Abstract
MicroRNA (miRNA)-17-92 cluster (miR-17-92), containing seven individual miRNAs, is frequently amplified and overexpressed in lymphomas and various solid tumors. We have found that it is also frequently amplified and the miRNAs are aberrantly overexpressed in mixed lineage leukemia (MLL)-rearranged acute leukemias. Furthermore, we show that MLL fusions exhibit a much stronger direct binding to the locus of this miRNA cluster than does wild-type MLL; these changes are associated with elevated levels of histone H3 acetylation and H3K4 trimethylation and an up-regulation of these miRNAs. We further observe that forced expression of this miRNA cluster increases proliferation and inhibits apoptosis of human cells. More importantly, we show that this miRNA cluster can significantly increase colony-forming capacity of normal mouse bone marrow progenitor cells alone and, particularly, in cooperation with MLL fusions. Finally, through combinatorial analysis of miRNA and mRNA arrays of mouse bone marrow progenitor cells transfected with this miRNA cluster and/or MLL fusion gene, we identified 363 potential miR-17-92 target genes that exhibited a significant inverse correlation of expression with the miRNAs. Remarkably, these potential target genes are significantly enriched (P < 0.01; >2-fold) in cell differentiation, hematopoiesis, cell cycle, and apoptosis. Taken together, our studies suggest that overexpression of miR-17-92 cluster in MLL-rearranged leukemias is likely attributed to both DNA copy number amplification and direct up-regulation by MLL fusions, and that the miRNAs in this cluster may play an essential role in the development of MLL-associated leukemias through inhibiting cell differentiation and apoptosis, while promoting cell proliferation, by regulating relevant target genes.
Keywords: cell apoptosis and viability, colony-forming/replating assay, MLL binding, gene regulation, miRNA target
MicroRNAs (miRNAs, miRs) are endogenous ≈22 nucleotides (nt) noncoding RNAs that play important regulatory roles in animals and plants by binding with the 3′ UTRs of messenger RNAs (mRNAs) of target genes, leading to mRNA cleavage/degradation or translational repression (1 –4). Rapidly accumulating evidence has revealed that miRNAs are strongly associated with cancer (3, 5 –7). Recent studies suggest that a cluster of miRNAs, the miR-17-92 polycistron located at 13q31 [containing seven individual miRNAs including miR-17-5p (now named miR-17), miR-17-3p (now named miR-17*), miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1], may function as an oncogene (8 –12). Particularly, He et al. (8) showed that enforced expression of the miR-17-92 cluster cooperated with Myc expression to accelerate tumor development in a mouse model of human B-cell lymphoma. Its oncogenic function is further supported by the finding that miRNAs from this cluster are overexpressed in lung, breast, colon, pancreas, prostate tumors, and chronic leukemias, and that overexpression of these miRNAs is usually correlated with the amplification of its genomic locus (i.e., 13q31 locus) (9, 10, 12 –14). Furthermore, these miRNAs can enhance cell proliferation and inhibit cell differentiation (9 –12, 15). However, the involvement of this miRNA cluster in the development of acute leukemia remains unclear.
Recently, in a large-scale, genome-wide study of miRNA expression profiling in acute leukemia samples, we observed that the miRNAs from the miR-17-92 cluster are frequently overexpressed in MLL (mixed lineage leukemia)-rearranged acute myeloid leukemias (AMLs) (16). The MLL gene, located at 11q23, is frequently involved in cytogenetic abnormalities in both AML and acute lymphoblastic leukemia (ALL), occurring in 5–6% of patients with AML, 7–10% of ALL, 60–70% of all acute leukemias in infants, and in the majority of patients with t-AML/t-ALL secondary to therapy that targets topoisomerase II (like etoposide) (17, 18). MLL-rearranged leukemia is classified as a disease of poor prognosis (19, 20), and cure rates for ALL patients with MLL rearrangements remain dismal (typically ≈20% and only one-third of AML patients with MLL rearrangements will survive longer than 5 years) (21). More than 60 different loci have been identified to translocate to the MLL gene locus (22 –25). The critical feature of these chromosomal rearrangements is the generation of a chimeric transcript consisting of 5′ MLL and 3′ sequences of the gene on the partner chromosome. Experimental data shows that MLL fusions inhibit hematopoietic differentiation in serial replating assays, a critical surrogate parameter for transformation activity (26, 27). Although MLL-associated leukemias have been intensively studied, how miRNAs contribute to their development and how they can contribute or trigger the clonogenic capacity of MLL-fusion dependent clones are largely unknown.
We previously reported that the individual miRNAs of the miR-17-92 cluster were overexpressed in the majority of MLL-rearranged AML (16, 28). In the present study, we demonstrate that the miR-17-92 cluster is highly expressed not only in MLL-associated AML, but also in MLL-associated ALL. We further show that both 13q31 amplification and up-regulation by MLL fusions may contribute to the overexpression of the miR-17-92 cluster in MLL-rearranged leukemias. Moreover, we show that miR-17-92 can significantly increase cell viability, while inhibiting apoptosis of human HeLa cells and 293T cells, and significantly enhance proliferation of mouse normal bone marrow progenitor cells, resulting in transformation of these cells by miR-17-92 alone and particularly in cooperation with MLL fusions. Finally, we identified a group of potential target genes of this miRNA cluster and we found that they were significantly enriched in pathways related to cell differentiation, hematopoiesis, cell cycle, and apoptosis.
Results
The miR-17-92 Cluster Is Particularly Overexpressed in MLL-Rearranged Acute Leukemias.
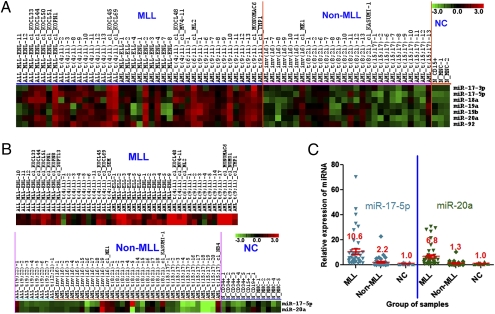
We have shown that the miR-17-92 cluster is particularly overexpressed in MLL-rearranged AMLs (16). Here, we also describe a similar pattern in MLL-rearranged ALL samples. As shown in Fig. 1A, the seven miRNAs in the miR-17-92 cluster (miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92) are all highly expressed in the majority of MLL-rearranged leukemia samples, including MLL-ELL/t(11;19)(q23;p13.1), MLL-ENL/t(11;19)(q23;p13.3), MLL-AF4/t(4;11), and MLL-AF9/t(9;11) in both ALLs and AMLs. We then performed a TaqMan quantitative real-time PCR (qPCR) to validate the expression of miR-17-5p and miR-20a (two representative members of the miR-17-92 cluster) in 97 samples, including 85 acute leukemia and 12 normal control samples. Of them, 63 (i.e., 67%) were independent samples that were not used in the bead-based miRNA profiling assay. The 85 leukemia samples included 44 MLL-rearranged (19 ALL and 25 AML) and 41 other [8 ALL, including 7 t(9;22) and one t(12;21), and 33 AML, including 11 each of inv (16), t(8;21), and t(15;17)] acute leukemias. The 12 normal control samples included 5 CD34+ hematopoietic stem/progenitor, one CD15+ myeloid progenitor, one CD19+ B-cell progenitor, and 5 mononuclear cell samples. As shown in Fig. 1B, both miR-17-5p and miR-20a are highly expressed in the majority of MLL-rearranged leukemia samples. In addition, the average expression level of miR-17-5p and miR-20a in 44 MLL-rearranged leukemia samples are 10.6- and 6.8-fold higher than those of the 12 normal control samples, respectively, whereas those in the 41 non-MLL-rearranged leukemia samples are only 1.3- to 2.2-fold higher than those of the normal controls (see Fig. 1C).
Fig. 1.
Overexpression of the miR-17-92 cluster in MLL-rearranged leukemias. (A) Expression profiling of the miRNA cluster in 72 acute leukemic and normal samples as detected by the bead-based method. (B) Expression profiling of the miRNA cluster in 85 leukemia and 12 normal samples as detected by qPCR assay. Expression data were mean-centered, and the relative value for each sample is represented by a color, with red representing a high expression and green representing a low expression (scale shown in the upper right). qPCR data are presented as ΔC T. cl_, cell line; N_, normal control; CD34+, CD34+ hematopoietic stem/progenitor cells; CD15+, CD15+ myeloid progenitor cells; CD19+, CD19+ B-cell progenitor cells; MNC_, mononuclear cells; MLL, MLL-rearranged acute leukemias; Non-MLL, non-MLL-rearranged acute leukemias; NC, normal control. (C) Relative expression level of miR-17-5p and miR-20a in MLL-rearranged, nonMLL-rearranged, and normal control samples. The relative expression level is a fold change (2−ΔΔCt) in expression level of miR-17-5p or miR-20a in a given sample compared with the average expression level in normal controls. Mean ± SE values are shown.
Overexpression of the miR-17-92 Cluster in MLL-Rearranged Leukemias Is at Least Partly Associated with DNA Copy Number Amplification, but Not with DNA Methylation.
To gain insights into the mechanism that underlies the overexpression of the miR-17-92 cluster in MLL-rearranged leukemias, we first examined the DNA copy number of the miRNA cluster locus at 13q31.3 in 46 samples, including 31 (17 MLL- and 14 non-MLL-rearranged) primary leukemia samples, 13 (10 MLL- and 3 non-MLL-rearranged) leukemic cell lines, and 2 normal control samples. The average DNA copy number of the locus of this miRNA cluster is significantly greater (Mann–Whitney test, two-tailed, P = 0.018) in MLL-rearranged leukemia samples than the others. Specifically, among the 27 MLL-rearranged leukemia samples, 17 (63%) have over a 2-fold amplification of the genomic DNA locus relative to that of the normal controls; in contrast, only 41% (7 of 17) of non-MLL-rearranged leukemia samples have >2-fold increase of gene dosage (Fig. S1A). The difference of DNA copy number between MLL-rearranged leukemias and the other samples is still significant (Mann–Whitney test, two-tailed, P = 0.031) even after excluding all of the cell line samples. We further investigated the relationship between DNA copy number and expression level of this miRNA cluster. As shown in Fig. S1B, the average expression level of the seven miRNAs was significantly positively correlated (correlation coefficient r = 0.48; two-tailed, P = 0.0008; Spearman’s Rank Correlation test) with the average DNA copy number of the miR-17-92 locus. Thus, these results suggest that the overexpression of the miR-17-92 cluster in MLL-rearranged leukemias is associated, at least partly, with the genomic locus amplification.
In addition, we also investigated whether differential expression of the miRNAs in this cluster was associated with epigenetic regulation. There is a large CpG island with 218 CpG dinucleotides located upstream (within 2 kb) of the miR-17-92 cluster. We analyzed the DNA sequences of this CpG island by pyrosequencing after bisulfite treatment of genomic DNA from 24 samples, including 12 MLL-rearranged leukemia, 9 non-MLL-rearranged leukemia, and 3 normal control samples. As shown in Fig. S2, the average methylation level of the CpG island showed no significant difference (P = 0.11; two-tailed t test) between MLL-rearranged (2.8%) and non-MLL-rearranged (3.3%) samples, suggesting that the overexpression of miR-17-92 in MLL-rearranged leukemia is not a consequence of this CpG island demethylation.
MLL and, Particularly, MLL Fusions Bind to the Promoter Region of miR-17-92 and Promote Expression of the miRNAs Directly.
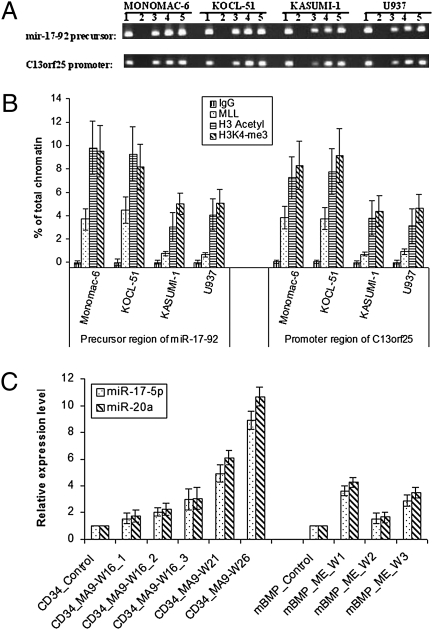
MLL belongs to the Trithorax Group (trx-G) of proteins, which antagonize the repressive function of the Polycomb Group (Pc-G) proteins and are responsible for positive maintenance of gene expression during development. To determine whether MLL can bind the locus of the miR-17-92 cluster and promote its expression directly, we performed a conventional chromatin immunoprecipitation (ChIP) assay. As shown in Fig. 2A, localization of MLL, histone H3 acetylation, and H3K4 trimethylation was observed in the promoter region of the miR-17-92 host gene (i.e., C13orf25) as well as directly upstream (0-500 bp) of the precursor region of the miRNA cluster in cell lines with and without MLL fusion genes. Furthermore, based on qPCR assays, we observed a substantial increase in the levels of MLL binding (4- to 6-fold), histone H3 acetylation (2- to 3-fold), and H3K4 trimethylation (1.5- to 2-fold) in cell lines with MLL fusions (i.e., MONOMAC-6 and KOCL-51) compared to those with wild-type MLL (i.e., KASUMI-1 and U937; Fig. 2B), which is similar, albeit relatively less than the regulation of HOXA9 by wild-type MLL and MLL fusion proteins reported by others (29). Thus, our ChIP assay results suggest that expression of the miR-17-92 cluster might also be regulated directly by wild-type MLL and, particularly, MLL fusion proteins through direct binding and chromatin modification. Consistent with the above ChIP analysis results, the expression levels of miR-17-5p and miR-20a were significantly increased 16–26 weeks after transfection of MLL-AF9 (MA9) fusion gene into human CD34+ cells (1.5 to 11-fold) or 1–3 weeks after transfection of MLL-ELL (ME) into mouse bone marrow progenitor (mBMP) cells (1.5- to 4-fold) (Fig. 2C), suggesting that MLL fusions can upreguate expression of miR-17-92 directly.
Fig. 2.
MLL and, particularly, MLL fusions bind to the miR-17-92 cluster locus and up-regulate expression of the miRNAs directly. (A) ChIP analysis of MLL, histone H3 acetylation, and histone H3K4 trimethylation at the miR-17-92 cluster locus. Purified ChIP DNA was amplified by PCR and detected by ethidium bromide staining after electrophoresis on 2.5% agarose gels. PCR templates used are as follows: 1, input chromatin DNA (5% of total chromatin DNA); 2, negative control DNA bound by antibodies against IgG; 3, DNA bound by antibodies against MLL N-terminal; 4, DNA bound by antibodies against H3 acetylation; 5, DNA bound by antibodies against H3K4 trimethylation. Two sets of primers were used to amplify a 100- to 190-bp region directly upstream (0-500 bp) of the miR-17-92 precursor region, and directly upstream (0–500 bp 5′ to the first exon) of the host gene C13orf25, respectively. (B) Purified ChIP DNA was amplified by quantitative real-time PCR using the same primer pairs. (C) Direct up-regulation of miR-17-5p and miR-20a by MLL fusions (MA9, MLL-AF9; ME, MLL-ELL) in human CD34+ umbilical cord blood cells (CD34_) and mouse bone marrow progenitor (mBPG_) cells, respectively, at different time points (W, week; h, hour) after retroviral transduction (for human CD34+ and mouse bone marrow progenitor cells). W16_1, 2, and 3 are three different transduction at 16 weeks after transduction.
miR-17-92 Inhibits Apoptosis and Increases Cell Viability.
To examine the functional role of the miR-17-92 cluster in human cells, we performed gain- and loss-of-function experiments in human cell lines. We observed that forced expression of the miR-17-92 cluster inhibited apoptosis (Fig. S3A) but increased cell viability (Fig. S3B) of HeLa (a human cervical cancer cell line) and 293T (a variant of human embryonic kidney 293 cell line) cells significantly. Forced expression of the individual miRNAs after transfection was confirmed by qPCR (Fig. S3C).
miR-17-92 Enhances the Colony Forming Capacity of Mouse Normal Bone Marrow Progenitor Cells Alone and, Particularly, in Cooperation with MLL Fusions.
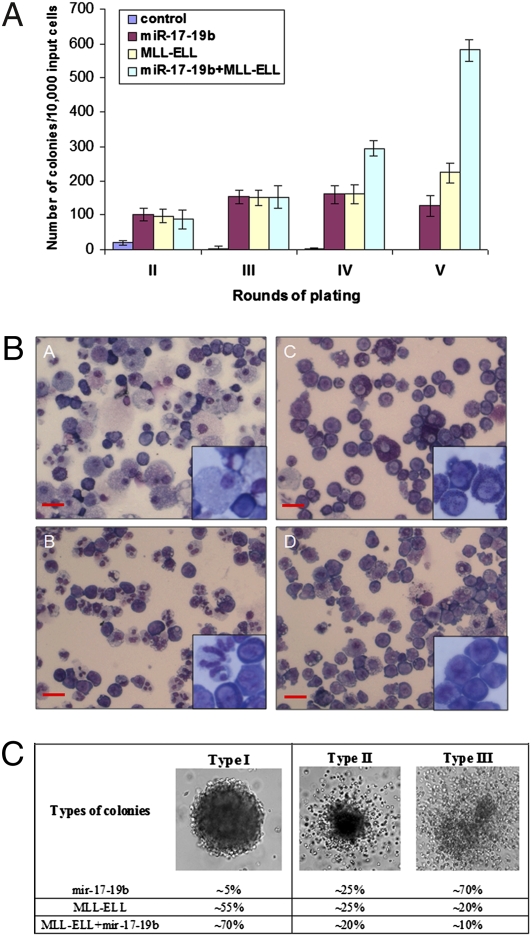
To evaluate the role of the miR-17-92 cluster in leukemogenesis further, we conducted colony-forming/replating (i.e., immortalization) assays. Mouse bone marrow progenitor cells were retrovirally transduced with (i) MSCVpuro or MSCVneo empty vector (i.e., control), (ii) MSCVpuro-miR-17-19b (i.e., miR-17-19b; a functional truncated form of the miR-17-92 cluster that only lacks miR-92; ref. 8), (iii) MSCVneo-MLL-ELL (i.e., MLL-ELL), and (iv) MSCVpuro-miR-17-19b and MSCVneo-MLL-ELL (i.e., miR-17-19b+MLL-ELL), respectively (Materials and Methods). As shown in Fig. 3A, forced expression of miR-17-19b alone could result in a significant number (>100) of colonies. Remarkably, cotransfection of miR-17-19b plus MLL-ELL fusion gene resulted in many more colonies (300–500 vs. 100–200; Fig. 3A) and almost twice as many cells per colony than transduction of each alone after four rounds of plating (i.e., in the fourth and fifth round of plating). Their difference is not significant in the second and third rounds of plating, which likely is owing to a much lower efficiency of cotransduction of miR-17-19b plus MLL-ELL than that of transduction of each alone. Transduction of the miR-17-92 entire cluster alone or together with MLL-ELL yielded a similar result. The forced expression of the miRNAs and/or MLL-ELL was confirmed in relevant cells by qPCR. As shown in Fig. 3B, cells of the secondary colonies with empty vector (i.e., control; plot A) were mostly differentiated; whereas cells of the tertiary colonies with miR-17-19b (plot B) were partially differentiated with some granulocytes, while cells of the tertiary colonies either with MLL-ELL alone (plot C) or with MLL-ELL plus miR-17-19b (plot D) displayed a very immature morphology (basophilic cytoplasm, large nucleus, evident nucleolus, and lacy chromatin). Regarding to colony morphology, the majority (≈70%) of miR-17-19b tertiary colonies were diffuse-type colonies of migrating cells (i.e., Type III), whereas the majority of tertiary colonies of MLL-ELL (≈80%), and particularly of MLL-ELL plus miR-17-19b (≈90%) were compact and dense-type colonies (i.e., Types I and II) (27) (see Fig. 3C). These results suggest that miR-17-92 can only partially block cell differentiation, whereas MLL-ELL can strongly block cell differentiation as reported (26). Taken together, our data indicate that there is a synergistic effect between miR-17-92 and MLL-ELL in both enhancing cell proliferation and blocking cell differentiation.
Fig. 3.
In vitro colony-forming/replating assays. (A) Numbers of colonies per dish (≥50 cells/colony; 1 × 104 input cells) of the second through the fifth replating are shown (mean ± SD). (B) Morphology of cells of secondary or tertiary colonies (i.e., colonies formed at the second or third round of plating). Cells of secondary colonies with empty vector (Plot A); cells of tertiary colonies with miR-17-19b (Plot B); cells of tertiary colonies with MLL-ELL (Plot C); and cells of tertiary colonies with miR-17-19b and MLL-ELL (Plot D). Cells were stained with Wright-Giemsa. (Scale bars: 10 μm.) (C) Morphological types and proportions of colonies. According to Lavau et al. (27), there were three morphological types of secondary or tertiary colonies generated by MSCV-MLL-fusion-infected hematopoietic cells. Similarly, we have also found three types of such tertiary colonies: (I) extremely compact and resembled colonies; (II) colonies with a dense center surrounded by a halo of migrating cells; and (III) diffuse colonies of migrating cells. Lavau et al. (27) found that colonies of types I and II consisted of immature cells whereas the colonies of types III also included more differentiated cells.
Potential Target Genes of the miR-17-92 Cluster and Their Relevant Pathways in Leukemogenesis.
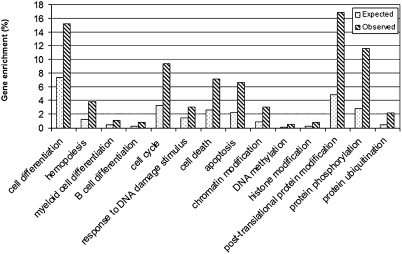
To identify potential critical target genes of this miRNA cluster in the colony-forming/replating assays, we assessed expression profiles of both miRNAs and mRNAs in the cells transduced with this miRNA cluster, an MLL fusion gene, both of them, and empty vector, by using microarrays. By use of Pearson Correlation test, we identified 363 potential target genes of the miR-17-92 cluster that exhibited significant negative correlation (correlation coefficient r <−0.5; P < 0.01) of expression with the corresponding individual miRNAs of the cluster (Table S1). All of the 363 genes are potential direct targets of miR-17-92 as predicted by at least one of four major miRNA-target prediction programs (i.e., PITA, TargetScan, Miranda, and miRbase Targets). In analysis of GO process enrichment, we found that these potential targets are significantly enriched (2- to 6-fold higher than expected by chance) in cell differentiation, particularly hematopoiesis (including both myeloid and B cell differentiation), cell cycle, response to DNA damage stimulus, cell death and apoptosis, chromatin modification (including DNA methylation and histone modification), and posttranslational protein modification (including protein phosphorylation and protein ubiquitination) (see Fig. 4). The relevant pathways/networks of the miR-17-92 potential target genes are enriched in cell differentiation, hematopoiesis, cell cycle, and apoptosis are shown in Fig. S4.
Fig. 4.
Representatives of GO processes in which the miR-17-92 putative target genes are significantly enriched. An expected enrichment in a given GO process is the proportion of the 27,792 genes (i.e., the entire set of mouse genes that are predicted miRNA targets) enriched in that GO process, whereas the observed enrichment is the proportion of the 363 potential miR-17-92 targets enriched. All of the observed proportions are significantly (P < 0.01) greater than the corresponding expected proportions.
Discussion
Although large-scale, global miRNA expression profiling assays have reported the correlation of signatures of many miRNAs with cytogenetic and molecular subtypes of acute leukemia, as well as patient response to treatment (16, 30 –37), the mechanisms underlying the deregulation of and the function of individual miRNAs in acute leukemia are largely unknown (7).
MLL-rearranged acute leukemias are a type of very aggressive disease. In the present study, we first showed that the individual miRNAs of the miR-17-92 cluster were aberrantly expressed in MLL-rearranged acute leukemias (see Fig. 1). We then investigated the potential mechanisms underlying their overexpression in MLL-rearranged acute leukemias. The overexpression of this miRNA cluster in various cancers such as lung, breast, colon, pancreas, and prostate tumors is usually correlated with the DNA amplification of its genomic locus (i.e., 13q31 locus) (9, 10, 12 –14). As expected, we have also observed that overexpression of the miR-17-92 cluster in MLL-rearranged leukemias is also, at least partly, associated with the genomic locus amplification (Fig. S1).
In addition, because MLL is a well-known transcription factor, we further investigated whether MLL and, particularly, MLL fusions could up-regulate expression of the miR-17-92 cluster directly. Indeed, MLL belongs to the Trithorax Group (trx-G) of proteins, which antagonizes the repressive function of the Polycomb Group (Pc-G) proteins and positively regulates gene expression through chromatin association and modification. The best studied downstream targets of MLL and trx function are the homeobox (HOX) genes such as HOXA9, as well as the HOX cofactor MEIS1 (25). MLL is believed to function at the level of chromatin organization through its C-terminal SET domain that possesses intrinsic histone methytransferase activity specific for H3K4; trimethylation of H3K4 is a long-lasting marker associated with an active gene (38). Because MLL fusion proteins lack the SET domain, it was previously thought that they should prevent methylation of H3K4 (25). However, recent studies from others (29, 39) showed that leukemogenic MLL fusion proteins bind across a broad region of the Hoxa9 locus, and enforced expression of MLL fusions is associated with increased levels of MLL binding (5- to 15-fold), histone H3 acetylation (2- to 5-fold) and H3K4 methylation (1- to 2-fold) at target genes, such as Hoxa9 and Meis1. As a consequence, MLL fusion proteins forced persistent expression of target genes, including Hoxa9 and Meis1, which are critical for leukemogenesis (40). The mechanism underlying the finding that Hoxa9 and other targets (e.g., Meis1) show high, rather than low level of H3K4 trimethylation in the presence of MLL fusions is not clear; It is probably through recruitment of wild-type MLL by MLL fusion proteins to transcriptionally active loci (29) and/or due to the function of MLL partner genes such as AF4, AF9, ENL, and ELL. In an independent study, Guenther et al. (41) performed a genome-wide ChIP-on-chip analysis by using a human promoter DNA microarray and found that MLL functions as a human equivalent of yeast Set1; like Set1, MLL binds near the transcriptional start sites of most PolII occupied genes (including the locus of the miR-17-92 cluster), and this binding seems to correlate with gene expression. Consistent with the finding from this ChIP-on-chip assay (41), our conventional ChIP assay clearly demonstrated that wild-type MLL and, particularly, MLL fusion proteins bind directly to the locus of the miR-17-92 cluster (see Fig. 2 A and B) and promote the expression of the individual miRNAs directly, as evidenced by our findings regarding miR-17-5p and miR-20a shown in Fig. 2C.
Epigenetic regulation such as DNA methylation and histone modification also play an essential role in regulating expression of some miRNAs in acute leukemias (7), such as miR-128 (30) and miR-126 (16). Nonetheless, we found that the average methylation level of the CpG island located upstream (within 2 kb) of the miR-17-92 cluster showed no significant difference between MLL-rearranged (2.8%) and non-MLL-rearranged (3.3%) or normal control samples (Fig. S2). Therefore, our studies indicate that overexpression of the miR-17-92 cluster in MLL-rearranged leukemias is likely attributed to both 13q31 amplification and up-regulation by MLL fusions, but not DNA demethylation.
We have shown that forced expression of the miR-17-92 cluster could significantly enhance viability and inhibit apoptosis of human HeLa and 293T cells (Fig. S3), and more importantly, could significantly increase proliferation and inhibit differentiation of mouse normal bone marrow progenitor cells alone and especially, in cooperation with MLL fusions, leading to transformation of the cells (Fig. 3). Indeed, in normal hematopoiesis, miR-17-92 has been reported to play an essential role in monocytopoiesis (42) and megakaryocytopoiesis (43), and in B cell development (44 –46). miR-17-92 is down-regulated during monocytopoiesis and megakaryocytopoiesis, and its forced expression represses monocytopoiesis (42) and megakaryocytopoiesis (43). miR-17-92 also inhibits B cell development at the pro-B to pre-B transition (44 –46). Together, our data and those of others suggest that aberrant overexpresion of miR-17-92 in MLL-rearranged acute leukemia would promote cell proliferation and inhibit normal hematopoiesis and, thereby, may play an important role in the development of MLL-rearranged leukemias.
Revealing the critical target genes and relevant pathways/networks is pivotal to understanding the mechanisms by which the miRNA cluster plays a role in leukemogenesis. A group of target genes of miR-17-92 have been reported such as E2F1, E2F2 and E2F3 (47 –49), PTEN (14, 44 –46), BIM (BCL2L11), (44 –46), AML1 (42), TGFBR2 (TβRII) (11, 50), NCOA3 (AIB1) (51), RBL2 (P130) (10), THBS1 (TSP1) and CTGF (52) in other types of cancers or during normal cell development. In acute leukemias, we reported that RASSF2 and RB1 are direct targets of miR-17-92 (28). In the present study, we performed a systematic study through combinatorial analysis of miRNA and mRNA arrays of mouse bone marrow progenitor cells transduced with this miRNA cluster, MLL fusion gene, both of them, or empty vector. We identified 363 potential target genes that exhibit a significant inverse correlation of expression with the miRNAs. Remarkably, the miR-17-92 potential targets are significantly enriched in cell differentiation, hematopoiesis, cell cycle, and apoptosis. (Fig. 4 and Fig. S4), which is consistent with the critical role of miR-17-92 in normal hematopoiesis and its oncogenic role in tumorigenesis.
In sum, our studies suggest that overexpression of the miR-17-92 cluster in MLL-rearranged leukemias is attributable to both DNA copy number amplification and a direct up-regulation by MLL fusions, but not through DNA demethylation. In addition, the miRNAs in the miR-17-92 cluster may play an essential role in the development of MLL-rearranged acute leukemia likely through negatively regulating targets that are positive regulators of cell differentiation (hematopoiesis) and apoptosis, or negative regulators of cell proliferation.
Materials and Methods
See SI Text for more details on materials and methods used.
Leukemic and Normal Control Samples.
All of the patient samples were obtained at the time of diagnosis or relapse and with informed consent at the University of Chicago or other hospitals (see characteristics of patients in Table S2). Normal control cells were purchased from AllCells. (Emeryville, CA).
Bead-Based miRNA Expression Profiling Assay, qPCR Assays, and Bisulfite Genomic Sequencing.
The bead-based expression assay, data filtering, and normalization were described (16, 30). qPCR assays for miRNAs or target genes, and for genomic DNA copy numbers were performed as described (16, 28, 30), and so did bisulfite genomic sequencing (30).
Chromatin Immunoprecipitation (ChIP) Assay.
ChIP assay was performed with Upstate Biotechnology ChIP assay kit (Lake Placid, NY) by following the manufacturer’s protocol with some modifications. Purified ChIP DNA was then amplified by regular and real-time qPCR.
Transduction of MLL-AF9 into Human Normal CD34+ Cells and Maintenance of the Cells.
This experiment has been reported (53). Cells were harvested at different time points for the qPCR assay of miR-17 and miR-20a.
Cell Apoptosis and Viability Assays.
ApoONE Homogenous Caspase 3/7 Assay (Promega) and CellTiter-Blue Reagent (Promega) were used for the assays of cell apoptosis and viability, respectively.
Colony-Forming and Replating Assay.
In vitro colony-forming (i.e., immortalization) assays were performed as described (16, 26) with some modifications.
miRNA Array and Gene Array Assays.
A group of 18 samples (Table S3) obtained from the in vitro colony-forming/replating assays were included in the miRNA array and gene (i.e., mRNA) array assays. The miRNA array assay was performed by Exiqon using the miRCURY LNA arrays (v.10.0). Gene arrays were performed by using Affymetrix GeneChip Mouse Gene 1.0 ST Array. Partek Genomics Suite (Partek) was used for the analysis of the normalized data.
Identification of Potential Target Genes of miR-17-92 and Analysis of Their Enrichment in GO Processes and Pathways
PITA (54) (version 6), TargetScan (55) (release 5.1), Miranda (56) (version released in September 2008), and miRbase Targets (version 5; www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) were used to identify potential direct targets of the miRNAs. GeneGo MetaCore software was used in the analysis of the enrichment of target genes in GO processes and pathways.
Supplementary Material
Acknowledgments
The authors thank Drs. Y. Sato and K. Sugita at Yanamashi University, Japan, for kindly providing KOPN1, KOCL33, KOCL44, KOCL45, KOCL48, KOCL50, KOCL51, and KOCL69 cell lines. This work was supported in part by the National Institutes of Health (NIH) CA127277 (to J.C.), University of Chicago Cancer Center Pilot Grant (to J.C.), and CA118319 Sub-Award (to J.C.M. and J.C.), the G. Harold and Leila Y. Mathers Charitable Foundation (J.C.), Leukemia and Lymphoma Society Translational Research Grant (to J.D.R.), the Spastic Paralysis Foundation of the Illinois, Eastern Iowa Branch of Kiwanis International (J.D.R.), the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (to P.P.L.), and Leukemia and Lymphoma Society Specialized Center of Research (to M.J.T.), NIH PO1CA105049 (M.J.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914900107/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Odenike O, Rowley JD. Leukemogenesis: More Than Mutant Genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 2007;98:1482–1490. doi: 10.1111/j.1349-7006.2007.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venturini L, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2255–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloonan N, et al. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: Overview report. Genes Chromosomes Cancer. 2002;33:331–345. doi: 10.1002/gcc.10040. [DOI] [PubMed] [Google Scholar]
- 18.Pui CH, et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia. 2003;17:700–706. doi: 10.1038/sj.leu.2402883. [DOI] [PubMed] [Google Scholar]
- 19.Rowley JD. Seminars from the University of Minnesota. Chromosome translocations: Dangerous liaisons. J Lab Clin Med. 1998;132:244–250. doi: 10.1016/s0022-2143(98)90036-1. [DOI] [PubMed] [Google Scholar]
- 20.Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 21.Slany RK. When epigenetics kills: MLL fusion proteins in leukemia. Hematol Oncol. 2005;23:1–9. doi: 10.1002/hon.739. [DOI] [PubMed] [Google Scholar]
- 22.Ziemin-van der Poel S, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 24.Thirman MJ, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 25.Popovic R, Zeleznik-Le NJ. MLL: How complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 26.Lavau C, Luo RT, Du C, Thirman MJ. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci USA. 2000;97:10984–10989. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69:1109–1116. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 30.Mi S, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 32.Garzon R, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcucci G, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcucci G, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 36.Dixon-McIver A, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schotte D, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 38.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 39.Milne TA, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeisig BB, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guenther MG, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontana L, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 43.Garzon R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koralov SB, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 47.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 48.Sylvestre Y, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 49.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 50.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei J, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 55.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 56.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.