| BW372 | nth (endonuclease III) | B. Weiss, Atlanta, USA |
| BW527 | nfo (endonuclease IV) | B. Weiss, Atlanta, USA |
| BW9091 | xthA (exonuclease III) | B. Weiss, Atlanta, USA |
| pBW21 | nfo (endonuclease IV) | B. Weiss, Atlanta, USA |
All strains are derivatives of E. coli K-12.
2.3. Growth Medium
Bacterial cells were grown overnight at 37°C, with shaking in LB medium [40]. A starting culture for experiments was taken from overnight samples, and the cells were grown in the same medium up to exponential phase (1-2 × 108 cells/ml); harvested by centrifugation (2.940 × g; 15 min; 4°C), washed twice and suspended in 0.9% NaCl.
2.4. Survival of Escherichia coli
Bacterial cultures in mid-log growth phase, suspended in 0.9% NaCl, were divided in two fractions, each containing 2 ml. One of them was illuminated with UVA (Phillips lamp, 125 W, emitting more than 95% in 365 nm) for 1 hour, at 25 J/m2/s, totaling 90 kJ/m2. During irradiation, a glass filter was used to cut off lower wavelengths. Following, 1 ml of each fraction (illuminated or not) was taken and incubated with SnCl2 (25 μg/ml), during 1 hour, with shaking. After this, it was obtained four samples: control (not treated), treated with SnCl2, UVA-irradiated, and pre-illuminated plus SnCl2 incubation.
Aliquots from each fraction were taken every 20 minutes, appropriately diluted in 0.9% NaCl solution and then spread onto LB medium. After 18 hours of incubation at 37°C, the colonies were counted and the number of viable cells (N), obtained for the various incubation times, was matched to that observed for time zero (N0), as a measure of survival fractions. All experiments were done at least three times and data plotted represent the mean values with their respective standard deviations. Untreated controls did not show significant variation (P > .05) in cell number along the time (data not shown). Data collected were analyzed by ANOVA, followed by the Tukey's multiple comparison test, using GraphPad Prism software, version 4.0, with a 5% significance level. Before ANOVA statistical analysis, the normal distribution of data was ensured by Kolgomorov-Smirnov test.
2.5. Alkaline Gel Electrophoresis
All E. coli strains assayed with this methodology were prepared as described above, in survival experiments. Being the volume of cultures increased to 6 ml and the SnCl2-treatment time reduced for 40 minutes. Following treatments, 1.5 ml from each fraction was centrifuged and the protocol described by De Mattos et al. 2008 [39] was done with the aim to detect DNA strand breaks induced by SnCl2, UVA, and pre-illumination with UVA, followed by SnCl2 incubation. The E. coli residual fractions (4.5 ml) were centrifuged, suspended in fresh LB medium and incubated with shaking for 90 minutes, at 37°C. Aliquot (1.5 ml) from each sample was taken every 30 minutes, in order to verify the lesions repair induced by the agents used in this work. Briefly, collected aliquots were centrifuged, suspended in cold TE and mixed with low melting point agarose. The mixture was placed in acrylic container to form squared blocks. They were incubated in lysing solution and placed in alkaline normal agarose gel. It was submitted to electrophoresis, neutralized, and then stained in ethidium bromide. Alkaline electrophoresis assays were carried out at least three times to confirm the obtained data and the best gels were selected. The images obtained from alkaline electrophoresis were analyzed using the software Image J 1.33u version to quantify the DNA strand breaks. The collected data were submitted to chi-square test (χ2), adopting 5% as significance level.
3. Results and Discussion
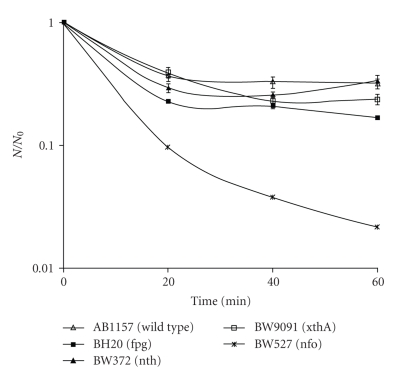
Survival experiments have shown, for all strains tested, that the UV dose used in this work (90 kJ/m2) was not able to induce bacterial inactivation, being considered sublethal (data not shown). Experiments carried out with SnCl2 (25 μg/mL) have shown no significant differences (P > .05) among inactivation levels produced in AB1157 strain (wild type), BW9091 (xthA), and BW372 (nth) mutants. However, a significant difference (P < .001) was observed among wild type, BW527 (nfo) and BH20 (fpg) strains (Figure 1), being nfo mutant the most sensitive. These results indicate that endonuclease IV and Fpg proteins must be important in the repair of SnCl2-induced lesions. A significant difference (P < .001) can be observed, in the same figure, between fpg and nth mutants. This result was expected, as Friedberg and coworkers, 2006 [26] reported that the Fpg enzyme recognizes oxidative lesions, such as 8-oxodGuo. Moreover, SnCl2 may be able to induce this kind of damage, as supported by Cabral et al. 1998 [14]. It can also be ascertained, in the same figure, that endonuclease IV enzyme could be more important than xthA gene product. This assertion is based on the fact that, in our experimental conditions, BW9091 was less sensitive than BW527 to the treatment, which could be due to nfo gene product performance that works in the first one.
Figure 1.
Survival fraction of E. coli AB1157, BH20, BW372, BW9091, and BW527 treated with SnCl2. Mid-log phase cells suspended in 0.9% NaCl were treated with 25 μg/mL SnCl2 for different incubation times and the specific survival fraction. Survival fraction values are means ± standard deviations of the three isolated experiments.
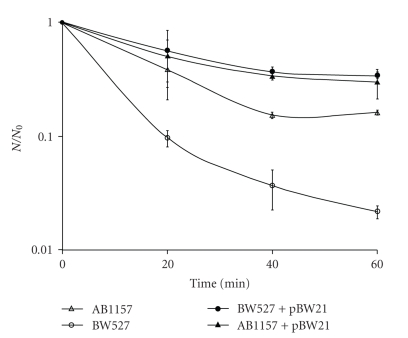
Regarding the UVA pre-treatment, followed by SnCl2 incubation, it was found that this association produced a synergistic lethal effect in all strains tested, when compared to SnCl2 alone (P < .001). Figure 2 presents the results obtained with wild type (AB1157) strain and BH20, BW372, BW9091, and BW527 mutants, pre-illuminated with UVA, followed by SnCl2 incubation. In this case, a significant difference (P < .001) was observed among these. As it had occurred with the SnCl2 treatment, BW527 (nfo) mutant had the lowest survival fraction in response to the association of UVA with SnCl2. This finding corroborates, furthermore, that the product of nfo gene plays an important role in the repair of the damage induced, not only by SnCl2, but also by the association of this agent with UVA. In these treatment conditions, BH20 (fpg) strain was more sensitive than BW372 (nth) mutant, suggesting, again, that Fpg enzyme is more efficient than Nth to recognize and remove lesions produced by the association of both agents.
Figure 2.
Survival fraction of E. coli AB1157, BH20, BW372, BW9091, and BW527 treated with UVA and SnCl2. Mid-log phase cells suspended in 0.9% NaCl were illuminated with 90 kJ/m2 UVA, followed by SnCl2 (25 μg/mL), for different incubation times and the specific survival fractions. Survival fraction values are means ± standard deviations of the three isolated experiments.
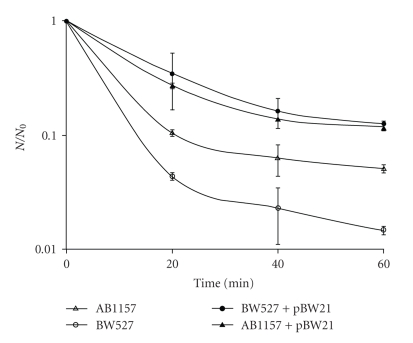
In order to ascertain the actual role of endonuclease IV in the E. coli survival to SnCl2 and UVA + SnCl2 treatments, BW527 (nfo) strain was transformed with pBW21 plasmid carrying the nfo gene, and submitted to bacterial inactivation experiments. The wild type strain (AB1157) was also transformed with pBW21 plasmid. The results demonstrated, for SnCl2 treatment, that BW527 + pBW21 presented the same inactivation level (P > .05) of AB1157 + pBW21, whereas a significant difference was obtained between these strains and BW527 (P < .001) (Figure 3). Similar results were obtained when the strains were pre-illuminated with UVA, and then incubated with SnCl2 (Figure 4). So, this data demonstrated that Nfo enzyme accounts for E. coli survival and reinforces the fact that the lesions induced by SnCl2 and by its association with UVA constitute Nfo protein targets.
Figure 3.
Survival fraction of E. coli AB1157, AB1157+pBW21, BW527, and BW527 + pBW21 treated with SnCl2. Mid-log phase cells suspended in 0.9% NaCl were treated with 25 μg/mL SnCl2 for different incubation times and the specific survival fraction. Survival fraction values are means ± standard deviations of the three isolated experiments.
Figure 4.
Survival fraction of E. coli AB1157, AB1157+pBW21, BW527, and BW527 + pBW21 treated with UVA and SnCl2. Mid-log phase cells suspended in 0.9% NaCl were illuminated with 90 kJ/m2 UVA, followed by SnCl2 (25 μg/mL), for different incubation times and the specific survival fractions. Survival fraction values are means ± standard deviations of the three isolated experiments.
The results found in this work are similar to those obtained by Levin and Demple 1996 [41] when studying the bleomicine effect on wild type (AB1157) and nfo mutant (BW527) E. coli cells. These authors introduced a plasmid DNA containing the nfo gene in AB1157 strain and observed that this strain was able to repair the bleomicine-induced lesions. So, it was shown that Nfo overexpression, in vivo, contributes to the elimination of bleomicine-induced DNA lesions. In this manner, it was clear that endonuclease IV would be more important than exonuclease III in bleomicine-induced damage repair, since its level is the same as in the nfo mutant and in the strain overexpressing Nfo protein.
To better understand the damage and repair mechanisms induced by the agents used in this work, alkaline gel electrophoresis was assayed to detect strand breaks in bacterial genome. Results obtained with this procedure have shown that SnCl2, UVA, and its association with SnCl2, in pre-illumination condition, were able to produce DNA strand breaks in all tested strains. Furthermore, the UVA association with SnCl2 generated a higher number of DNA strand breaks, when compared to SnCl2 treatment (P < .0001).
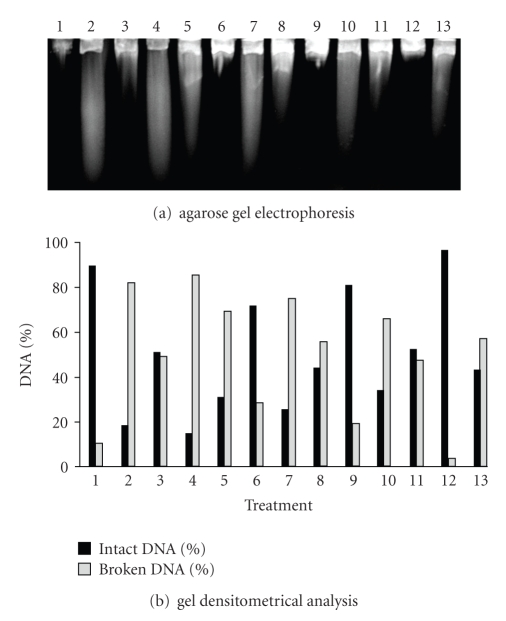
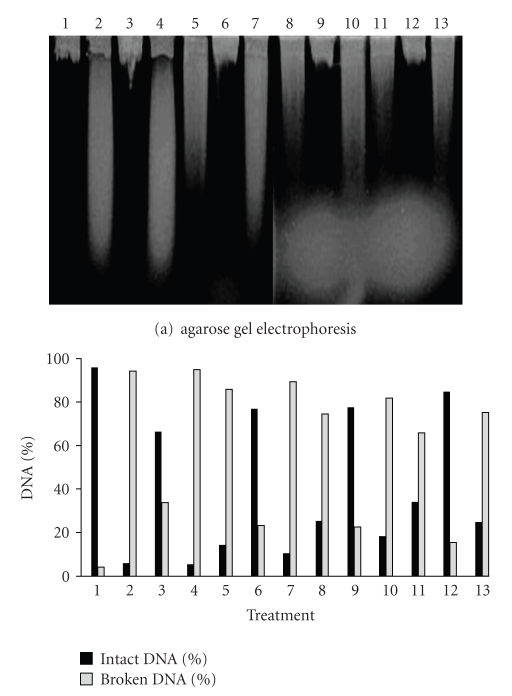
The gel densitometrical analysis revealed, for UVA treatment, that AB1157 (wild type) was the strain presenting the highest efficiency in lesions-induced repair by this radiation (Figure 5). This result was expected, since this strain possesses all active repair mechanisms.
Figure 5.
Alkaline gel electrophoresis of E. coli AB1157 treated with SnCl2 (25 μg/mL), UVA (90 kJ/m2) and pre-illuminated with UVA, followed by SnCl2 incubation. Lanes: (1) control; (2) SnCl2; (3) UVA; (4) UVA + SnCl2; (5) SnCl2 30 minutes; (6) UVA 30 minutes; (7) UVA + SnCl2 30 minutes; (8) SnCl2 60 minutes; (9) UVA 60 minutes; (10) UVA + SnCl2 60 minutes; (11) SnCl2 90 minutes; (12) UVA 90 minutes; (13) UVA + SnCl2 90 minutes.
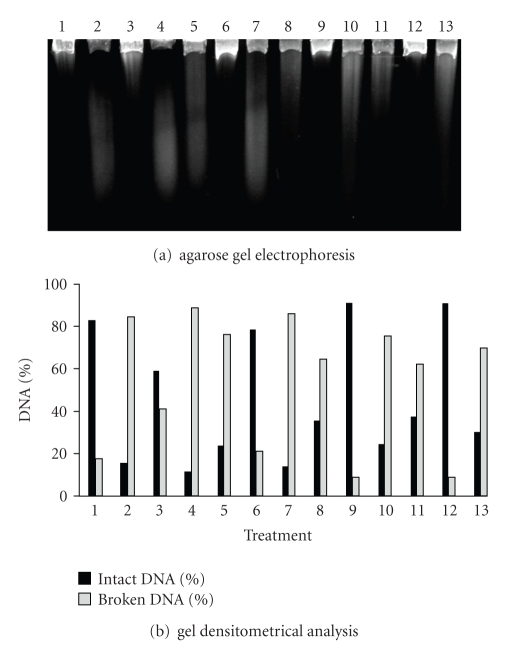
When comparing the simple mutants (BH20, BW372, BW527, and BW 9091), it was observed that BH20 (fpg) strain showed the slowest DNA strand break repair kinetics (Figure 6). This result agrees with the literature, since the fpg gene product is able to recognize 8-oxodGuo lesions [26, 27, 42]. As exposed previously, UVA radiation can induce ROS generation which can lead to 8-oxodGuo formation [43, 44]. The BH20 strain display delay in lesion restoration upon a lack of Fpg.
Figure 6.
Alkaline gel electrophoresis of E. coli BH20 treated with SnCl2 (25 μg/mL), UVA (90 kJ/m2) and pre-illuminated with UVA, followed by SnCl2 incubation. Lanes: (1) control; (2) SnCl2; (3) UVA; (4) UVA + SnCl2; (5) SnCl2 30 minutes; (6) UVA 30 minutes; (7) UVA + SnCl2 30 minutes; (8) SnCl2 60 minutes; (9) UVA 60 minutes; (10) UVA + SnCl2 60 minutes; (11) SnCl2 90 minutes; (12) UVA 90 minutes; (13) UVA + SnCl2 90 minutes.
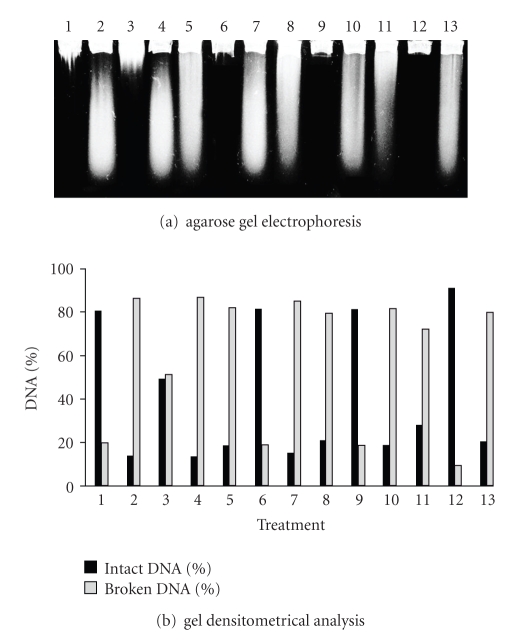
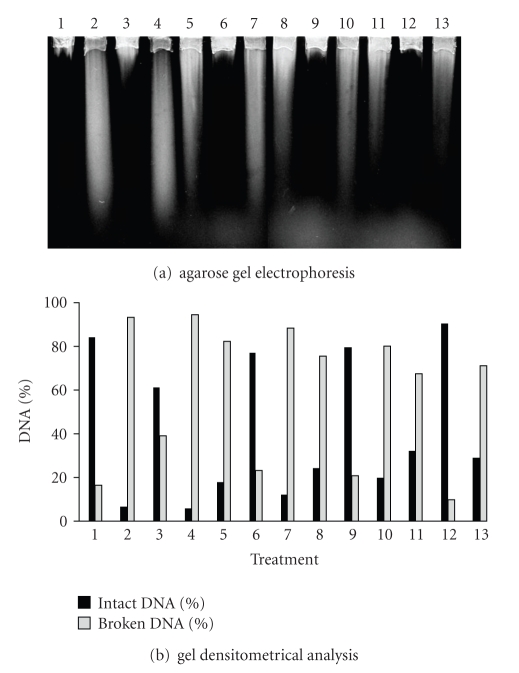
The BW372 (Figure 7), BW527 (Figure 8), and BW9091 (Figure 9) strains were statistically identical, regarding UVA-induced lesion repair capacity. However, these mutants were more efficient in lesion restoration than BH20 (P < .05), a fact which can be explained by Fpg protein functioning. This data supports the hypothesis that Fpg enzyme could be important in recognizing UVA-induced damages.
Figure 7.
Alkaline gel electrophoresis of E. coli BW372 treated with SnCl2 (25 μg/mL), UVA (90 kJ/m2) and pre-illuminated with UVA, followed by SnCl2 incubation. Lanes: (1) control; (2) SnCl2; (3) UVA; (4) UVA + SnCl2; (5) SnCl2 30 minutes; (6) UVA 30 minutes; (7) UVA + SnCl2 30 minutes; (8) SnCl2 60 minutes; (9) UVA 60 minutes; (10) UVA + SnCl2 60 minutes; (11) SnCl2 90 minutes; (12) UVA 90 minutes; (13) UVA + SnCl2 90 minutes.
Figure 8.
Alkaline gel electrophoresis of E. coli BW527 treated with SnCl2 (25 μg/mL), UVA (90 kJ/m2) and pre-illuminated with UVA, followed by SnCl2 incubation. Lanes: (1) control; (2) SnCl2; (3) UVA; (4) UVA + SnCl2; (5) SnCl2 30 minutes; (6) UVA 30 minutes; (7) UVA + SnCl2 30 minutes; (8) SnCl2 60 minutes; (9) UVA 60 minutes; (10) UVA + SnCl2 60 minutes; (11) SnCl2 90 minutes; (12) UVA 90 minutes; (13) UVA + SnCl2 90 minutes.
Figure 9.
Alkaline gel electrophoresis of E. coli BW9091 treated with SnCl2 (25 μg/mL), UVA (90 kJ/m2), and pre-illuminated with UVA, followed by SnCl2 incubation. Lanes: (1) control; (2) SnCl2; (3) UVA; (4) UVA + SnCl2; (5) SnCl2 30 minutes; (6) UVA 30 minutes; (7) UVA + SnCl2 30 minutes; (8) SnCl2 60 minutes; (9) UVA 60 minutes; (10) UVA + SnCl2 60 minutes; (11) SnCl2 90 minutes; (12) UVA 90 minutes; (13) UVA + SnCl2 90 minutes.
Nevertheless, the results pointed to Nth, Nfo, and XthA protein participation in a given repair step, since the strains deficient in these enzymes were not so efficient, as the wild type strain (AB1157), in lesion repair. In this case, it should be remembered that Nth protein can also attack different residues generated by oxidative agents [26]. In agreement, it was shown here that Fpg is the preferential enzyme in recognizing UVA-induced lesions, since BH20 (fpg) displayed a delay in lesion repair when compared to BW372 (nth). This assertion is based on the fact that, during BH20 repair kinetics, a decrease was not observed in DNA strand break number between the times 30 and 60 minutes of repair (P > .05—Figure 6, lanes 6 and 9). In contrast, in BW372 strain, the maximum repair level was obtained in 60 minutes (Figure 7, lane 9).
The residues left at the lesion site by Nth or Fpg enzymes action need to be processed by AP endonucleases to complete BER repair. These residues constitute exonuclease III protein targets [45], and for this fact, BW9091 strain (xthA) was not able to complete the repair (Figure 9) as efficiently as in the wild type. As described by Levin et al. 1988 [35] endonuclease IV (Nfo) protein can also take part in the ROS lesion-induced repair, since this enzyme possesses a similar substratum to that of exonuclease III [26]. Thus, in the absence endonuclease IV (BW527), restoration was not successfully completed after 90 minutes (Figure 8), when compared with the wild type.
Comparing BW9091 (xthA) and BW527 (nfo) strains, it was observed that the latter was slower in the repair process than the former, since the DNA strand breaks number in BW527 remains the same between the times 30 and 60 minutes (P > .05—Figure 8, lanes 6 and 9). This data could suggest that Nfo enzyme is more important than XthA protein in UVA-induced lesions repair. Such idea was supported by gel densitometrical analysis of double mutants lacking endonuclease IV which were least efficient in the repair (data not shown).
As cited above, the endonuclease IV is the only inducible BER enzyme [37] and the UVA dose employed in this study is known to induce growth delay in E. coli [46]. Thus, this could explain the least efficiency of BH20 (fpg) in UVA-induced lesion repair in that, even possessing Nfo enzyme, its synthesis would be partially inhibited by the growth delay mechanism. Another evidence of growth delay is based on the same behavior presented by BW527 (nfo), BW9091 (xthA), BW372 (nth), and BW434 (xthA-nth—data not shown) strains. The mutants which possess the Nfo protein were not more efficient in repairing the damages than BW527, suggesting that nfo gene translation could be inhibited by the growth delay.
The gel densitometrical analysis revealed, for SnCl2 treatment, that AB1157 (Figure 5) was the strain which repaired the SnCl2-induced lesions with the most efficiency, as expected for a wild type strain.
The simple mutant analysis showed that BW527 (nfo) strain presented the most delay in the SnCl2-lesion-induced repair (Figure 8), being followed by BW9091 (xthA—Figure 9), BH20 (fpg—Figure 6), and BW372 (nth—Figure 7) strains. These results point to the important roles developed by AP endonucleases (Nfo and XthA) in processing AP sites, generated by DNA glicosylases enzymes (Fpg and Nth).
When BH20 (Figure 6) and BW372 (Figure 7) strains were compared, it was observed that BH20 (fpg) showed the least capacity in restoring SnCl2-induced damages. As literature reports, SnCl2 is able to induce 8-oxodGuo lesions [14], which constitute Fpg targets [26, 42]. So, despite the description that endonuclease III (Nth) could recognize and remove oxidative damages [26], the data obtained in this study with BH20 and BW372 mutants suggest that SnCl2 would be producing 8-oxodGuo, thereby reinforcing the results of Cabral et al. 1998 [14]. In addition, Fpg enzyme would also be required in processing this lesion.
The comparison between BW527 (Figure 8) and BW9091 (Figure 9) revealed that the Nfo deficient strain (BW527) remained with the most DNA strand breaks number after repair kinetics, suggesting that Nfo enzyme would play a more important role than XthA in SnCl2-induced damage repair.
Literature data showed that, among BER enzymes, exonuclease III was required to restore SnCl2-induced lesions [14]. However, in this study, this fact was not observed with the same intensity, even when using xthA deleted strain (BW9109—data not shown). Thus, SnCl2 could be producing damages which would be preferentially recognized by endonuclease IV, as occurring with the chemical agents tert-butyl hydroperoxide and bleomicine, which produce lesions not recognized by exonuclease III, but only by endonuclease IV [36].
It is important to emphasize that despite the fact that endonuclease IV represents only 10% of total AP endonuclease activity [34] it is the sole inducible BER protein [37]. Its expression may be increased 20-fold, when treatments with oxidative agents, such as paraquat, which leads to superoxide radical production, are performed [47]. Thus, in this work, it could be suggested that SnCl2 would be inducing an increase in Nfo levels, since this salt is able to produce ROS [9, 11]. Then, upon a lack of Nfo, as occurs in BW527 strain, the cell would find it more difficult to repair SnCl2-induced lesions. It is known that endonuclease IV increase is depedent on SoxRS system induction [37], in order to ascertain whether SnCl2 would be able to induce the SoxRS system and, consequently, endonuclease IV synthesis, SOS chromotest assays were performed. The results obtained were positive (Asad, LMBO, personal communication), corroborating the hypothesis presented in this study.
The Nfo role in SnCl2-damage induced repair was reinforced by double mutant analysis (data not shown), in which the strains lacking Nfo protein showed the slowest repair kinetics, when compared to the simple mutants.
When the bacterial strains were pre-illuminated with UVA, followed by SnCl2 incubation, a higher DNA strand break number was observed than that secured with SnCl2 treatment alone. Once more, the wild type (AB1157) strain presented the highest efficiency in repairing the lesions induced by UVA+SnCl2 association (Figure 5).
As occurring with SnCl2 treatment, BW527 (nfo) strain once more showed the least efficiency in restoring damages produced by UVA plus SnCl2 (Figure 8). Following BW527, was the BH20 (fpg) strain with the lowest ability in repairing UVA + SnCl2-induced lesions (Figure 6) than the other mutants. Moreover, when BH20 was compared to BW372 (nth), the latter showed the smallest DNA strand break number at the repair kinetics end (Figure 7, lane 13). This data suggests that UVA and SnCl2 association could be inducing oxidative damages, such as 8-oxodGuo, as previously described [14, 43, 44] and Fpg enzyme would be preferential in recognizing and removing this damage.
The BW527 (Figure 8) and BW9091 (Figure 9) gel analysis reinforced the idea that endonuclease IV would be more important than exonuclease III in restoring UVA + SnCl2-induced lesions, in that, at the repair kinetics end, BW527 still presented a higher DNA strand break number (Figure 8, lane 13). This evidence was corroborated by double mutant analysis (data not shown) in which endonuclease IV deficient strains showed the least efficiency in repairing the UVA and SnCl2 induced lesions.
The repair efficiency profiles of the strains treated with both agents follow different patterns from those revealed by the only UVA illuminated strains. Thus, even if the pretreatment with this radiation may induce the growth delay phenomenon, the constitutive existence of Nfo protein will still warrant BW372, BW9091, and BH20 strains major efficiency in repairing the lesions induced by UVA and SnCl2 association than that of nfo mutant strain (BW527).
Moreover, in addition to its action on AP sites, endonuclease IV is able to recognize and cleave DNA on the 5′ side of various oxidatively damaged bases via NIR, generating the proper ends for DNA repair synthesis [38]. As demonstrated by Ischenko and colleagues, 2006 [48], NIR is essential for handling lethal oxidative DNA lesions. Therefore, our study also could suggest Nfo participation, via NIR, in removing damages induced by SnCl2 and UVA + SnCl2.
Taken together, our results point to endonuclease IV activity via BER and possible NIR in protecting E. coli cells against damages induced by SnCl2 and UVA + SnCl2.
Acknowledgments
The authors are grateful to Dr Bernard Weiss for kindly providing E. coli strains and plasmid and to Carlos Brown Scavarda for English language support. The study was supported by CAPES and FAPERJ (E-26/152.018/2004; E-26/171.332/2006).
References
- 1.Bayir H. Reactive oxygen species. Critical Care Medicine. 2005;33(12, supplement):S498–S501. doi: 10.1097/01.ccm.0000186787.64500.12. [DOI] [PubMed] [Google Scholar]
- 2.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 4th edition. New York, NY, USA: Oxford University Press; 2007. [Google Scholar]
- 3.Hallas LE, Cooney JJ. Tin and tin resistant microorganisms in Chesapeake Bay. Applied and Environmental Microbiology. 1981;41(2):466–471. doi: 10.1128/aem.41.2.466-471.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLean JRN, Blakey DH, Douglas GR, Kaplan JG. The effect of stannous and stannic (tin) chloride on DNA in Chinese hamster ovary cells. Mutation Research. 1983;119(2):195–201. doi: 10.1016/0165-7992(83)90129-x. [DOI] [PubMed] [Google Scholar]
- 5.Rader JI. Anti-nutritive effects of dietary tin. Advances in Experimental Medicine and Biology. 1991;289:509–524. doi: 10.1007/978-1-4899-2626-5_34. [DOI] [PubMed] [Google Scholar]
- 6.White DJ. A “return” to stannous fluoride dentifrices. Journal of Clinical Dentistry. 1995;6:29–36. [PubMed] [Google Scholar]
- 7.Hladik WB, III, Saha GB, Study KT. Essentials of Nuclear Medicine Science. Baltimore, Md, USA: Williams and Wilkins; 1987. [Google Scholar]
- 8.Saha GB. Fundamentals of Nuclear Pharmacy. New York, NY, USA: Springer; 1992. [Google Scholar]
- 9.Dantas FJS, Moraes MO, Carvalho EF, Valsa JO, Bernardo-Filho M, Caldeira-de-Araujo A. Lethality induced by stannous chloride on Escherichia coli AB1157: participation of reactive oxygen species. Food and Chemical Toxicology. 1996;34(10):959–962. doi: 10.1016/s0278-6915(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 10.Dantas FJS, De Mattos JCP, Moraes MO, et al. Genotoxic effects of stannous chloride (SnCl2) in K562 cell line. Food and Chemical Toxicology. 2002;40(10):1493–1498. doi: 10.1016/s0278-6915(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 11.Caldeira-de-Araujo A, Dantas FJS, Moraes MO, Felzenszwalb I, Bernardo-Filho M. Stannous chloride participates in the generation of reactive oxygen species. Free Radical Research in Latin America. 1996;48(1-2):109–113. [Google Scholar]
- 12.Dantas FJS, Moraes MO, De Mattos JCP, et al. Stannous chloride mediates single strand breaks in plasmid DNA through reactive oxygen species formation. Toxicology Letters. 1999;110(3):129–136. doi: 10.1016/s0378-4274(99)00126-5. [DOI] [PubMed] [Google Scholar]
- 13.De Mattos JCP, Dantas FJS, Bezerra RJAC, et al. Damage induced by stannous chloride in plasmid DNA. Toxicology Letters. 2000;116(1-2):159–163. doi: 10.1016/s0378-4274(00)00213-7. [DOI] [PubMed] [Google Scholar]
- 14.Cabral REC, Leitão AC, Lage C, et al. Mutational potentiality of stannous chloride: an important reducing agent in the Tc-99m-radiopharmaceuticals. Mutation Research. 1998;408(2):129–135. doi: 10.1016/s0921-8777(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 15.Pungartnik C, Viau C, Picada J, Caldeira-de-Araujo A, Henriques JAP, Brendel M. Genotoxicity of stannous chloride in yeast and bacteria. Mutation Research. 2005;583(2):146–157. doi: 10.1016/j.mrgentox.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Eisenstark A. Mutagenic and lethal effects of near-ultraviolet radiation (290–400 nm) on bacteria and phage. Environmental and Molecular Mutagenesis. 1987;10(3):317–337. doi: 10.1002/em.2850100311. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Rosenstein BS, Wang Y, Lebwohl M, Mitchell DM, Wei H. Induction of 8-oxo-7,8-dihydro-2′-deoxyguanosine by ultraviolet radiation in calf thymus DNA and HeLa cells. Photochemistry and Photobiology. 1997;65(1):119–124. doi: 10.1111/j.1751-1097.1997.tb01886.x. [DOI] [PubMed] [Google Scholar]
- 18.Caldeira de Araujo A, Favre A. Near ultraviolet DNA damage induces the SOS responses in Escherichia coli. The EMBO Journal. 1986;5(1):175–179. doi: 10.1002/j.1460-2075.1986.tb04193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozmin S, Slezak G, Reynaud-Angelin A, et al. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(38):13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pouget JP, Douki T, Richard MJ, Cadet J. DNA damage induced in cells by γ and UVA radiation as measured by HPLC/GC-MS and HPLC-EC and comet assay. Chemical Research in Toxicology. 2000;13(7):541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 21.Favre A, Hajnsdorf E, Thiam K, Caldeira de Araujo A. Mutagenesis and growth delay induced in Escherichia coli by near-ultraviolet radiations. Biochimie. 1985;67(3-4):335–342. doi: 10.1016/s0300-9084(85)80076-6. [DOI] [PubMed] [Google Scholar]
- 22.De Mattos JCP, Lage C, Dantas FJS, et al. Interaction of stannous chloride leads to alteration in DNA, triphosphate nucleotides and isolated bases. Molecular and Cellular Biochemistry. 2005;280(1-2):173–179. doi: 10.1007/s11010-005-8908-6. [DOI] [PubMed] [Google Scholar]
- 23.Courdavault S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutation Research. 2004;556(1-2):135–142. doi: 10.1016/j.mrfmmm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42(30):9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 25.Rochette PJ, Therrien JP, Drouin R, et al. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Research. 2003;31(11):2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedberg EC, Walker GC, Sied W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edition. Washington, DC, USA: ASM Press; 2006. [Google Scholar]
- 27.Berti PJ, McCann JAB. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chemical Reviews. 2006;106(2):506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 28.Guedes AP, Cardoso VN, De Mattos JCP, et al. Cytotoxic and genotoxic effects induced by stannous chloride associated to nuclear medicine kits. Nuclear Medicine and Biology. 2006;33(7):915–921. doi: 10.1016/j.nucmedbio.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Shennan MG, Palmer CM, Schellhorn HE. Role of fapy glycosylase and UvrABC excinuclease in the repair of UVA (320–400 nm)-mediated DNA damage in Escherichia coli. Photochemistry and Photobiology. 1996;63(1):68–73. doi: 10.1111/j.1751-1097.1996.tb02993.x. [DOI] [PubMed] [Google Scholar]
- 30.Palmer CM, Serafini DM, Schellhorn HE. Near ultraviolet radiation (UVA and UVB) causes a formamidopyrimidine glycosylase-dependent increase in G to T transversions. Photochemistry and Photobiology. 1997;65(3):543–549. doi: 10.1111/j.1751-1097.1997.tb08602.x. [DOI] [PubMed] [Google Scholar]
- 31.Serafini DM, Schellhorn HE. Endonuclease III and endonuclease IV protect Escherichia coli from the lethal and mutagenic effects of near-UV irradiation. Canadian Journal of Microbiology. 1999;45(7):632–637. [PubMed] [Google Scholar]
- 32.Kim KJ, Chakrabarty I, Li GZ, Grösch S, Kaina B, Rünger TM. Modulation of base excision repair alters cellular sensitivity to UVA1 but not to UVB. Photochemistry and Photobiology. 2002;75(5):507–512. doi: 10.1562/0031-8655(2002)075<0507:mobera>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 33.Demple B, Halbrook J, Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. Journal of Bacteriology. 1983;153(2):1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. The Journal of Biological Chemistry. 1977;252(9):2808–2814. [PubMed] [Google Scholar]
- 35.Levin JD, Johnson AW, Demple B. Homogeneous Escherichia coli endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. The Journal of Biological Chemistry. 1988;263(17):8066–8071. [PubMed] [Google Scholar]
- 36.Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. Journal of Bacteriology. 1986;168(3):1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends in Biotechnology. 2001;19(3):109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 38.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415(6868):183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 39.De Mattos JCP, Motta ES, Oliveira MBN, Dantas FJS, Caldeira-de-Araujo A. Alkaline gel electrophoresis assay to detect DNA strand breaks and repair mechanisms in Escherichia coli. Brazilian Archives of Biology and Technology. 2008;51:121–126. [Google Scholar]
- 40.Luria SE, Burrous JW. Hybridization between Escherichia coli and Shigella. Journal of bacteriology. 1957;74(4):461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin JD, Demple B. In vitro detection of endonuclease IV-specific DNA damage formed by bleomycin in vivo. Nucleic Acids Research. 1996;24(5):885–889. doi: 10.1093/nar/24.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aruoma OI, Halliwell B, Dizdaroglu M. Iron ion-dependent modification of bases in DNA by the superoxide radical-generating system hypoxanthine/xanthine oxidase. The Journal of Biological Chemistry. 1989;264(22):13024–13028. [PubMed] [Google Scholar]
- 43.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutation Research. 2005;571(1-2):19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 44.Rünger TM, Kappes UP. Mechanisms of mutation formation with long-wave ultraviolet light (UVA) Photodermatology Photoimmunology & Photomedicine. 2008;24(1):2–10. doi: 10.1111/j.1600-0781.2008.00319.x. [DOI] [PubMed] [Google Scholar]
- 45.Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutation Research. 1990;236(2-3):173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 46.Aragão BR, Caldeira-de-Araujo A, De Mattos JC, Favre A, Alcantara-Gomes R. Near-ultraviolet illumination inhibits the liquid holding recovery in Escherichia coli B. Brazilian Journal of Medical and Biological Research. 1990;23(12):1263–1266. [PubMed] [Google Scholar]
- 47.Chan E, Weiss B. Endonuclease IV of Escherichia coli is induced by paraquat. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(10):3189–3193. doi: 10.1073/pnas.84.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishchenko AA, Deprez E, Wlaksimenko A, Brochon JC, Tauc P, Saparbaev MK. Uncoupling of the base excision and nucleotide incision repair pathways reveals thier respective biological roles. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2564–2569. doi: 10.1073/pnas.0508582103. [DOI] [PMC free article] [PubMed] [Google Scholar]