Abstract
Regulation of gene expression by small RNAs (∼20–30 nucleotides in length) plays an essential role in developmental pathways and defense responses against genomic parasites in eukaryotes. MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) commonly direct the inactivation of cognate sequences through a variety of mechanisms, including RNA degradation, translation inhibition, and transcriptional repression. Recent studies have provided considerable insight into the biogenesis and the mode of action of miRNAs and siRNAs. However, relatively little is known about mechanisms of quality control and small RNA decay in RNA interference (RNAi) pathways. Here we show that deletion of MUT68, encoding a terminal nucleotidyltransferase in the alga Chlamydomonas reinhardtii, results in elevated miRNA and siRNA levels. We found that MUT68 plays a role in the untemplated uridylation of the 3′ ends of small RNAs in vivo and stimulates their degradation by the RRP6 exosome subunit in vitro. Moreover, RRP6 depletion also leads to accumulation of small RNAs in vivo. We propose that MUT68 and RRP6 cooperate in the degradation of mature miRNAs and siRNAs, as a quality control mechanism to eliminate dysfunctional or damaged small RNA molecules.
Keywords: exosome, RNA interference, miRNA quality control
RNA-mediated silencing is an evolutionarily conserved mechanism(s) by which small RNAs (sRNAs) induce the inactivation of cognate sequences, although sRNAs may also participate in the activation of gene expression (1 –3). At least two major classes of small RNAs have been identified in many eukaryotes: microRNAs and small interfering RNAs (1, 2). miRNAs originate from endogenous noncoding RNA transcripts or introns that fold into imperfect stem loop structures and often modulate the expression of genes with roles in development, physiological processes, or stress responses (1 –3). siRNAs are produced from long, near-perfect complementarity double-stranded RNAs (dsRNAs) of diverse origins, including the transcripts of long inverted repeats, the products of convergent transcription or RNA-dependent RNA polymerase activity, viral RNAs, or dsRNAs experimentally introduced into cells (1 –5). These siRNAs play various roles in posttranscriptional regulation of gene expression, suppression of viruses and transposable elements, and/or heterochromatin formation (1 –5). Hairpin and long dsRNAs are processed into sRNAs by an RNaseIII-like endonuclease named Dicer (1, 2). These small RNAs are then incorporated into multisubunit effector complexes, such as the RNA-induced silencing complex (RISC) (1, 2). Argonaute proteins, which include two main subfamilies of polypeptides named after Arabidopsis thaliana ARGONAUTE1 (AGO1) and Drosophila melanogaster PIWI, are core components of the RISC and some function as sRNA-guided endonucleases (1, 2, 6, 7). Recent evidence suggests that a siRNA duplex is first loaded into RISC and AGO cleaves one of the siRNA strands (the passenger strand) (1, 2). Ribonucleases, such as Trax or QIP, then promote RISC activation by removing the passenger strand cleavage products (8, 9). Activated RISC uses the remaining single-stranded siRNA as a guide to identify homologous RNAs, ultimately triggering transcript degradation and/or translation repression (1 –3).
The biogenesis and the mode of action of sRNAs have attracted great attention (1–3, 10), but much less is known about mechanisms of miRNA/siRNA turnover and their role in small RNA function. Degradation of mature miRNAs in Caenorhabditis elegans, mediated by the 5′-to-3′ exoribonuclease XRN2, has recently been shown to modulate miRNA accumulation in vivo (11). A conserved nuclease from C. elegans and Schizosaccharomyces pombe, Eri-1, degrades siRNA duplexes with 2-nucleotide 3′ overhangs in vitro and reduces the efficiency of RNAi in vivo (12, 13). Another family of 3′-to-5′ exoribonucleases, encoded by the SMALL RNA DEGRADING NUCLEASE (SDN) genes, has been implicated in mature miRNA turnover in A. thaliana (14). In C. elegans and mammalian cells, Lin-28 (a stem-cell-specific regulator) binds the precursor of the let-7 miRNA in the cytoplasm and promotes its 3′ end uridylation by a poly(U) polymerase (15 –17). This leads to precursor RNA degradation and downregulation of the let-7 miRNA. In contrast, terminal uridylation of mature miR-26 in mammalian cells appears to impart functional differences that attenuate miRNA-targeted repression without obvious changes in miRNA steady-state levels (18). In A. thaliana, 3′ end uridylation has been postulated, although not demonstrated yet, to stimulate the decay of small RNAs (19, 20). Indeed, the enzymes involved in untemplated nucleotide additions to the 3′ ends of mature sRNAs and the functional significance of these modifications remain largely uncharacterized in most eukaryotes. Moreover, plant miRNAs and siRNAs and animal siRNAs and PIWI-interacting RNAs (piRNAs) have a 2′-O-methyl group on their 3′ termini, introduced by the RNA methyltransferase HEN1, which seems to protect them against nucleotide additions and/or exonucleolytic shortening (19 –22). However, the putative enzyme(s) involved in the proposed 3′-to-5′ degradation of unmethylated and uridylated sRNAs is not known because Arabidopsis SDN1 is inhibited by 3′ terminal uridylation (14).
We report here on the characterization of a C. reinhardtii mutant (Mut-68) that is defective in RNAi and provides further insight on the mechanisms of mature miRNA/siRNA degradation. Mut-68, which lacks the MUT68 nucleotidyltransferase (23), displays elevated levels of miRNAs and siRNAs. The MUT68 enzyme is involved in the untemplated uridylation of the 3′ termini of small RNAs in vivo and in stimulating their degradation by the RRP6 exosome subunit in vitro. Moreover, RRP6 depletion by RNAi also results in the accumulation of miRNAs and siRNAs in vivo. We hypothesize that MUT68, in association with RRP6, is part of a quality control mechanism for the degradation of functionally defective small RNAs.
Results
Mut-68 Mutant Shows Enhanced Levels of Small RNAs and of the ARGONAUTE3 (AGO3) Protein.
We have previously reported that MUT68 is required for the addition of untemplated nucleotides to the 5′ RNA fragments produced by RISC cleavage of target transcripts and for their efficient decay (23). Interestingly, a mutant strain, deleted for the cognate gene, also shows higher levels of several miRNAs (Fig. 1A and Fig. S1), relative to the wild-type (CC-124) and the parental transgenic strain (Maa7-IR44). Likewise, Mut-68 displays enhanced accumulation of siRNAs produced from inverted repeat transgenes (Fig. S2). These changes are paralleled by increased levels of AGO3 (Fig. 1B), one of the three C. reinhardtii Argonaute proteins (4), and this effect is likely the result of translational or posttranslational regulation because transcripts for the three AGO genes are present at similar amounts in Mut-68 and the control strains (Fig. S3A).
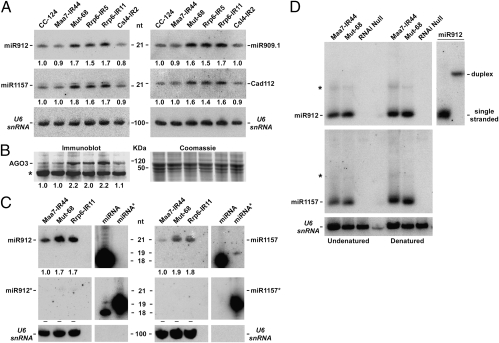
Fig. 1.
Mut-68 and strains depleted for the RRP6 exosome subunit show increased levels of miRNAs and of the AGO3 protein. (A) Northern blot analyses of sRNAs isolated from the indicated strains and detected with probes specific for Chlamydomonas miRNAs. Cad112, candidate miRNA 112 (31). The numbers below the blots indicate the relative abundance of the miRNAs. CC-124, wild-type strain; Maa7-IR44, CC-124 transformed with an IR transgene designed to induce RNAi of MAA7 (encoding tryptophan synthase β subunit); Mut-68, MUT68 deletion mutant; Rrp6-IR5 and Rrp6-IR11, Maa7-IR44 transformed with IR transgenes inducing RNAi of two distinct genes encoding the RRP6 exosome subunit; Csl4-IR2, strain transformed with an IR transgene triggering RNAi of CSL4 (encoding a core exosome subunit). (B) Immunoblot analysis of AGO3 protein levels. The specificity of the indicated AGO3 band was verified by peptide blocking assays. The asterisk shows a cross-reacting antigen. Coomassie-blue staining of an equivalent gel is shown as a control for similar loading of the lanes (Right). (C) Northern blot analysis of sRNAs isolated from the indicated strains and detected using probes specific for the guide strand (miRNA) or the passenger strand (miRNA*) of Chlamydomonas miR912 or miR1157. Control hybridizations to synthetic oligonucleotides are shown on the Right of each panel. (D) Northern blot analysis of sRNAs from the indicated strains separated by nondenaturing polyacrylamide gel electrophoresis. Isolated RNA was resuspended in a nondenaturing buffer (undenatured) or in formamide and denatured by heating (denatured) before loading the gel. The asterisks indicate uncharacterized RNA forms that are present in both Maa7-IR44 and Mut-68 although, due to reduced sample loading, they are less noticeable in the Mut-68 lanes. Because the intensity of these bands was not affected by denaturation, they do not appear to correspond to dsRNA. RNAi null, uncharacterized Chlamydomonas mutant lacking miRNAs. Synthetic oligoRNAs corresponding to miR912 and its miRNA* were used to demonstrate that an annealed duplex is stable under our extraction and electrophoretic conditions (Right). Annealed dsRNAs were resuspended in formamide and denatured by heating (single-stranded lane) or resuspended in nondenaturing buffer (duplex lane) before loading on the same gel as for the miR912 samples.
To define the step of small RNA biogenesis at which MUT68 may act to influence sRNA levels, we determined whether the amounts of the miRNA* [the strand complementary to a guide miRNA (1, 2)] were also affected by depletion of this enzyme. For two miRNA tested, miR912 and miR1157, the miRNA strand showed 70–90% higher accumulation in the mutant strain (relative to the controls) whereas the miRNA* was barely detectable (Fig. 1C). In contrast, if the MUT68 deletion were to stabilize simultaneously and to the same degree both strands of a miRNA/miRNA* duplex, clearly detectable amounts of the miRNA* would be expected, given the observed enhancement in miRNA levels. An analysis of sRNAs by high-throughput sequencing is also consistent with a lack of an effect of MUT68 on miRNA*s (see below). Indeed, miR912 and miR1157 appear to be predominantly in a single-stranded conformation in both Mut-68 and its parental strain. When sRNAs were separated by nondenaturating polyacrylamide gel electrophoresis, with or without prior denaturation of the RNA samples, miRNA/miRNA* duplexes were undetectable (Fig. 1D). This is in contrast to the accumulation of nicked duplex sRNAs caused by depletion of the QIP nuclease (9), which removes AGO-cleaved passenger strands in Neurospora crassa. Moreover, because no single-stranded guide sRNA seems to be produced before Argonaute association and RISC activation (2, 21, 24), our observations imply that mature miRNAs and siRNAs appear to be stabilized by the MUT68 deletion. This interpretation is also consistent with the increase in AGO3 protein levels in Mut-68 (Fig. 1B).
Recombinant MUT68 Functions as a Terminal Nucleotidyltransferase on OligoRNA Substrates.
To begin addressing the role of MUT68 on sRNA stability, we tested whether the enzyme displayed terminal nucleotidyltransferase activity on oligoRNAs. Recombinant, His-tagged MUT68 was incubated with a synthetic C15A10 RNA, 32P-labeled at its 5′ end, in the presence of different ribonucleotides or deoxyribonucleotides. His-MUT68 used preferentially ATP and UTP to add an untemplated oligonucleotide tail to the RNA primer (Fig. 2A). In contrast, CTP, GTP, and deoxyribonucleotides were incorporated very poorly or not at all (Fig. 2A). Two point mutations (D68A and D70Q) in the putative catalytic site resulted in a recombinant protein with abolished activity [Fig. 2A, MUT68(DADQ)]. His-MUT68 was also active on a single-stranded RNA corresponding in sequence to miR912 (Fig. 2B). However, the enzyme was unable to use a 2′-O-methylated miR912 oligoRNA as a substrate (Fig. 2B). Similarly to higher plant sRNAs (20), Chlamydomonas siRNAs and miRNAs appear to have a monophosphate group at their 5′ ends (Fig. S4A) and a 2′-O-methyl group at their 3′ ends (25), as suggested by their resistance to periodate oxidation and β elimination (Fig. S4B). Hence, MUT68 may only be able to modify in vivo newly processed or damaged siRNAs/miRNAs, lacking a terminal 2′-O-methyl group.
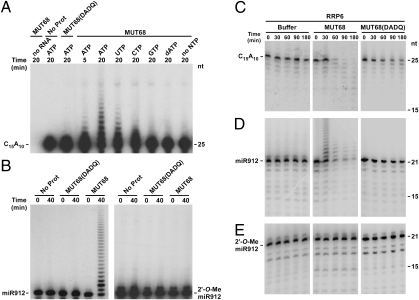
Fig. 2.
MUT68 acts as a terminal nucleotidyltransferase and promotes the degradation of oligoribonucleotides by the RRP6 exosome subunit in vitro. (A) Recombinant MUT68 was incubated with a 32P-labeled oligoRNA and the indicated nucleotide triphosphates for 5 or 20 min. Products were separated on a denaturing polyacrylamide/urea gel and analyzed by autoradiography. Controls included the omission of RNA, protein, or nucleotides as well as the substitution of MUT68 for the catalytically inactive MUT68(DADQ). (B) MUT68 activity, in the presence of UTP, on synthetic oligoRNAs corresponding in sequence to miR912, either unmodified or 2′-O-methylated on the 3′ terminal ribose. (C) 5′-end-labeled C15A10 oligoRNA was incubated with affinity-purified RRP6 alone (buffer) or with the addition of MUT68 or the catalytically inactive MUT68(DADQ). Reactions were stopped at the indicated times, separated by denaturing PAGE, and analyzed by autoradiography. (D) Enzymatic activity of RRP6, RRP6/MUT68, or RRP6/MUT68(DADQ) on an unmodified miR912 oligoRNA. (E) Enzymatic activity of RRP6, RRP6/MUT68, or RRP6/MUT68(DADQ) on miR912 with a 3′ terminal 2′-O-methyl group.
MUT68 Promotes the Degradation of Small RNAs by the RRP6 Exosome Subunit.
In Saccharomyces cerevisiae, the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complex has been implicated in stimulating the exosome, a 3′-to-5′ multisubunit exonuclease, during the degradation of misfolded tRNAs and several noncoding RNAs (26 –28). TRAMP is a nuclear complex that includes a nucleotidyltransferase (Trf4/5) weakly related to MUT68 (23, 26–28). However, MUT68 is found predominantly in the cytosol, in some cells with distinct perinuclear localization (Fig. S5), and appears to cooperate with the cytoplasmic exosome in the turnover of RISC-generated 5′ RNA cleavage products (23). Indeed, RNAi-mediated knockdown of core exosome subunits, such as CSL4 or RRP40, results in stabilization of the 5′ RNA products from RISC-cleaved transcripts (Fig. S6A), but the peripheral RRP6 exosome subunit does not appear to participate in this decay pathway (Fig. S6B). Given these observations, we examined whether the exosome might be involved in the degradation of sRNAs. Depletion of CSL4 had no discernible effect on the levels of miRNAs (Fig. 1A and Fig. S1). Likewise, mature miRNA amounts were not affected by defects in core exosome subunits in A. thaliana (29). In contrast, RNAi-mediated suppression of RRP6 (Fig. S3B) resulted in enhanced sRNA levels, comparable to those in Mut-68 (Fig. 1A and Figs. S1 and S2E). The Rrp6-IR strains also showed higher accumulation of the AGO3 protein (Fig. 1B).
RRP6 is related to bacterial RNase D and can act as a distributive 3′-to-5′ hydrolytic exonuclease (30). To test directly whether MUT68 could stimulate its activity, we obtained recombinant His-tagged RRP6 expressed in baculovirus-infected Spodoptera frugiperda cells. However, this protein, under the conditions used, had very little to no activity on several oligoRNA substrates (Fig. 2 C–D). Interestingly, the addition of MUT68 to recombinant RRP6 significantly enhanced the decay of unmethylated RNA substrates (Fig. 2 C–D and Fig. S7). The C15A10 oligoribonucleotide was degraded faster than the miR912 RNA, with the latter showing a transient buildup of products with untemplated oligonucleotide tails before their turnover (Fig. 2D). The catalytically inactive MUT68(DADQ) had virtually no effect on the RRP6 activity (Fig. 2D) or, possibly, a very minor stimulation of the C15A10 decay (Fig. 2C). Intriguingly, yeast Trf4 has been demonstrated to play a role in the turnover of certain transcripts that is independent of its polyadenylation activity (26, 27). In contrast, 2′-O-methylated miR912 was resistant to degradation under all of our in vitro conditions (Fig. 2E). Thus, our results suggest that MUT68 and RRP6 cooperate in the degradation of unmethylated sRNAs.
Mut-68 Mutant Displays Reduced 3′ Terminal Uridylation of miRNAs and siRNAs.
To gain further insight into the in vivo role of MUT68, we characterized the small RNA populations in two biological replicates of Mut-68 and Maa7-IR44 by high-throughput sequencing. During library construction each sample was given a unique 2-nt barcode: GA, UC, AG, and CU for the Mut-68–1, Mut-68–2, Maa7-IR44–1 and Maa7-IR44–2 libraries, respectively. To minimize any potential bias these barcodes might have on the sequenced small RNA populations, Mut-68–1 was compared to Maa7-IR44–1 and Mut-68–2 to Maa7-IR44–2. Only changes consistent between these two datasets are presented. As previously reported (25, 31), most sequenced sRNAs fell into the 20- to 22-nt size class (Fig. 3A and Fig. S8A). However, the size distribution was slightly shifted toward longer sRNAs in Mut-68 (Fig. 3A and Fig. S8A). The datasets also showed a decrease in the relative abundance of transposon siRNAs (76% for Mut-68–1 and 82% for Mut-68–2) and an increase in the relative abundance of miRNAs (50% for Mut-68–1 and 95% for Mut-68–2) in the mutant strain (Table S1 in Dataset S1).
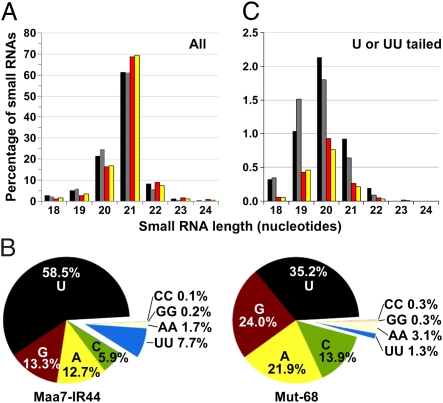
Fig. 3.
Analysis of endogenous small RNA sizes and their 3′ untemplated nucleotide additions in Mut-68 and the parental strain Maa7-IR44. (A) Size distribution of redundant small RNAs, after subtracting putative degradation products and normalization. Two independent libraries were examined for each strain. Redundant sRNA sequences from Maa7-IR44 are represented by black (Maa7-IR44-1) or gray (Maa7-IR44-2) bars. Redundant sRNA sequences from Mut-68 are represented by red (Mut-68-1) or yellow (Mut-68-2) bars. (B) Frequency of the predominant untemplated nucleotides added to the 3′ ends of small RNAs (relative to all sRNAs with 3′ nucleotide additions). (C) Size distribution of redundant small RNAs with an untemplated 3′ terminal U or UU (after removing the tails). Two independent libraries were examined for each strain and the bars are colored as indicated above (A).
Interestingly, ∼7.3% of the sRNAs had 3′ untemplated nucleotides in Maa7-IR44 but this fraction was reduced to ∼4.9% in Mut-68 (Table S2 in Dataset S1). Moreover, uridylation, the predominant addition to the 3′ termini of small RNAs, was markedly lower in the mutant (Fig. 3B and Fig. S9). Consistent with the possibility that U-tailed RNAs may be degradation intermediates, their average size was smaller than that of the sRNAs in the entire population, which is dominated by untailed sequences (Fig. 3C and Fig. S8B). Different classes of AGO-associated sRNAs (phased siRNAs, transposon siRNAs, miRNAs) followed these trends (Tables S3–S5 in Dataset S1). In contrast, putative RNA degradation products (including those matching rRNAs, tRNAs, and chloroplast and mitochondria RNAs) showed a fairly similar distribution and abundance of 3′ untemplated sequences in the two strains examined (Tables S3 and S4 in Dataset S1). Likewise, the miRNA* population was not influenced by the MUT68 deletion (Tables S3 and S4 in Dataset S1). Overall these observations are consistent with an in vivo role of MUT68 in the 3′ terminal uridylation and subsequent degradation of certain, but not all, miRNAs and siRNAs. However, uridylation of sRNAs was not entirely abolished in the deletion mutant, likely reflecting redundant activities from other nucleotidyltransferases encoded in the Chlamydomonas genome (32).
Discussion
Deletion of MUT68 in Chlamydomonas results in five defects linked to RNAi: (i) increased accumulation of miRNAs and siRNAs (Fig. 1 and Fig. S2), (ii) reduced 3′ untemplated uridylation of small RNAs (Fig. 3B and Fig. S9), (iii) lack of oligoadenylation of RISC generated 5′ RNA cleavage products (23), (iv) stabilization of partly degraded 5′ RNA fragments from target transcripts (23), and (v) enhanced levels of uncleaved, full-length target transcripts (ref. 23 and Fig. S2B). The first four defects in the Mut-68 mutant can be explained by the function of MUT68 as a terminal nucleotidyltransferase (Fig. 2 A and B) that promotes the exonucleolytic degradation of miRNAs/siRNAs and of the 5′ RNA fragments resulting from RISC cleavage. MUT68 appears to collaborate with the RRP6 exosome subunit in the decay of small RNAs (Fig. 1 and Fig. 2 C and D) and with the core exosome in the turnover of long RNAs (ref. 23 and Fig. S6). Interestingly, in vivo, MUT68 seems to carry out preferentially uridylation of small RNAs (Fig. 3B and Fig. S9) and adenylation of RISC-cleaved transcripts (23). The basis for this differential specificity is presently unclear but nucleotidyltransferases with context-dependent nucleotide preferences have been previously described. For instance, human TUTase1 has been reported to adenylate mitochondrial mRNAs and to uridylate histone transcripts (33, 34). Moreover, recombinant S. pombe Cid1 can add either uridine or adenosine to RNA in vitro and its specificity is likely conferred by associated proteins because Cid1 complexes immunoprecipitated from cells only add uridine (35). In the context of sRNA degradation, 3′ terminal adenylation, unlike uridylation, has recently been proposed to lead to selective stabilization of mammalian miRNAs (36).
The last deficiency in Mut-68, increased levels of full-length siRNA-targeted transcripts (ref. 23 and Fig. S2B), is suggestive of a defect in RISC activity. Indeed, this mutant was originally identified as being deficient in RNAi (23) and the Rrp6-IR strains also show diminished RNA interference triggered by another IR transgene. Because Mut-68 contains enhanced levels of sRNAs in single-stranded conformation, which correlate with higher amounts of AGO3, RISC assembly appears to occur normally. As already mentioned, siRNAs seem to be loaded onto RISC as duplexes and then AGO cleaves the passenger strands triggering their dissociation from the complex and the concomitant maturation of RISC (1, 2, 37). Similarly, miRNA/miRNA* duplexes are loaded onto AGO and rapidly unwound by a poorly characterized mechanism (2, 37). No single-stranded siRNA or miRNA appears to be produced before this maturation step (2, 21, 24) and, thus, the single-stranded sRNAs detected in Chlamydomonas Mut-68 likely correspond to those associated with Argonautes. However, the function of a significant fraction of these RISC complexes may be compromised if the associated guide sRNAs are dysfunctional and/or damaged (see below), resulting in the sequestration of AGO proteins into inactive complexes. Although alternative explanations are possible, this interpretation for the diminished RNAi activity in Mut-68 (and in the Rrp6-IR strains) is consistent with a role for MUT68/RRP6 as a quality control mechanism for the removal of functionally defective sRNAs in Chlamydomonas. Moreover, this process may be operative in other eukaryotes because a recent RNAi screen to identify genes involved in miRNA/siRNA pathways in D. melanogaster revealed that depletion of an RRP6 homolog resulted in an RNAi defect (38). In addition, the C. elegans nucleotidyltransferase CDE-1 is required for the uridylation of siRNAs bound to a specific Argonaute protein (CSR-1) and in the CDE-1 absence these siRNAs accumulate to inappropriate levels, accompanied by defects in an RNAi pathway involved in chromosome segregation (39).
Untemplated nucleotide additions to sRNAs have been observed in animals, plants, and now algae (19, 36, 39 –42). The role of these modifications is poorly understood but our findings and those of others (18, 36, 39) suggest that they may influence the stability and/or the function of mature small RNAs. MUT68 depletion resulted in a defect in 3′ terminal uridylation of certain small RNAs and an alteration in their steady-state levels. We hypothesize that the MUT68 nucleotidyltransferase activity may be required for the degradation of sRNAs that are dysfunctional in a RISC environment (Fig. 4). Indeed, recent results suggest that RISC-bound sRNAs can be subfunctional. For instance, changing the 5′ uracil residue of the let-7a miRNA did not affect the formation of a complex with human AGO2 but reduced significantly the association of this complex with a target mRNA (43). In A. thaliana, it has been proposed that HEN1 methylates sRNA duplexes before their loading onto RISC (1, 20). In contrast, the D. melanogaster HEN1 appears to methylate single-stranded piRNAs and siRNAs already associated with certain AGO-PIWI proteins (1, 21, 22). Thus, some sRNAs lacking 2′-O-methyl groups are loaded onto RISC in animals and, conceivably, this may also happen for at least a fraction of the small RNAs in Chlamydomonas. In these cases, the MUT68/RRP6 machinery may act as a quality control mechanism in competition with HEN1 (Fig. 4A). Functional guide sRNAs (with respect to their interactions with AGO) may be protected by HEN1-mediated 3′ end methylation whereas subfunctional or dysfunctional sRNAs may be preferentially degraded by MUT68/RRP6. Kinetic competition with HEN1 may also result in the decay of a fraction of functional sRNAs. In the absence of MUT68, these small RNAs would be methylated by HEN1 and stabilized, consistent with the fact that miRNAs and siRNAs are resistant to periodate oxidation and β elimination in the mutant background (Fig. S4B).
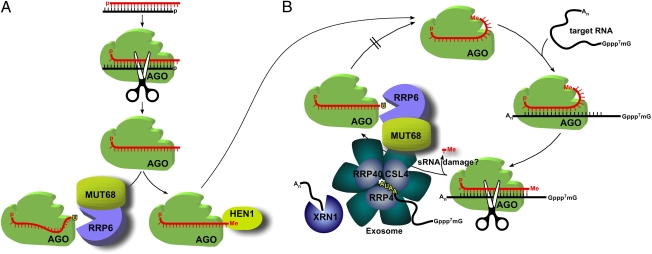
Fig. 4.
Proposed model for the role of MUT68 and RRP6 in the degradation of mature miRNAs and siRNAs (see text for details). (A) MUT68/RRP6 may eliminate dysfunctional or subfunctional guide small RNAs in kinetic competition with the methyltransferase HEN1 (after cleavage and dissociation of the passenger strand during RISC loading of siRNAs or perfectly complementary miRNA duplexes). (B) MUT68/RRP6 may also degrade damaged guide sRNAs during the multiple cycles of RISC activity.
In degradative RNAi, RISC functions as a multiple turnover enzyme (2, 44) and a quality control mechanism(s) may also be necessary to assess the integrity of guide siRNAs after each round of target RNA cleavage (Fig. 4B). In mature RISC, the 3′ end of the guide siRNA is bound by the AGO PAZ domain but, when the siRNA forms an extensive duplex with a target RNA, its 3′ terminus is released from the PAZ pocket (6, 7, 37) (Fig. 4B). After RISC-mediated endonucleolytic cleavage, the target RNA products are released and degraded by exoribonucleases (23, 45). At this step, the 3′ end of the guide siRNA may become accessible to the MUT68/RRP6 machinery before rebinding to the PAZ domain (Fig. 4B). We speculate that MUT68/RRP6 may also operate here, as a quality control mechanism to degrade damaged sRNAs lacking 2′-O-methyl groups. However, addressing the molecular details of the proposed mechanism (Fig. 4) will require further experimentation because the nature of the putative dysfunctional or damaged small RNAs is not clear.
Materials and Methods
Transgenic Strains, Mutants, and Culture Conditions.
The isolation of Mut-68 has been reported (23) and transgenic strains containing inverted repeat constructs homologous to CSL4, RRP40, RRP6a, or RRP6b were generated as previously described (46). The Chlamydomonas genome contains two genes encoding RRP6 homologs: RRP6a (511862) and RRP6b (284930) (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). DNA fragments for building the IR constructs were generated by RT-PCR amplification with the following primers: for CSL4, Csl4-1 (5′-CGGATACATACGCTGCTGGAG-3′) and Csl4-2 (5′-CAACCGCACAACTACCTGCTC-3′); for RRP40, Rrp40-1 (5′-TCGCTTAAAGGCGGGGTATTA-3′) and Rrp40-2 (5′-GCCTAGCACGAAGCTGAACAA-3′); for RRP6a, Rrp6a-sca27-F1 (5′-ACATGGCCGCAGACAAGG-3′) and Rrp6a-sca27-R1 (5′-CTTGCGGTACAGCGTGAGG-3′); and for RRP6b, Rrp6b-sca11-F1 (5′-CCGGGACTACATCCTGGACT-3′) and Rrp6b-sca11-R1 (5′-CGGTAGGTCTTGAGGCACAG-3′). For all analyses, unless noted otherwise, C. reinhardtii cells were grown photoheterotrophically in Tris-acetate-phosphate medium (4, 46).
RNA Analyses.
Total cell RNA was purified with TRI reagent (Molecular Research Center). For Northern analysis, the isolated RNA was separated by agarose/formaldehyde gel electrophoresis, blotted onto nylon membranes, and hybridized with 32P-labeled probes (4, 46, 47). Small RNAs, fractionated through Microcon YM-100 centrifugal devices (Millipore), were resolved on 15% polyacrylamide/7 M urea gels, and electroblotted to Hybond-XL membranes (GE Healthcare) (23, 46, 47). Blots were hybridized with 32P-labeled DNA probes at 40 °C for 48 h using the High-Efficiency Hybridization System (Molecular Research Center). Specific miRNAs or miRNA*s were detected by hybridization with DNA oligonucleotides labeled at their 5′ termini with γ-[32P]ATP and T4 Polynucleotide Kinase (New England Biolabs). For the analysis of duplex RNA, total cell RNA was purified with TRI reagent, resuspended in TE buffer, and separated on a 16% native polyacrylamide gel (47). Parallel samples were resuspended in formamide and heated to 75 °C before loading onto the native gel. After electrophoresis, the gel was incubated for ∼10 min in 50 mM NaOH and electroblotted onto Hybond-XL membranes for Northern hybridization (46, 47). Control miR912/miR912* duplexes were generated by annealing synthetic oligoRNAs (miR912, UGGAUUGAUCCCAGCCAGGCG; miR912*, CCUGGCUGGGAUCAAUGCAAG) in 10 mM Tris-HCl (pH 7.5), 100 mM NaCl and 0.2 mM EDTA (47). The annealed duplexes were purified by the same protocol as total RNA, before gel loading, to ensure that our RNA extraction procedure preserves the integrity of small dsRNAs.
Immunoblot Analysis.
The Chlamydomonas AGO3 protein was immunodetected, following a standard protocol (46), by overnight incubation at 4 °C with a 1:10,000 dilution of a rabbit antibody raised against a C-terminal peptide (ASRSGRGAGAAEGG) conjugated to KLH (GenScript).
Terminal Nucleotidyltransferase Assays.
A DNA fragment corresponding to the coding sequence of MUT68 was subcloned into pET30c(+) and transformed into E. coli BL21(DE3), according to the manufacturer’s protocol (Novagen). To generate MUT68(DADQ), point mutations were introduced in the putative catalytic site (D68A and D70Q) of the wild-type sequence (23) by using the QuikChange XL mutagenesis kit (Stratagene). Recombinant proteins were purified under denaturing conditions on nickel-nitrilotriacetic acid agarose His-binding columns and refolded while attached to the solid matrix (48). These proteins were then used for poly(A) polymerase assays following a described protocol (26, 49). Briefly, reaction mixtures contained 20 mM Tris-HCl (pH 7.8), 50 mM KCl, 0.5 mM DTT, 0.7 mM MnCl2, 2.5 mM MgCl2, 200 μg/mL BSA, 0.3 μM synthetic 32P-labeled RNA substrates, and 0.5 mM of different nucleotide triphosphates. Reactions were incubated at 25 °C for 5–40 min, stopped by the addition of formamide/EDTA gel-loading buffer, and the products analyzed on 15% polyacrylamide/urea sequencing gels (47). Oligoribonucleotide substrates: C15A10, CCCCCCCCCCCCCCCAAAAAAAAAA; miR912, UGGAUUGAUCCCAGCCAGGCG; and 2′-O-Me miR912, UGGAUUGAUCCCAGCCAGGCmG.
RRP6 Exosome Subunit Assays.
Recombinant His-tagged RRP6 (PM/Scl100 human), expressed in baculovirus-infected S. frugiperda cells, was obtained from ProSpec. For the exoribonuclease assays, an empty pGBKT7 plasmid (Clontech) or pGBKT7 plasmids encoding MUT68 or MUT68(DADQ) were incubated with unlabeled methionine in the TNT T7 Quick Coupled Transcription/Translation System for 90 min at 30 °C, following the manufacturer’s directions (Promega). Recombinant RRP6 (∼50 ng) and 5′-32P-labeled synthetic oligoRNAs were then added to the reactions and the buffer adjusted to 10 mM Tris-HCl (pH 8.0), 10 mM DTT, 50 mM KCl, 5 mM MgCl2, 0.5 mM UTP, and 1 unit/μL RNase inhibitor (Ambion) (50). Incubation was continued at 30 °C and 5-μL aliquots were removed at the indicated times (Fig. 2 C–E and Fig. S7). Samples were quenched with formamide/EDTA gel-loading buffer and resolved on 15% polyacrylamide/urea sequencing gels (47). These gels were dried on blotting paper and the radioactivity detected with a PhosphorImager (Molecular Dynamics).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (H.C.) and the National Science Foundation (H.C. and P.J.G.). We also acknowledge the support of the Nebraska Experimental Program to Stimulate Competitive Research (EPSCoR).
Footnotes
The authors declare no conflict of interest.
Data deposition: The deep sequencing libraries reported in this paper have been deposited in NCBI GEO (accession no. GSE17815).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912632107/DCSupplemental.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 4.Casas-Mollano JA, et al. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics. 2008;179:69–81. doi: 10.1534/genetics.107.086546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 6.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiti M, Lee H-C, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans . Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 13.Iida T, Kawaguchi R, Nakayama J. Conserved ribonuclease, Eri1, negatively regulates heterochromatin assembly in fission yeast. Curr Biol. 2006;16:1459–1464. doi: 10.1016/j.cub.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 14.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis . Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehrbach NJ, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans . Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones MR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis . Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 22.Saito K, et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- 24.Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii . Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 26.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 27.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 28.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chekanova JA, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 30.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao T, et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii . Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer SL, Fei Z, Stern DB. Genome-based analysis of Chlamydomonas reinhardtii exoribonucleases and poly(A) polymerases predicts unexpected organellar and exosomal features. Genetics. 2008;179:125–136. doi: 10.1534/genetics.107.086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 34.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rissland OS, Mikulaslova A, Norbury CJ. Efficient RNA polyuridylation by non-canonical poly(A) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh T, et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 38.Zhou R, et al. Comparative analysis of Argonaute-dependent small RNA pathways in Drosophila . Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both microRNAs and their miRNA* strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felice KM, Salzman DW, Shubert-Coleman J, Jensen KP, Furneaux HM. The 5′ terminal uracil of let-7a is critical for the recruitment of mRNA to Argonaute2. Biochem J. 2009;422:329–341. doi: 10.1042/BJ20090534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 45.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohr J, Sarkar N, Balenger S, Jeong B-R, Cerutti H. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas . Plant J. 2004;40:611–621. doi: 10.1111/j.1365-313X.2004.02227.x. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW. Molecular Cloning – A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 48.Holzinger A, Phillips KS, Weaver TE. Single-step purification/solubilization of recombinant proteins: Application to surfactant protein B. Biotechniques. 1996;20:804–806. doi: 10.2144/96205bm16. [DOI] [PubMed] [Google Scholar]
- 49.Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: Identification of functional domains by mutational analysis. J Biol Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.