Abstract
Mutations of the inositol 5′ phosphatase oculocerebrorenal syndrome of Lowe (OCRL) give rise to the congenital X-linked disorders oculocerebrorenal syndrome of Lowe and Dent disease, two conditions giving rise to abnormal kidney proximal tubule reabsorption, and additional nervous system and ocular defects in the case of Lowe syndrome. Here, we identify two closely related endocytic proteins, Ses1 and Ses2, which interact with the ASH-RhoGAP–like (ASPM-SPD-2-Hydin homology and Rho-GTPase Activating Domain-like) domain of OCRL. The interaction is mediated by a short amino acid motif similar to that used by the rab-5 effector APPL1 (Adaptor Protein containing pleckstrin homology [PH] domain, PTB domain and Leucine zipper motif 1) APPL1 for OCRL binding. Ses binding is mutually exclusive with APPL1 binding, and is disrupted by the same missense mutations in the ASH-RhoGAP–like domain that also disrupt APPL1 binding. Like APPL1, Ses1 and -2 are localized on endosomes but reside on different endosomal subpopulations. These findings define a consensus motif (which we have called a phenylalanine and histidine [F&H] motif) for OCRL binding and are consistent with a scenario in which Lowe syndrome and Dent disease result from perturbations at multiple sites within the endocytic pathway.
Keywords: Dent disease, endocytosis, Lowe syndrome
Inositol phospholipids play a critical regulatory function in cell physiology, including membrane traffic. The interconversion of different phosphoinositide species via the action of kinases and phosphatases plays a key role in the maturation and progression of membranes through different stations of the exocytic and endocytic pathways, coordinating these processes with signaling reactions. Thus, the correct spatial and temporal regulation of these enzymes is of central importance (1, 2). As a consequence, not only the lack of these enzymes but also the disruption of their interactions can have profound effects on cell function and result in pathological conditions (2 –9).
Two human diseases are caused by mutations in an enzyme that selectively dephosphorylates the inositol ring at the 5′ position, using phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3], two phosphoinositides concentrated in the plasma membrane, as its preferred substrates (10). One such disease is oculocerebrorenal syndrome of Lowe (hence the enzyme name “OCRL”) (3, 11). This condition, also referred to as “Lowe syndrome,” is a severe X-linked disorder characterized by congenital cataracts (12), kidney readsorption defects caused by proximal tubule dysfunction, cognitive impairment, muscle hypotonia, and autism spectrum behavioral disorders (13, 14). The other condition is Dent disease, an X-linked disorder involving kidney defects very similar to those associated with Lowe syndrome but, for reasons not yet known, few other dysfunctions (15 –19).
OCRL comprises an N-terminal Pleckstrin Homology (PH) domain followed in sequence by a central inositol 5′-phosphatase domain, an ASPM-SPD-2-Hydin (ASH) domain, and a catalytically inactive RhoGAP (GTPase Activating Protein)–like domain (20). OCRL interacts with several endocytic proteins, including clathrin (20 –23), the clathrin adaptor AP2 (21, 24), and several endocytic (e.g., Rab5) (25, 26) and nonendocytic Rab GTPases (20, 26, 27). It has a broad distribution on endosomes (24, 25) and also is present at late-stage clathrin-coated pits (20, 22–24) and in the Golgi complex area (23, 24, 28). Through these localizations OCRL is thought to remove PI(4,5)P2 and PI(3,4,5)P3 from membranes that have been internalized and to prevent inappropriate accumulation of 5′-phosphorylated inositol phospholipids on internal membranes.
Disease-causing mutations occur in all regions of the OCRL coding sequence. Some mutations produce absence of the protein or either truncations or functional disruption of its inositol 5′ phosphatase domain (http://research.nhgri.nih.gov/lowe/). However, other mutations are truncations or missense mutations in the C-terminal ASH-RhoGAP–like module, indicating that this region, which is important for several protein interactions, is critical for function (29, 30) and that aberrant binding properties of missense mutations can compound the deficit caused by reduction in levels of protein expression commonly found in these mutations (17). Importantly, fibroblasts from Lowe syndrome patients display an increase in the lipid substrate PI(4,5)P2, regardless of the kind of mutation (31 –33). This finding supports the idea that not only inositol 5′ phosphatase activity, but also proper regulation and localization of this activity via protein–protein interactions, is required for OCRL to act on specific phosphoinositide pools.
Recent work has identified the Rab5 effector APPL1 (Adaptor protein containing PH domain, PTB domain and Leucine Zipper motif 1) as a ligand of the ASH-RhoGAP–like domain (20, 29). APPL1 resides on a subset of peripheral OCRL-positive endosomes (20, 34) that represent the earliest stations of endocytic traffic for a subset of endocytic vesicles (35). This subset includes vesicles derived from clathrin-coated pits that internalize growth factor receptors and macropinosomes that originate from the activation of growth factor receptors and/or their downstream signaling machinery (35). APPL1 binding was shown to be the only known OCRL interaction that is abolished by several mutations of the ASH-RhoGAP–like domain leading to Lowe syndrome (20, 29), suggesting its key importance to OCRL function. However, APPL1 is present only in the vertebrate lineage, whereas the OCRL ASH-RhoGAP–like module is conserved throughout evolution, and the majority of amino acids required for APPL1 binding are preserved in the Drosophila ASH-RhoGAP–like domain. Here we describe a small, conserved protein, which we named “Ses” (from the word “sesquipedalian,” meaning an unnecessarily long description of a simple thing). Ses, which arises in the invertebrate lineage, occurs as two isoforms in mammals and utilizes the same ligand-binding motif as APPL1 to bind OCRL. We show that Ses proteins reside on an OCRL-positive endocytic compartment downstream of APPL1 endosomes and that the associations of APPL1 and Ses with OCRL are mutually exclusive. These data suggest that patient mutations may affect OCRL function at multiple stations of the endocytic pathway by disturbing interaction with different ligands of the same binding sites in OCRL.
Results
C-Terminal Region of Ses Binds the ASH-RhoGAP-Like Domain of OCRL.
APPL1 is a protein that is unique to the vertebrate lineage. Because of the strong level of evolutionary conservation between the ASH-RhoGAP–like modules of invertebrate and vertebrate OCRL (Fig. S1), including the conservation of those amino acids whose mutations are responsible for Lowe syndrome (29 –31, 36 –38, and this paper), it seemed reasonable to assume that this module originally had evolved to bind proteins other than APPL1. In scanning the literature, it became apparent that one candidate interactor of OCRL had been isolated previously in a genome-wide yeast two-hybrid screen of Drosophila melanogaster proteins (39). This small 26-kDa protein, CG12393, which we named “Ses,” is retained throughout evolution to Homo sapiens. In the same way that the invertebrate OCRL locus duplicates to encode the vertebrate proteins inositol-1,4,5-trisphosphate 5-phosphatase (INPP5B) and OCRL, so does the single invertebrate Ses locus duplicate to encode the vertebrate proteins Ses1/FAM109a and Ses2/FAM109b.
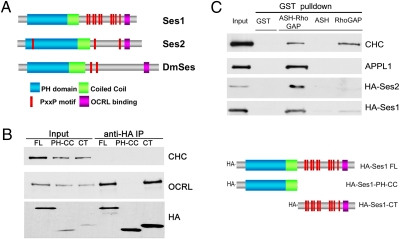
The N-terminal portion of Ses1/2, which comprises a PH domain followed by a predicted coiled-coil domain (Fig. 1A), is well conserved across species and is similar in the two isoforms. The C-terminal portion, which is less conserved, is predicted to be primarily unfolded and to contain multiple putative SRC homology 3 domain-binding proline-rich (PxxP) motifs (Fig. 1A).
Fig. 1.
(A) Domain cartoon of Ses proteins. Vertebrates have two Ses genes, Ses1/FAM109a and Ses2/Fam109b, whereas Drosophila (DmSes, CG12393) and all invertebrates examined to date have a single Ses gene. Ses proteins harbor a PH domain immediately followed by a putative coiled-coil region and then by a predicted unfolded C-terminal portion containing several PxxP motifs and a short highly conserved sequence (OCRL-binding site) close to the C terminus. (B ) The C-terminal portion of Ses1 is necessary and sufficient for OCRL binding. HA-tagged fragments of Ses1 (Right) were overexpressed in Cos7 cells and immunoprecipitated (IP) from cell extracts (Input) with antibodies directed against the HA tag. The Western blots shown (Left) demonstrate that endogenous OCRL coprecipitated with both full-length (FL) Ses1 and the C-terminal (CT) region of Ses1. Clathrin heavy chain (CHC) is used as a negative control. (C) Western blots of GST pulldowns from Cos7 cells expressing either HA-tagged human Ses1 or HA-tagged Ses2. Both HA-Ses1 and HA-Ses2, like endogenous APPL1 (20), strongly interact with a GST fusion of the ASH-RhoGAP–like domain of OCRL but not with the GST-ASH domain alone, the GST-RhoGAP–like domain alone, or GST. Both the ASH-RhoGAP–like domain and the RhoGAP–like domain alone bind clathrin, as expected (20).
To confirm the interaction of Ses1/2 with OCRL and to establish whether this interaction also occurs in primates, coimmunoprecipitation and GST pulldown experiments were performed using extracts of Cos7 cells expressing HA-tagged human Ses1 (Fig. 1 B and C) or HA-Ses2 (Fig. 1C), corresponding to the expressed sequences NM_175474.3 and BC104175, respectively. Anti-HA immunoprecipitates generated from these extracts were enriched for endogenous (monkey) OCRL, as revealed by Western blotting (but not for clathrin, used as a control), supporting conservation of the interaction (Fig. 1B). Binding was mediated by the C-terminal region of Ses1, as shown by analysis of anti-HA immunoprecipitates from cells expressing the HA-tagged N-terminal PH-coiled-coil portion or the C-terminal portion of Ses1, respectively (Fig. 1B). Conversely, GST pulldowns from HA-Ses1 or -Ses2–expressing cells using OCRL fragments as bait showed that HA-tagged Ses1 and Ses2 bound the ASH-RhoGAP–like domain of human OCRL (Fig. 1C). Binding required both the presence of the ASH domain and of the RhoGAP-like domain (Fig. 1C), in agreement with the previous report that these two domains are linked together tightly to form a single functional unit (20).
Consistent with the similarity of the ASH-RhoGAP–like domains of OCRL and INPP5B, Ses1 also was pulled down by the INPP5B ASH-RhoGAP–like domain (Fig. S2).
OCRL and Ses Colocalize on Endosomes.
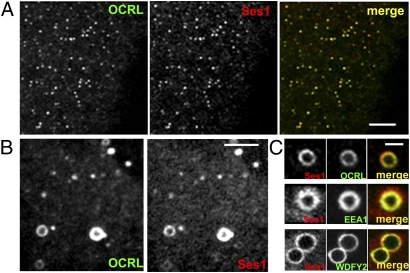
To determine whether the biochemical interaction reflects physical proximity in cells, colocalization of the two proteins was explored by immunofluorescence. GFP-tagged human OCRL and Ses1 or Ses2 N-terminally tagged with the photostable red fluorescent protein variant TagRFP-T (40) were coexpressed in Cos7 cells. Striking colocalization was observed for both Ses1 and Ses2 with small OCRL-positive puncta previously shown to reflect predominantly endosomes (Fig. 2A and Fig. S3) (20, 23). Colocalization also was observed on larger vesicles (Fig. 2B), which also were positive for phosphatidylinositol-3-phosphate (PI3P)-binding endosomal markers such as early endosomal antigen 1 (EEA1) (41) and WD repeat and FYVE domain containing 2 (WDFY2) (35, 42, 43) (Fig. 2C). These organelles may have resulted from the coalescence and fusion of smaller endosomes or from the maturation of macropinosomes, given the known presence of OCRL on these organelles. Accordingly, GFP-OCRL and TagRFP-T-Ses1 also colocalized on macropinosomes generated by cotransfection with the dominantly active Ras, HRasV12G (Fig. S4).
Fig. 2.
Ses1 colocalizes with OCRL on endocytic structures. (A) Spinning disk confocal image showing a section of a Cos7 cell expressing TagRFP-T-Ses1 and GFP-OCRL. (Scale bar, 5 μm.) (B) TagRFP-T-Ses1 and GFP-OCRL also colocalize on macropinosomes. (Scale bar, 4 μm.) (C) TagRFP-T-Ses1–positive macropinosomes are positive for GFP-EEA1 and GFP-WDFY2, two markers of PI3P-positive, Rab5-positive endosomes (35, 41). (Scale bar, 2 μm.)
Ses Binds to the ASH-RhoGAP–Like Domain via a Conserved Amino Acid Motif.
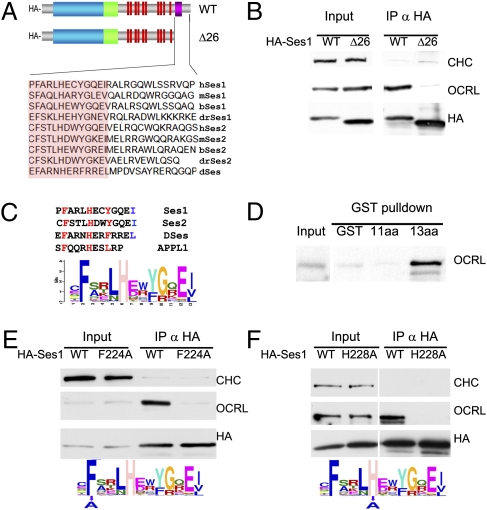
Patient mutations in the ASH-RhoGAP–like domain of OCRL are thought to produce disease by disrupting the interaction of OCRL with other proteins. To understand better the interaction of Ses with OCRL, we further investigated the minimal OCRL-binding region in Ses1 and Ses2. The most heavily conserved amino acid stretch in the poorly conserved C terminus of the Ses proteins corresponds to the C-terminal 26 amino acids (Fig. 3A and Fig. S5). As shown by anti-HA immunoprecipitation from cells expressing the HA-tagged truncated protein, deletion of this region abolished OCRL coprecipitation, suggesting that it was necessary for the interaction (Fig. 3B). Within this region, several amino acids are well conserved in all Ses proteins from Drosophila to man (Fig. 3A). Interestingly, these amino acids define a motif that is reminiscent of the OCRL-binding motif in APPL1 (20), including a phenylalanine essential for this interaction (Fig. 3C).
Fig. 3.
Minimal OCRL-binding sequence of Ses1 and similarity with the minimal OCRL-binding peptide of APPL1. (A) The C-terminal 26 amino acids of human Ses1 contain a sequence (pink box) well conserved among Ses proteins. (B) Deletion of this sequence abolishes the ability of HA-Ses1 to immunoprecipitate endogenous OCRL from Cos7 cells. Clathrin was used as a negative control. (C) Weighted matrix of a conserved 13 amino acid peptide (highlighted in orange in A) recovered from the C-termini of the 55 Ses proteins found in sequenced genomes. Inspection of the peptide reveals heavily conserved phenylalanine, histidine, and hydrophobic residues (red) at positions that also are conserved in the minimal OCRL-binding peptide of APPL1. The last two positions, which include a conserved hydrophobic residue in Ses proteins (blue), are not conserved and in the APPL1 peptide are not needed for OCRL binding. (D) A GST fusion of the conserved 13 amino acid peptide sequence of human Ses1 (C), but not the same peptide lacking the last two amino acids, is sufficient to bind OCRL from a rat brain extract. (E and F) Mutations to alanine of the conserved phenylalanine at position 224 and histidine at position 228 of full-length HA-tagged human Ses1 abolishes OCRL binding. The figure shows Western blots for the HA epitope, endogenous OCRL, and endogenous clathrin (as a control) of anti-HA immunoprecipitates (and starting extracts) from transfected Cos7 cells.
GST pulldowns from brain extracts using fragments of human Ses1 fused to GST demonstrated that the C-terminal portion of Ses1 also was sufficient for the interaction and mapped the minimal binding peptide to a sequence of 13 amino acids that contained the motif conserved between APPL1 and Ses1. Ses binding, however, critically required two additional C-terminal hydrophobic amino acids relative to the minimal APPL1-binding peptide (Fig. 3D), which comprises only 11 amino acids (Fig. 3C) that are preserved nearly identically from fish to man (20).
Conservation in this 13 amino acid peptide among 55 Ses proteins sequenced from different species thus far was assessed by creating a weighted matrix of this region using the gapless multiple alignment of multiple sequences (GLAM) motif-generating engine (Fig. 3C) (44). This analysis revealed high conservation at four positions, namely an obligate F-x-x-x-H-x-x-Φ motif, which also is conserved in APPL1, as well as a Ses-specific hydrophobic residue at the 13th amino acid position in the peptide. Given that APPL1 binding is known to depend critically on the conserved phenylalanine in this peptide (20), we examined if conserved phenylalanine also is required for Ses1 binding by performing anti-HA immunoprecipitation from cells expressing WT and mutant HA-Ses1 in which the phenylalanine of the Ses1 minimal OCRL binding motif was changed to alanine (HA-Ses1-F224A). Despite being expressed at equivalent levels to WT HA-Ses1, HA-Ses1-F224A did not coprecipitate endogenous OCRL, confirming that F224 is critical for OCRL binding (Fig. 3E). Similar experiments were performed using an HA-Ses1 mutant harboring a mutation of the conserved histidine present in both Ses and APPL1. This mutation resulted in complete loss of OCRL binding as well (Fig. 3F), confirming the importance of the F-x-x-x-H-x-x-Φ consensus motif (hereafter defined as a F&H motif) identified in APPL1 and Ses1 for OCRL binding.
Ses1 and APPL1 Binding Are Abolished by the Same Subset of Patient Mutations.
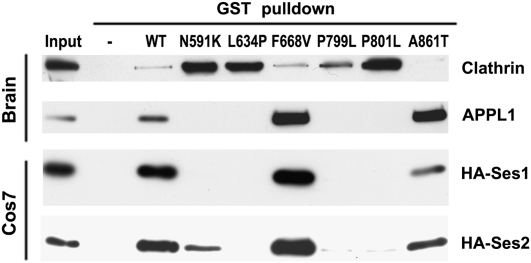
We previously reported that disease-causing missense mutations in the ASH-RhoGAP–like domain of OCRL abolish APPL1 binding but not other interactions of this domain (20, 29). Given the striking similarity of the Ses1/2 and APPL1 sequences required for binding to the ASH-RhoGAP–like domain, we examined whether disease-causing mutations affect APPL1 and Ses1/2 binding in the same way. GST pulldowns from brain extracts and transfected Cos7 cells using WT and mutant ASH-RhoGAP–like domains as bait [including the three previously unpublished mutations F668V, P799L, and P801L and the recently published N591K (37), L634P, and A861T (36)] revealed that mutations that disrupt APPL1 binding also disrupt Ses1/2 binding (Fig. 4 and Fig. S2). Importantly, two of the previously untested Lowe syndrome mutations (F668V and A861T) preserve both APPL1 and Ses1 binding.
Fig. 4.
APPL1 and Ses1 binding to the ASH-RhoGAP–like domain is similarly abolished in a subset of mutations that cause Lowe syndrome. Western blots of GST pulldowns from mouse brain extracts (APPL1 and clathrin; Upper Panels) and transfected Cos7 cells (HA-Ses1 and HA-Ses2; Lower Panels) on GST fusions of WT and mutant human ASH-RhoGAP–like domain. In common with all other Lowe syndrome mutations of the ASH-RhoGAP–like domain tested (20, 29), the recently discovered patient mutations N591K, L634P, P799L, and P801L abolish APPL1 binding while preserving clathrin binding. P799L mutation causes Dent disease. All mutants deficient for APPL1 binding also are deficient for HA-Ses1 and HA-Ses2 binding; detectable but strongly decreased Ses2 binding occurred for the N591K mutant, including all mutants previously tested negative for APPL1 binding (Fig. S2) (20, 29). Two mutations (F668V and A861T) preserve both APPL1 and Ses1/2 binding. Collectively, these results indicate that APPL1 and Ses proteins use a binding modality similar to that of the OCRL ASH-RhoGAP–like domain.
Notably, pulldowns from brain extracts revealed that clathrin binding relative to WT is enhanced by mutations that impair APPL1/Ses binding but not by mutations that preserve the APPL1–Ses interaction. Steric hindrance may prevent clathrin binding when APPL1 or Ses is bound to the ASH-RhoGAP–like domain. Alternatively, disease-causing mutations of this region of OCRL may cause misfolding and favor peptide-mediated interactions (the interaction with clathrin) over those requiring a folded domain (the interaction with APPL1/Ses).
APPL1 and Ses Cannot Bind Simultaneously to OCRL.
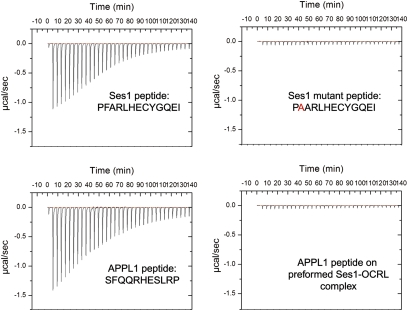
The similar properties of the interaction of APPL1 and Ses proteins with OCRL prompted us to characterize the direct binding of the Ses peptide to the ASH-RhoGAP–like domain and the potential competition between the APPL1- and the Ses-binding peptides. Affinity measurements were performed using isothermal titration calorimetry (ITC). The affinity of the 13 amino acid minimal binding peptide of Ses1 for the OCRL ASH-RhoGAP–like domain was found to be 0.7 ± 0.08 μM, whereas, under the same experimental conditions, the affinity of the minimal binding peptide of APPL1 was found to be 12 ± 2μM, a value that compares well with the previously determined value of 20 μM (20) (Fig. 5). Furthermore, the F224A mutation abolished binding, as predicted by immunoprecipitation experiments involving full-length Ses1 (Fig. 3E).
Fig. 5.
Direct binding of the Ses1 minimal 13 amino acid peptide to the OCRL ASH-RhoGAP–like domain. ITC was used to measure the affinity of the minimal OCRL-binding peptides of APPL1 (11 amino acids;12 ± 2 μM affinity) and Ses1 (13 amino acids; 0.7 ± 0.08 μM). As in the case of the APPL1 peptide (20), Ses1 binding critically depended on a conserved phenylalanine residue. The similarity between Ses- and APPL1-binding properties suggested a competitive mechanism for OCRL binding. Accordingly, a Ses peptide:OCRL ASH-RhoGAP complex saturated for Ses binding did not bind the APPL1 peptide.
To test if APPL1 and Ses1 binding are mutually exclusive, the affinity of APPL1 for a preformed complex of the ASH-RhoGAP–like domain with the Ses1 peptide was measured. No interaction was detected between APPL1 and this complex, suggesting that Ses1 binding occludes APPL1 binding to OCRL. This finding is consistent with APPL1 and the Ses proteins being competitive ligands of the OCRL ASH-RhoGAP–like domain.
APPL1 and Ses Associate with Distinct Endocytic Organelles.
OCRL has a broad distribution on endosomes, including PI3P-positive endosomes, and already associates with endocytic vesicles at the internalization step from the plasma membrane (20–22, 35). In contrast, we have shown that APPL1 associates with endosomes at early postinternalization stages (in both clathrin-coated pit-derived vesicles and macropinosomes) and then dissociates as endocytic vesicles become PI3P positive and thus acquire PI3P binding proteins such as WDFY2 and EEA1 (35). Acute loss of PI3P, either by the acute recruitment of an inositol-3′-phosphatase to the endosomal surface or by treatment with the PI3P synthesis inhibitor wortmannin, perturbs this reaction. Under these conditions, the transition from the APPL1 to the WDFY2 and EEA1 stage is arrested, and WFDY2 and EEA1 are shed from vesicles which instead reacquire APPL1 (35).
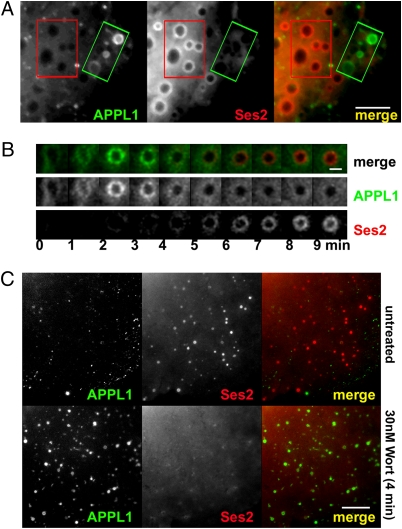
The colocalization of Ses with EEA1 and WDFY2 (Fig. 2) suggested that, in spite of their similar mode of binding to OCRL, APPL1 and Ses proteins are not localized on the same endocytic compartments. To address this question, GFP-APPL1 and TagRFP-T-Ses2 were cotransfected along with OCRL in Cos7 cells and were imaged live by widefield or spinning disk confocal microscopy. Because macropinosomes can be followed easily from frame to frame, we focused our observations on these organelles. These experiments showed that APPL1 and Ses2 indeed were localized on distinct populations of vesicles (Fig. 6A) and that many APPL1 macropinosomes converted to Ses-positive organelles (Fig. 6B and Movie S1). Furthermore, wortmannin treatment resulted in the loss of Ses2 and in the replacement of Ses2 with APPL1 on the very same vesicles (Fig. 6C and Movie S2). These results, which are consistent with the competition of Ses and APPL1 for the same binding site in OCRL, demonstrate that APPL1 and Ses associate sequentially with endosomes and that Ses proteins are localized on a PI3P-positive compartment.
Fig. 6.
APPL1 and Ses2 are localized at different sites along the endocytic pathway. (A) Widefield microscopy of a transfected Cos7 cell showing the different localization of GFP-APPL1–positive and TagRFP-T-Ses2–positive spontaneously occurring macropinosomes. Rectangles define the same regions, primarily occupied by APPL1 vesicles and Ses2 vesicles, respectively, in the two channels. The corresponding movie (Movie S1) shows that GFP-APPL1–positive micropinosomes disappear or mature to become TagRFP-T-Ses2 positive. (Scale bar, 8 μm.) (B) APPL1 precedes Ses2 on the same macropinosome. Selected frames at 1-min intervals of a spinning disk-confocal movie. The micropinosome acquired APPL1, then shed it and acquired Ses2. APPL1 and Ses2 did not overlap. (Scale bar, 2 μm.) (C) The association of Ses2 with endosomes requires the presence of PI3P. Inhibition of PI 3′ kinases with wortmannin induces the dissociation of Ses2 from endosomes which then acquire APPL1. (Scale bar, 10 μm.)
Discussion
We have characterized an interactor of OCRL, Ses, which is expressed as two very similar isoforms, Ses1 and Ses2, in mammalian cells. These proteins harbor an OCRL-binding motif in their C-terminal regions which shares common features with the previously described binding peptide of the endocytic adaptor APPL1 (20). Together, the binding peptides of APPL1 and Ses define a consensus, referred to as “F&H,” for OCRL binding. Both these proteins, like APPL1, are localized on endosomes, but in a mutually exclusive manner that correlates with different and sequential stages of progression along the endocytic pathway.
Studies of APPL1 had shown that APPL1 and OCRL are recruited to endosomes independently of each other, at least in part through the binding of both proteins to Rab5 (20, 26, 29, 34). Thus, the APPL1–OCRL interaction is likely to have a synergistic or regulatory function. We speculate that, likewise, the recruitment of Ses to endosomes requires more than the interaction with OCRL. As suggested by the shedding of Ses upon PI3P depletion, Ses may receive multiple protein and lipid cues that may help the vectorial progression of endosomes from an APPL1-positve to a Ses-positive stage. Ses itself has a PH domain whose structure is highly similar to the 3′ phosphoinositide-binding protein, B-cell adaptor molecule of 32 kDa (BAM32)/(45, 46). However, no lipid binding of the Ses PH domain has been observed so far. The interaction between OCRL and APPL1 and the interaction of OCRL with Ses proteins may mediate regulatory mechanisms that are specific to two distinct endosomal stations.
Our study has implications for the understanding of Dent disease and Lowe syndrome pathology. We now have tested 12 different missense mutations of the OCRL ASH-RhoGAP–like domain, 11 of which cause Lowe syndrome and one of which, P799L, causes Dent disease. Ten of these mutants abolish both APPL1 and Ses binding, consistent with previous reports demonstrating loss of APPL1 binding of some of these mutants (20, 29). Because one such mutant is the Dent disease mutant P799L, these findings indicate that a selective loss of APPL1/Ses binding to OCRL cannot explain differences between Lowe syndrome and Dent disease. However, A861T (36) and F668V (this study), which are both Lowe syndrome mutations, do not affect the binding of OCRL to either APPL1 or Ses1/2 and thus to the newly identified F&H consensus motif. It remains possible that such mutations may cause little or no protein expression. For example, the A861T mutation is both a missense and a splice-site mutation (32), which may affect protein expression by generating a non-sense transcript. Alternatively, these two mutations may impair other interactions of the ASH-RhoGAP–like domain of OCRL. Identification of these interactions, which could be either intermolecular or intramolecular, will be useful in determining which properties of the ASH-RhoGAP–like domain must be coordinated to evoke correct OCRL function.
The identification of a consensus binding motif for the ASH-RhoGAP–like domain of OCRL suggests that there may be additional ligands for this domain, besides APPL1 and Ses1/2. The ASH-RhoGAP–like domain of OCRL-like 5′phosphatases is well conserved in many organisms, including the model organisms Caenorhabtidis elegans and Dictyostelium discoideum, where Ses-like and APPL1-like proteins are absent. These considerations raise the possibility that there may be at least one, even more ancient interactor of the ASH-RhoGAP–like domain to be identified. Unbiased scans of different genomes with the F&H consensus binding motif (Fig. 3) produced candidate hits in several proteins that participate in trafficking reactions in which OCRL has been implicated, such as traffic to and from endosomes. An important priority for future studies will be to validate these potential interactions and assess whether these proteins are involved in Lowe syndrome and Dent disease pathology.
Ses proteins are evolutionarily older than APPL1 and originally may have served some of the functions now fulfilled by the APPL1–OCRL interaction. Both APPL1 and Ses1/2 have PH domains, and their general structure suggests adaptor protein function. Intriguingly, Drosophila Ses and fish Ses1 proteins have clathrin-binding motifs (Esox lucius LIDL, Tetraodon nigroviridus LVDLD, Danio rerio LIDL, Drosophila melanogaster LIQL) whereas clathrin-binding motifs are absent from Ses proteins in higher organisms, suggesting that perhaps parts of the ancestral role of Ses proteins are now distributed over several proteins.
These results suggest that the OCRL protein may use its ASH-RhoGAP–like binding surface at successive points in the endocytic pathway, binding a progression of different endocytic proteins via the same motif. In this manner one can speculate that Lowe syndrome and Dent disease may be caused, not by a malfunction at the level of a single endosome, but by cumulative defects in the endocytic cycle as OCRL fails to act at subsequent points on the endocytic pathway.
Methods
Plasmids.
Human Ses1 and Ses2 ORFs (with stop codons) were amplified from plasmids encoding the mRNA sequences NM_175474.3 and BC104175, respectively (Open Biosystems) and subcloned into pcDNA3.1 HA via BamHI/XhoI to produce N-terminally tagged HA-Ses1 and HA-Ses2. Ses1 and Ses2 subsequently were excised from these constructs with BamHI/XbaI and ligated to dam negative DNA from pmTagRFP-T-C1 (40) to produce N-terminally TagRFP-tagged Ses1 and Ses2. Site-directed mutagenesis (Stratagene QuikChange II XL) was used to generate the mutant constructs HA-Ses1-PH-CC and HA-Ses1Δ26 (by introducing stop codons at position G123 and F224, respectively) and the point mutants HA-Ses1 F224A and HA-Ses1 H228A. The truncated construct HA-Ses1-CT, which comprises amino acids 132 to stop, was cloned into pcDNA3.1 HA via BamHI/XhoI as above. GST fusions of the Ses1 minimal OCRL-binding sequence, which comprise amino acids 223–233 followed by a stop codon (GST-Ses1 11mer) and amino acids 223–235 followed by a stop codon (GST-Ses1 13mer), were cloned into pGex6P-1 using BamHI/XhoI. All constructs were fully sequenced.
GFP-OCRL is described elsewhere (20), as are GFP-EEA1, GFP-APPL1, GFP-WDFY2, and HrasV12G (35). Six mutations of the GST-OCRL ASH-RhoGAP were previously described (20, 29). The previously uncharacterised mutants of the ASH-RhoGAP-like domain were introduced in the corresponding WT construct by the Stratagene QuikChange II XL kit. GST-ASH and GST-RhoGAP–like encode amino acids 564–678 and 678–stop of human OCRL. cDNAs were transfected in cultured cells via electroporation (Neucleofector; Fisher Scientific).
Antibodies.
Mouse anti-clathrin heavy chain (TD1), mouse anti-human OCRL, rabbit anti-mouse OCRL, and rabbit anti-APPL1 were described previously (20, 29). Peroxidase Anti-HA (3F10) was purchased from Roche Diagnostics.
Immunoprecipitation.
Cos7 cells were extracted 24–36 h after transfection in chilled immunoprecipitation buffer [25 mM Hepes (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.5 mM EDTA plus protease inhibitors (Complete Mini EDTA-free; Roche Diagnostics)]. The extract was sonicated, cleared by 15 minutes of 16,000 x g centrifugation at 4 °C in a table-top microcentrifuge and incubated with either GST fusion proteins bound to glutathione-Sepharose 4B (GE Healthcare Biosciences) or with anti-HA antibodies conjugated to agarose beads (anti-HA Affinity Matrix; Roche Diagnostics). Rat brain lysates were prepared as described (20, 29). GST fusion proteins of the OCRL ASH-RhoGAP–like domain or mutants thereof were purified over glutathione Sepharose in 10% glycerol, 50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, adjusted to pH 8.0. Protein-coupled beads were kept at 4 °C and never frozen before use.
Imaging.
Transfected cells were seeded in glass-bottomed 35-mm dishes (MatTek Corporation) and imaged at 0.2 Hz at 37 °C by either spinning disk confocal (Perkin-Elmer) or widefield (CTR6000; Leica Microsystems) microscopy. Images were processed using ImageJ (47).
Isothermal Titration Calorimetry.
Experiments were performed at the Keck Research Facility at Yale University. In each experiment, 3-μL aliquots of peptide solution (2 mM) were injected into a calorimetric cell preloaded with 1.4267 mL of the C-terminal region of OCRL (30 μM) using a rotating stirrer syringe (250-μL vol) every 240 s at 30 °C. Both the peptide and the protein solutions contained 20 mM Hepes (pH 7.0), 20 mM NaCl and were degassed before use. To perform measurements on the preformed OCRL:Ses peptide complex, 1.4267 mL of a Ses peptide:OCRL ASH-RhoGAP–like domain mixture (138:27.9 μM; i.e., with a molar excess of the Ses peptide) was equilibrated in 20 mM Hepes (pH 7.0), 20 mM NaCl, and 3-μL aliquots of 2 mM of APPL1 peptide were injected as above.
To estimate a blank heat effect associated with dilution and mechanical phenomena, peptide injections into degassed buffer were performed before the titration with protein. ITC measurements were performed using a Microcal VP-ITC isothermal titration calorimeter equipped with a PC running VPViewer software (http://www.info.med.yale.edu/wmkeck/prochem/). Dissociation constant values were obtained with Origin software (MicroCal).
Supplementary Material
Acknowledgments
We thank Ewa Folta-Stogniew, Emily Yin, Helen Xu, Massimiliano Stagi, and Frank Wilson for assistance and Danny Balkin for discussion. This work was supported in part by National Institutes of Health Grants NS36251, DK082700, DK45735, and DA018343 and by the Lowe Syndrome Association (P.D.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914658107/DCSupplemental.
References
- 1.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 2008;27:2457–2470. doi: 10.1038/emboj.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attree O, et al. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- 4.Laporte J, et al. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 5.Bolino A, et al. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet. 2000;25:17–19. doi: 10.1038/75542. [DOI] [PubMed] [Google Scholar]
- 6.Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby M, et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat Genet. 2009;41:1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- 8.McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooms LM, et al. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem J. 2009;419:29–49. doi: 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Jefferson AB, Auethavekiat V, Majerus PW. The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc Natl Acad Sci USA. 1995;92:4853–4856. doi: 10.1073/pnas.92.11.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schurman SJ, Scheinman SJ. Inherited cerebrorenal syndromes. Nat Rev Nephrol. 2009;5:529–538. doi: 10.1038/nrneph.2009.124. [DOI] [PubMed] [Google Scholar]
- 12.Delleman JW, Bleeker-Wagemakers EM, van Veelen AW. Opacities of the lens indicating carrier status in the oculo-cerebro-renal (Lowe) syndrome. J Pediatr Ophthalmol. 1977;14:205–212. [PubMed] [Google Scholar]
- 13.Kleta R. Fanconi or not Fanconi? Lowe syndrome revisited. Clin J Am Soc Nephrol. 2008;3:1244–1245. doi: 10.2215/CJN.02880608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenworthy L, Charnas L. Evidence for a discrete behavioral phenotype in the oculocerebrorenal syndrome of Lowe. Am J Med Genet. 1995;59:283–290. doi: 10.1002/ajmg.1320590304. [DOI] [PubMed] [Google Scholar]
- 15.Dent CE, Friedman M. Hypercalcuric rickets associated with renal tubular damage. Arch Dis Child. 1964;39:240–249. doi: 10.1136/adc.39.205.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho HY, et al. Renal manifestations of Dent disease and Lowe syndrome. Pediatr Nephrol. 2008;23:243–249. doi: 10.1007/s00467-007-0686-9. [DOI] [PubMed] [Google Scholar]
- 17.Hoopes RR, Jr, et al. Dent disease with mutations in OCRL1. Am J Hum Genet. 2005;76:260–267. doi: 10.1086/427887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Utsch B, et al. Novel OCRL1 mutations in patients with the phenotype of Dent disease. Am J Kidney Dis. 2006;48(6):942–956. doi: 10.1053/j.ajkd.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Shrimpton AE, et al. OCRL1 mutations in Dent 2 patients suggest a mechanism for phenotypic variability. Nephron Physiol. 2009;112:p27–p36. doi: 10.1159/000213506. [DOI] [PubMed] [Google Scholar]
- 20.Erdmann KS, et al. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Y, et al. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 2009;28:1831–1842. doi: 10.1038/emboj.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem. 2009;284:9965–9973. doi: 10.1074/jbc.M807442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhury R, et al. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell. 2005;16:3467–3479. doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ungewickell A, Ward ME, Ungewickell E, Majerus PW. The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc Natl Acad Sci USA. 2004;101:13501–13506. doi: 10.1073/pnas.0405664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin HW, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyvola N, et al. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 2006;25:3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Olivos-Glander IM, Jänne PA, Nussbaum RL. The oculocerebrorenal syndrome gene product is a 105-kD protein localized to the Golgi complex. Am J Hum Genet. 1995;57:817–823. [PMC free article] [PubMed] [Google Scholar]
- 29.McCrea HJ, et al. All known patient mutations in the ASH-RhoGAP domains of OCRL affect targeting and APPL1 binding. Biochem Biophys Res Commun. 2008;369:493–499. doi: 10.1016/j.bbrc.2008.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Addis M, Loi M, Lepiani C, Cau M, Melis MA. OCRL mutation analysis in Italian patients with Lowe syndrome. Hum Mutat. 2004;23:524–525. doi: 10.1002/humu.9239. [DOI] [PubMed] [Google Scholar]
- 31.Lichter-Konecki U, Farber LW, Cronin JS, Suchy SF, Nussbaum RL. The effect of missense mutations in the RhoGAP-homology domain on ocrl1 function. Mol Genet Metab. 2006;89:121–128. doi: 10.1016/j.ymgme.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Kawano T, Indo Y, Nakazato H, Shimadzu M, Matsuda I. Oculocerebrorenal syndrome of Lowe: Three mutations in the OCRL1 gene derived from three patients with different phenotypes. Am J Med Genet. 1998;77:348–355. [PubMed] [Google Scholar]
- 33.Lin T, et al. Spectrum of mutations in the OCRL1 gene in the Lowe oculocerebrorenal syndrome. Am J Hum Genet. 1997;60:1384–1388. doi: 10.1086/515471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miaczynska M, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 35.Zoncu R, et al. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136:1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bockenhauer D, et al. Renal phenotype in Lowe syndrome: A selective proximal tubular dysfunction. Clin J Am Soc Nephrol. 2008;3:1430–1436. doi: 10.2215/CJN.00520108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuksel A, Karaca E, Albayram MS. Magnetic resonance imaging, magnetic resonance spectroscopy, and facial dysmorphism in a case of Lowe syndrome with novel OCRL1 gene mutation. J Child Neurol. 2009;24:93–96. doi: 10.1177/0883073808321047. [DOI] [PubMed] [Google Scholar]
- 38.Monnier N, Satre V, Lerouge E, Berthoin F, Lunardi J. OCRL1 mutation analysis in French Lowe syndrome patients: Implications for molecular diagnosis strategy and genetic counseling. Hum Mutat. 2000;16:157–165. doi: 10.1002/1098-1004(200008)16:2<157::AID-HUMU8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Giot L, et al. A protein interaction map of Drosophila melanogaster . Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 40.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 42.Fritzius T, et al. A WD-FYVE protein binds to the kinases Akt and PKCzeta/lambda. Biochem J. 2006;399:9–20. doi: 10.1042/BJ20060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayakawa A, et al. The WD40 and FYVE domain containing protein 2 defines a class of early endosomes necessary for endocytosis. Proc Natl Acad Sci USA. 2006;103:11928–11933. doi: 10.1073/pnas.0508832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frith MC, Hansen U, Spouge JL, Weng Z. Finding functional sequence elements by multiple local alignment. Nucleic Acids Res. 2004;32:189–200. doi: 10.1093/nar/gkh169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall AJ, et al. A novel B lymphocyte-associated adaptor protein, Bam32, regulates antigen receptor signaling downstream of phosphatidylinositol 3-kinase. J Exp Med. 2000;191:1319–1332. doi: 10.1084/jem.191.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowler S, Currie RA, Downes CP, Alessi DR. DAPP1: A dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem J. 1999;342:7–12. [PMC free article] [PubMed] [Google Scholar]
- 47.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.