Abstract
This work describes a genetic approach to isolate small, artificial transmembrane (TM) proteins with biological activity. The bovine papillomavirus E5 protein is a dimeric, 44-amino acid TM protein that transforms cells by specifically binding and activating the platelet-derived growth factor β receptor (PDGFβR). We used the E5 protein as a scaffold to construct a retrovirus library expressing ∼500,000 unique 44-amino acid proteins with randomized TM domains. We screened this library to select small, dimeric TM proteins that were structurally unrelated to erythropoietin (EPO), but specifically activated the human EPO receptor (hEPOR). These proteins did not activate the murine EPOR or the PDGFβR. Genetic studies with one of these activators suggested that it interacted with the TM domain of the hEPOR. Furthermore, this TM activator supported erythroid differentiation of primary human hematopoietic progenitor cells in vitro in the absence of EPO. Thus, we have changed the specificity of a protein so that it no longer recognizes its natural target but, instead, modulates an entirely different protein. This represents a novel strategy to isolate small artificial proteins that affect diverse membrane proteins. We suggest the word “traptamer” for these transmembrane aptamers.
Keywords: bovine papilloma virus, E5 protein, expression libraries, protein engineering, traptamers
It is thought that up to 30% of all proteins span cell membranes (1). Most membrane-spanning protein segments adopt an α-helical conformation that is largely independent of their amino acid sequence and minimally influenced by extramembraneous protein domains. Although transmembrane (TM) domains were once thought to be rather nonspecific protein segments whose primary function was to anchor proteins in membranes, they can participate in highly-specific, inter- and intramolecular side-by-side interactions that control important biological processes (2).
The 44-amino acid bovine papillomavirus type 1 (BPV) E5 protein is essentially an isolated TM domain. It contains a hydrophobic central segment and exists in the intracellular membranes of transformed cells as a type II TM homodimer stabilized by disulfide bonds in its carboxy-terminus (3–5). The E5 dimer binds to the TM domains of two molecules of the platelet-derived growth-factor β receptor (PDGFβR) tyrosine kinase, resulting in receptor dimerization and activation, leading to cell transformation (6–10). The E5 protein does not bind to other TM domains or to certain PDGFβR TM mutants (11).
Based on our analysis of the E5 protein, we proposed that it might be possible to construct small, artificial TM proteins that regulate a variety of viral and cellular TM proteins in trans (11, 12). To identify small proteins with different TM sequences that transform cells, we constructed expression libraries in which the TM domain of the E5 protein was replaced with randomized sequences of primarily hydrophobic amino acids and isolated clones from these libraries that induced focus formation or growth-factor independence in mouse cells (13–16). Although the active clones had diverse TM sequences, all of the proteins analyzed induced cell transformation by specifically activating the PDGFβR. These experiments established that the E5 protein could be used as a scaffold to construct active small proteins with unique TM domains, with the potential limitation that this approach may be restricted to proteins that target the PDGFβR.
Here, we isolated artificial TM proteins that activate the human erythropoietin (EPO) receptor (hEPOR), a 508-amino acid cytokine receptor unrelated to the PDGFβR. The hEPOR exists primarily as a preformed TM dimer in unstimulated cells and undergoes a conformational change upon ligand binding that results in intracellular signaling required for proliferation and erythroid differentiation (17, 18). We used the E5 protein as a scaffold to construct small TM proteins that bear no sequence similarity to EPO but, nevertheless, specifically activate the hEPOR and drive proliferation and erythroid differentiation of human hematopoietic cells in vitro. These experiments demonstrate that sequence changes restricted to the TM segment of a viral oncogene product can reprogram it to an entirely different protein target and describe an approach to construct and isolate small, biologically active proteins not found in nature. This approach may be applicable to a wide variety of viral and cellular TM targets and is likely to identify proteins with important research and possibly clinical uses.
Results
A System to Isolate Transmembrane Activators of the hEPO Receptor.
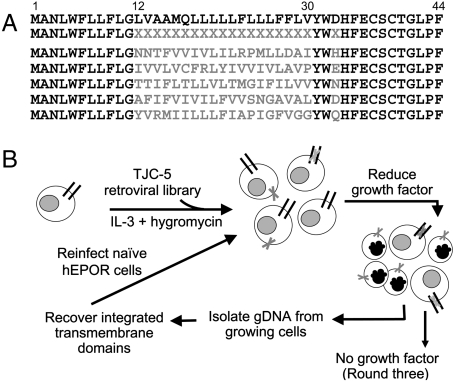
To isolate unique activators of the hEPOR, we constructed a retroviral library expressing many different small TM proteins and developed a genetic screen to select active clones from the library. Proteins in the library retained 24 amino acids of the wild-type E5 protein: the 11 N-terminal amino acids (positions 1–11), Tyr31, Trp32, and the 11 C-terminal amino acids (positions 34–44) (Fig. 1A). To randomize the TM domain, we used a degenerate oligonucleotide in which positions corresponding to codons 12–30 were synthesized with a mixture of nucleotides such that approximately 82% of the amino acids encoded by this segment were hydrophobic. This composition closely matches that of naturally-occurring TM domains (1). In addition, position 33 was randomized to encode six different hydrophilic amino acids because a hydrophilic amino acid is required at this position in the E5 protein (19). The randomized segment was amplified and cloned into the pT2H-F13 retrovirus vector to reconstitute an intact open reading frame, and the resulting mixture of plasmids was packaged into retrovirus particles to generate the TJC-5 library. Ultra-deep pyrosequencing of a portion of TJC-5 DNA and of TJC-5 proviruses recovered from transduced cells confirmed the expected composition of the randomized segment, identified genes encoding 120,000 different small TM proteins, and allowed us to estimate that the library in total encoded approximately 500,000 different 44-amino acid TM proteins. Detailed characterization of this library will be published elsewhere. Other than their overall hydrophobicity, the sequences of the TM domains of these proteins were essentially unrelated to each other or to the E5 protein (examples shown in Fig. 1A).
Fig. 1.
Construction and use of small TM protein libraries. (A) Amino acid sequence of the E5 protein (Top Line), the TJC-5 library with randomized positions indicated by Xs (Second Line), and five unselected clones from the library. Randomized amino acids shown in gray. (B) Diagram of the scheme to select small TM activators of the hEPOR. The Round Objects represent cells, those with Black Nuclei represent dying cells. The Dark Lines represent the hEPOR and the Gray X’s represent small TM proteins encoded by the library. See text for details.
We expressed the hEPOR and murine EPOR (mEPOR) in BaF3 cells, a murine cell line that grows in suspension and is strictly dependent on interleukin-3 (IL-3) for survival and proliferation, to generate BaF3/hEPOR and BaF3/mEPOR cells, respectively. Removal of IL-3 from the growth medium caused the cells to die within a few days, and E5 expression did not allow IL-3-independent proliferation (Fig. S1). However, addition of human recombinant EPO allowed BaF3/hEPOR and BaF3/mEPOR cells, but not parental BaF3 cells, to grow in the absence of IL-3 (Fig. S1), demonstrating that the ectopic EPOR could support cell proliferation.
Isolation of Small TM Proteins that Confer Growth-Factor Independence on Cells Expressing the hEPOR.
Fig. 1B is a schematic diagram of the selection used to isolate hEPOR activators. Twelve independent pools of clonal BaF3/hEPOR cells growing in IL-3 were infected in parallel with the TJC-5 library at a multiplicity of infection of 0.2, yielding 1.2 million infected cells. Because the library encoded approximately 500,000 different small TM proteins, it was, thus, screened with 2- to 3-fold redundancy. Beginning 2 d after infection, cells were selected in the presence of 1 mg/mL hygromycin B and 1/40 of the standard amount of IL-3. After two weeks, the cells were passaged into medium containing full-strength IL-3 for 3 d to expand the surviving population, and genomic DNA was purified from the growing cells. Library inserts were recovered from this DNA by PCR by using primers that recognized the invariant portions of the E5 gene and the flanking vector sequence. The mixtures of amplified inserts were cloned into pT2H-F13 and packaged into retrovirus to generate secondary libraries that were used to infect naïve BaF3/hEPOR cells. Cells were selected for growth in hygromycin and reduced IL-3 as above. The inserts were recovered from viable cells isolated by preparative sterile cell sorting, cloned, and packaged into retrovirus to generate tertiary libraries. After a third round of infection of BaF3/hEPOR cells with the individual tertiary libraries and selection in reduced IL-3, the cells were transferred to medium devoid of IL-3 or EPO. Genomic DNA was purified from two pools of cells that grew robustly in the absence of exogenous growth factors, and inserts were recovered, cloned, and sequenced.
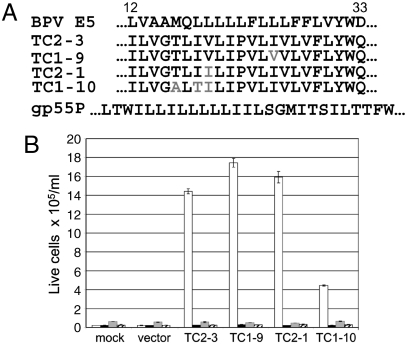
Each of the recovered clones encoded a 44-amino acid protein that retained the fixed portions of the E5 protein and a 19-amino acid hydrophobic stretch that showed no sequence similarity to the E5 protein (Fig. 2A). Remarkably, although the cells were initially infected with a library encoding many thousands of different TM proteins, the great majority of recovered clones encoded virtually identical proteins. The most abundant clone, TC2-3, was recovered multiple times, and the other clones differed from it by no more than three amino acid substitutions in the randomized segment.
Fig. 2.
Small TM proteins induce growth-factor independence in cells expressing the hEPOR. (A) Amino acid sequence of the TM domains of the E5 protein (positions 12–33) (Top Line), four clones recovered from growth-factor independent BaF3/hEPOR cells, and gp55P (Bottom Line). Sequence differences from TC2-3 in the recovered clones shown in Gray. The TM domains of the E5 protein and TC2-3 are thought to adopt a type II transmembrane orientation and are shown with their N termini to the Left, whereas gp55P, which adopts the opposite orientation, is shown with its N terminus to the Right. (B) Small TM proteins or empty CMMP vector were expressed in parental BaF3 cells (Striped Bars) or BaF3 cells expressing mPR (Gray Bars), hEPOR (White Bars), or mEPOR (Black Bars). After 2 d, IL-3 was removed, and viable cells were counted 6 d later. Cells expressing TC2-3 grew the fastest and at 6 d were dying due to overcrowding.
To test whether these proteins conferred growth-factor independence, each isolate was cloned into the bicistronic retroviral vector, pCMMP-IRES-GFP, which expresses small TM proteins at a higher level than the original vector. We tested the activity of these clones by infecting parental BaF3 cells, BaF3/hEPOR cells, BaF3/mEPOR cells, and BaF3/mPR cells (engineered to express the murine PDGFβR). Typically, > 90% of the cells were GFP-positive after infection. After 2 d, cells were washed and passaged into medium lacking IL-3 and EPO, and viable cells were counted for 6 d. As expected, all of the cell lines infected with the empty vector died (Fig. 2B), and the wild-type BPV E5 protein supported the growth of BaF3/mPR cells only (Fig. S1). The four clones selected from the library induced growth-factor-independent proliferation of cells expressing the hEPOR, but not parental Ba/F3 cells nor cells expressing the mEPOR or the PDGFβR (Fig. 2B). Thus, the recovered clones were active only in BaF3 cells expressing the hEPOR, and not in cells expressing other growth-factor receptors, including the closely related mEPOR. At lower expression levels, TC2-3 allowed robust BaF3/hEPOR cell proliferation in the absence of growth factors. TC1-9 allowed moderate proliferation, and TC2-1 and TC1-10 promoted cell survival but not proliferation (Fig. S2). Thus, the activity of TC2-3 can be affected by point mutations within its TM domain. Because TC2-3 was more active than the other clones, we focused on it for the remainder of this paper.
Expression and Dimerization of TC2-3.
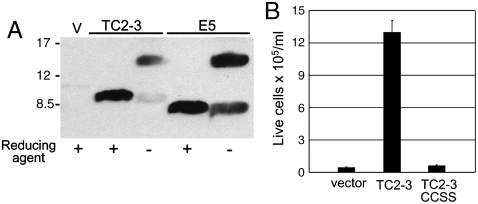
TC2-3 retained the C-terminal segment of the E5 protein that contains the two cysteines required for covalent dimerization as well as the epitope recognized by the anti-E5 antibody (αE5). To determine whether TC2-3 was expressed as a covalent dimer, we immunoprecipitated cell extracts with αE5. Samples were treated with reducing agents to disrupt disulfide bonds or left untreated, electrophoresed in the presence of SDS to dissociate non-covalent dimers, and immunoblotted with αE5. As shown in Fig. 3A, the reduced form of TC2-3 displayed similar mobility as the monomeric E5 protein in the presence of reducing agent. In the absence of reducing agent, TC2-3 and the E5 protein primarily migrated with the dramatically slower mobility expected for a dimer, indicating that TC2-3 formed a stable, disulfide-linked dimer in cells. This result implied that TC2-3, like the E5 protein (4), was in a type II TM orientation with the C-terminal cysteines located in the intralumenal or extracellular space rather than the reducing environment of the cytoplasm. To test whether the two cysteines were required for TC2-3 activity, both of them were simultaneously mutated to serine to generate TC2-3CCSS. In contrast to wild-type TC2-3, this mutant did not confer growth-factor independence in BaF3/hEPOR cells (Fig. 3B), suggesting that covalent dimerization of TC2-3 was required for biological activity. In addition, genetic experiments indicated that the TM domain of TC2-3 (amino acids 8–35, lacking the C-terminal cysteines) was able to form non-covalent dimers in bacterial membranes (Fig. S3), as is the case for the TM domain of the E5 protein (13, 20). The inability of TC2-3CCSS to induce growth-factor independence, even though the TM domain of TC2-3 appears to have intrinsic dimerization ability, suggests that the presence of disulfide bonds influences the conformation or stability of the TC2-3 dimer.
Fig. 3.
Dimerization of TC2-3. (A) RIPA extracts were prepared from BaF3/hEPOR cells expressing the empty CMMP vector (V), TC2-3, or BPV E5. After immunoprecipitation with αE5, samples were subjected to gel electrophoresis in the presence or absence of reducing agent and detected by immunoblotting with the same antibody. Size of protein markers (in kDa) is shown on Left. (B) BaF3/hEPOR cells expressing the empty T2H-F13 (Vector), TC2-3, or TC2-3CCSS were cultured in the absence of IL-3 and EPO. Viable cells were counted after 7 d.
TC2-3 Activates the hEPOR.
EPO treatment induces JAK2-mediated tyrosine phosphorylation of the EPOR (21). We conducted phosphotyrosine immunoblotting to determine whether TC2-3 induced biochemical activation of the hEPOR. We first expressed hEPOR containing an N-terminal HA-tag in BaF3 cells, to generate BaF3/HA-hEPOR cells. These cells proliferated in the presence of EPO or IL-3, or if TC2-3 was expressed, but not in the absence of growth factors (Fig. S1).
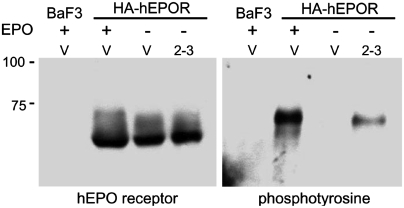
Cells grown in medium containing IL-3 were left untreated or stimulated briefly with EPO. After treatment with pervanadate, a phosphatase inhibitor, cells were harvested in radioimmunoprecipitation assay (RIPA) buffer. HA-hEPOR was immunoprecipitated from cell extracts with antibody recognizing the EPOR or the HA tag, and immunoblotting was performed with anti-EPOR or anti-phosphotyrosine antibody. No signal was detected in parental BaF3 cells lacking receptor expression, and HA-hEPOR migrated with an electrophoretic mobility of approximately 60 kDa, near its predicted size, with a smear extending to 75 kDa (Fig. 4, Left). TC2-3 expression did not affect the abundance or electrophoretic mobility of the hEPOR. After BaF3/HA-hEPOR cells were treated with EPO, the anti-HA antibody immunoprecipitated a tyrosine-phosphorylated protein with a mobility of ∼75 kD (Fig. 4, Right), similar to that of the top of the smear detected with the receptor antibody. This phosphorylated band was absent from parental BaF3 cells treated with EPO and from untreated BaF3/HA-hEPOR cells, and it was replaced by a phosphorylated band with much higher mobility in EPO-treated cells that expressed a truncated form of the hEPOR (Fig. S4), indicating that it was activated HA-hEPOR. Importantly, TC2-3 induced tyrosine-phosphorylation of HA-hEPOR in cells that were not treated with EPO, albeit to a lesser extent than in cells treated with EPO (Fig. 4, Right). These results showed that EPO treatment or TC2-3 expression caused tyrosine phosphorylation of a small fraction of the total hEPOR and provided biochemical evidence that TC2-3 activated the hEPOR.
Fig. 4.
Activation of the hEPOR. RIPA extracts were prepared from BaF3 cells or BaF3/HA-hEPOR cells expressing empty T2H-F13 vector (V) or TC2-3. Where indicated, cells were acutely stimulated with EPO (Plus Sign). Samples were immunoprecipitated and immunoblotted with C20 α-EPO receptor antibody (Left) or immunoprecipitated with anti-HA antibody and immunoblotted with 4G10 anti-phosphotyrosine antibody (Right). Half as much protein was immunoprecipitated for the EPO-stimulated lanes on the Right. Size of protein markers is shown on Left.
Mapping the Site of Interaction with the hEPOR.
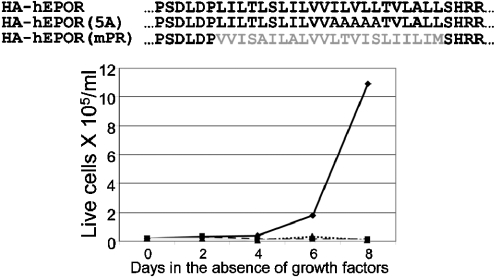
Because the E5 protein binds to the TM domain of the PDGFβR, and the TM domain of TC2-3 reprogrammed it from the PDGFβR to the hEPOR, it seemed likely that TC2-3 interacted with the TM region of the hEPOR. To investigate this possibility, we constructed two hEPOR variants: HA-hEPOR(5A) containing five consecutive alanines in the TM domain of the hEPOR, and HA-hEPOR(mPR), in which the TM domain of the hEPOR was replaced with the TM domain of the PDGFβR (Fig. 5, Top). These hEPOR mutants supported robust BaF3 cell proliferation in the presence of EPO (Fig. S5), demonstrating that the TM substitutions did not interfere with ligand-induced EPOR signaling.
Fig. 5.
The TM domain of the hEPOR is required for TC2-3 activity. (Top) Sequences of the TM domains and flanking sequences of the HA-hEPOR, HA-hEPOR(5A), and HA-hEPOR(mPR). PDGF receptor amino acids shown in Grey. (Bottom) BaF3 cells expressing HA-hEPOR (Solid Black Line), HA-hEPOR(5A) (Dashed Line), or HA-hEPOR(mPR) (Dotted Line) or parental BaF3 cells (Solid Gray Line) were infected with MSCV expressing TC2-3 and selected for hygromycin resistance. Drug-resistant cells were cultured in the absence of growth factors, and viable cells counted on the indicated days. Similar results were obtained in two independent experiments.
BaF3 cells expressing HA-hEPOR, HA-hEPOR(5A), or HA-hEPOR(mPR) were infected with the retrovirus expressing TC2-3. After selection in the presence of IL-3 and hygromycin, IL-3 was removed and viable cells were counted for 6 d. As expected, none of the cell types expressing the empty vector grew in the absence of growth factors (Fig. S1), and TC2-3 conferred growth-factor independence on cells expressing HA-hEPOR but not parental BaF3 cells (Fig. 5, Bottom). TC2-3 did not confer growth-factor independence in cells expressing HA-hEPOR(5A) or HA-hEPOR(mPR), indicating that the TM hEPOR activator TC2-3 required the intact TM domain of the hEPOR.
TC2-3 Induces Proliferation and Differentiation of Human Erythroid Precursors.
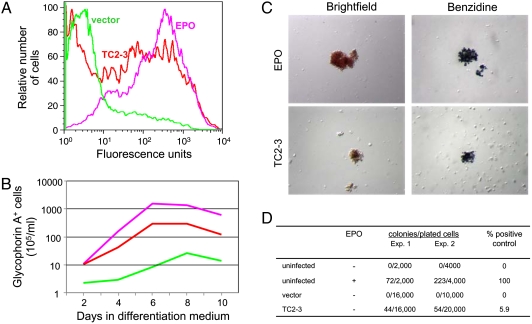
We also assessed the biological activity of TC2-3 in primary, multipotent, human hematopoietic progenitor cells (HPCs). These cells endogenously express the hEPOR and can be induced by EPO to undergo erythroid differentiation in vitro. Adult human CD34+ HPCs were infected with empty CMMP-IRES-GFP vector or the same vector expressing TC2-3, and GFP-positive cells were isolated by flow cytometry. Two days later, cells were cultured in serum-free differentiation medium that contains a combination of recombinant cytokines in the presence or absence of EPO. After various times, viable cells were counted and analyzed by flow cytometry for cell-surface expression of glycophorin A (GpA), a marker of late-stage erythroid differentiation. As expected, few cells infected with the empty vector proliferated or expressed GpA in the absence of EPO, whereas in medium supplemented with EPO, the cells grew robustly and > 80% of them expressed GpA (Fig. 6A and B). Strikingly, TC2-3 also induced the cells to proliferate and express cell-surface GpA in the absence of EPO. After 6 d in the absence of EPO, ∼36-fold more TC2-3 cells expressed GpA than did vector-infected cells. In addition, TC2-3 induced an approximately 6-fold increase in the level of β-globin mRNA in the absence of EPO, compared to cells expressing empty vector (Fig. S6).
Fig. 6.
TC2-3 stimulates EPO-independent proliferation and differentiation of HPCs. (A) CD34+ cells were infected with the CMMP-IRES-GFP vector or the same vector expressing TC2-3, sorted for GFP expression, and transferred to differentiation medium in the absence or presence of EPO. After 4 d in differentiation medium, expression of cell-surface GpA was assessed by immunostaining non-permeabilized cells and flow cytometry. Green, vector-infected cells minus EPO; Lavender, vector-infected cells plus EPO; and Red, TC2-3-expressing cells minus EPO. (B) Cells were handled as in part A. The total number of viable cells expressing cell-surface GpA (> 50 fluorescence units) was determined by immunostaining and flow cytometry at various times. The ability of TC2-3 to stimulate erythroid differentiation was observed in four independent experiments, although the magnitude of the effect was somewhat variable in these primary cells. (C) Cells were infected with CMMP-IRES-GFP (Top) or the same vector expressing TC2-3 (Bottom), sorted for GFP expression, and plated in replicate in methylcellulose. Cells infected with empty vector were also treated with EPO. After 14 d, colonies were visualized by brightfield microscopy (Left) or after staining with benzidine (Right). (D) Table showing the generation of dense erythroid colonies in methylcellulose. P-value for the difference between untreated cells infected with vector and TC2-3, < 0.001 (binomial test P-value).
We also assessed the ability of TC2-3 to induce the formation of colonies in methylcellulose. GFP-expressing human CD34+ cells were plated in methylcellulose in serum-free medium containing a cocktail of cytokines and colony formation was assessed after two weeks. In the absence of added EPO, cells infected with the empty vector formed no dense, red colonies, whereas addition of EPO allowed the formation of large, dense red colonies (Fig. 6C and D). Cells expressing TC2-3 also formed dense red colonies in the absence of EPO, but these colonies were usually smaller and less numerous than those that developed in the EPO-treated samples (Fig. 6C and D). These colonies contained heme, as demonstrated by staining with benzidine, which reacts with heme to produce a dark-blue color (Fig. 6C). Thus, as assessed by biochemical and physiological markers, TC2-3 can drive the proliferation and erythroid-specific differentiation of primary human HPCs.
Discussion
We constructed libraries expressing variants of the BPV E5 protein that contain unrelated TM sequences and used genetic selection to isolate short proteins from these libraries that specifically activate the hEPOR and not the closely related mEPOR. The ability of TC2-3 to stimulate erythroid differentiation of human HPCs in vitro, a process dependent on EPOR signaling, provided definitive proof that it activated the hEPOR. Thus, amino acid substitutions restricted to the TM region of BPV E5 can reprogram it to a new target, implying that TM interactions alone are sufficient to specify biological recognition in this system. These unique hEPOR activators are only 44-amino acids long, dimeric, and very hydrophobic and they presumably adopt a TM localization. In contrast, EPO is 165-amino acids long, hydrophilic, soluble, and monomeric. Furthermore, EPO binds to the extracellular domain of the EPOR, whereas our experiments imply that TC2-3 interacts with the TM domain of the hEPOR.
It is interesting to compare TC2-3 to gp55P, a ∼500 amino acid TM glycoprotein encoded by the Friend erythroleukemia virus. Cell proliferation is driven by the ability of gp55P to bind and activate the mEPOR, but it does not activate the hEPOR (22, 23). Thus, gp55P and TC2-3 display opposite species specificity. Genetic studies suggest that gp55P interacts directly with the TM domain of the mEPOR and that this interaction as well as an interaction with the extracellular domain of the mEPOR plays a role in pathogenicity. There is no amino acid sequence similarity between the TM domains of gp55P and TC2-3 (Fig. 2A).
By analogy to the BPV E5 protein, we propose that TC2-3 binds directly to the TM domain of the hEPOR and that this results in receptor activation. The finding that the TM sequence of TC2-3 is sufficient to redirect it from the TM domain of the PDGFβR to the hEPOR is consistent with this model, as is the inability of TC2-3 to activate hEPOR containing a mutant or foreign TM domain. In addition, the inability of TC2-3 to induce growth-factor independence in cells expressing the mEPOR indicates that TC2-3 does not induce the expression of EPO. However, attempts to demonstrate a specific physical interaction between TC2-3 and hEPOR have so far been unsuccessful, perhaps as a result of decreased stability of the complex in detergent micelles. Thus, we have not excluded the possibility that TC2-3 has an indirect mechanism of action that is dependent on the TM domain of the hEPOR.
The construction of proteins that specifically regulate the activity of native cellular proteins is usually complicated by three-dimensional interactions involving distant segments along the length of polypeptide chains, and high-resolution structural information is often required to identify accessible sites on the target protein. Furthermore, short protein segments excised from larger proteins usually do not adopt the stable conformations they exhibit in their native structure, and peptides are often unstructured and tend to aggregate. Active protein segments (aptamers) can be isolated from libraries of random amino acid sequences presented as solvent-exposed surface loops extending from a tightly-packed, soluble protein scaffold or phage particle. Our results demonstrate that it is also possible to construct and select biologically active TM proteins from libraries in which an α-helical segment in a hydrophobic membrane environment presents a random surface to potential targets. Because there are few known specific sequence constraints on the generation of stable TM α-helices, it may be possible to express a large variety of TM sequences from the E5 scaffold and hence display sequences that can bind to and possibly modulate most proteins that contain helical TM domains. This strategy may be widely applicable because it targets a structural element present in many proteins. We suggest the word “traptamer” to designate these transmembrane aptamers.
Traptamers may have important research or clinical uses. They may induce subtle differences in signaling compared to the natural ligand, or they may be more specific than the ligand. For example, TC2-3 activates the hEPOR but not the mEPOR, whereas human EPO activates both. It may also be possible to mutagenize TC2-3 to identify essential structural features; optimize its affinity or specificity; or fine-tune its activity. It may also be possible to isolate inhibitory derivatives of TC2-3 that force the hEPOR dimer to adopt a non-productive orientation, as has been reported for a peptide that binds to the extracellular domain of the hEPOR (24). Finally, traptamers may also serve as structurally simple templates that can inform the design of peptide or peptidomimetic reagents and drugs that specifically modulate a wide variety of cellular and viral transmembrane protein targets.
Methods
Retroviral Library Construction.
A library of genes with randomized TM domains was constructed by using a degenerate oligonucleotide (A Fwd in Table S1) containing E5 codons 10–30, in which codons 12–30 were randomized with mixture of nucleotides at all three positions. Within the randomized segment, the composition of A∶C∶G∶T was 1∶1∶1∶0.5 at the first position; 0.1∶0.25∶0.1∶1 at the second position; and 0∶1∶0.1∶0 at the third position. In addition, codon 33 was randomized by using a mixture of A∶C∶G (1∶1∶1) at the first position, A at the second position, and C∶G (1∶1) at the third position. A downstream oligonucleotide (A Rev in Table S1), complementary to codons 34–44 of the E5 gene, was annealed to the degenerate oligonucleotide, extended, and amplified to generate full-length, double-stranded DNA. This product was amplified with short internal primers, digested with AvrII and BamHI, and ligated into pT2H-F13, which expresses the first 13 residues of a modified E5 protein (E5P2A) and encodes hygromycin resistance (SI Text), to reconstitute an intact open reading frame. The ligation reaction was used to transform E. coli strain DH10β. Approximately 1.2 million ampicillin-resistant bacterial colonies were pooled, and plasmid DNA was harvested and named TJC-5. The library was characterized by sequencing randomly picked individual clones and by 454 pyrosequencing. Details of library construction are available from the authors upon request.
Cells and Viruses.
Vesicular stomatitis virus (VSV)-G pseudotyped retroviruses were prepared from human 293T cells. BaF3 cells were maintained in RPMI-1640 containing various supplements, including 6% WEHI-3B cell conditioned medium (a source of IL-3), unless otherwise noted. Adult human CD34+ cells were cultured in expansion medium, infected with CMMP-IRES-GFP or CMMP-IRES-GFP/TC2-3, and sorted for GFP expression. Cells were incubated 2 d later in differentiation medium [20 ng/mL recombinant human (rh)-stem cell factor (SCF), 5 ng/ml rh-IL-3, 0.2 μM β-Estradiol, and 2 μM dexamethasone in StemSpan serum-free medium] or methylcellulose medium (Methocult H4531, Stem Cell Technologies) containing 50 ng/mL rh-SCF, 20 ng/mL rh-IL-3, and 20 ng/mL rh-IL-6. Details for growing, manipulating, and analyzing all cells are presented in SI Text.
Untagged and HA-tagged hEPOR and mEPOR genes were recloned into retroviral expression vectors and used to generate hEPORs containing five alanines in the TM domain or the TM domain of the murine PDGFβR (SI Text). BaF3 cells expressing these receptors were generated by infection and drug selection. Clonal lines were established by serial dilution in RPMI medium lacking IL-3 but containing 0.5 units/mL of Epoetin-α recombinant human EPO (Amgen) (RPMI-EPO). Procedures used to select growth-factor independent BaF3/hEPOR cells and to recover and test individual clones are described in the Results and in detail in SI Text.
Biochemical and Immunological Analysis.
Standard procedures were used for immunoprecipitation and immunoblotting, flow cytometry, and quantitative real-time PCR (see SI Text for protocols and antibodies used).
Supplementary Material
Acknowledgments.
We thank Sara Marlatt, Yong Kong, Gregory Korbel, Bernard Forget, Stefan Constantinescu, Diane Krause, Milind Mahajan, Patrick Gallagher, Sherman Weissman, Murat Arcasoy, Kristina Talbert-Slagle, Edward Goodwin, and Bill Sugden for helpful advice and essential reagents; and Jan Zulkeski for assistance in preparing this manuscript. CD34+ cells were obtained from the Yale Center for Excellence in Molecular Hematology (NIH DK072442). T.J.C. was supported by a Warner Postdoctoral Fellowship and a training grant from the National Cancer Institute (NCI) (Grant CA09159). F.N.B. was supported by a postdoctoral fellowship from the Fundación Alfonso Martin Escudero. This work was supported by grants to D.D. from the NCI (Grant CA37157) and the Yale Skin Cancer SPORE (Grant CA121974; R. Halaban P.I.), and by a grant from the NIGMS to DME (Grant GM073857).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915057107/DCSupplemental.
References
- 1.Lehnert U, et al. Computational analysis of membrane proteins: Genomic occurrence, structure prediction and helix interactions. Q Rev Biophys. 2004;37:121–146. doi: 10.1017/s003358350400397x. [DOI] [PubMed] [Google Scholar]
- 2.Moore DT, Berger BW, and Degrado WF. Protein-protein interactions in the membrane: Sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlegel R, Wade-Glass M, Rabson MS, Yang Y-C. The E5 transforming gene of bovine papillomavirus encodes a small hydrophobic protein. Science. 1986;233:464–467. doi: 10.1126/science.3014660. [DOI] [PubMed] [Google Scholar]
- 4.Burkhardt A, Willingham M, Gay C, Jeang K-T, Schlegel R. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology. 1989;170:334–339. doi: 10.1016/0042-6822(89)90391-7. [DOI] [PubMed] [Google Scholar]
- 5.Surti T, Klein O, Aschheim K, DiMaio D, Smith SO. Structural models of the bovine papillomavirus E5 protein. Proteins. 1998;33:601–612. [PubMed] [Google Scholar]
- 6.Petti L, Nilson LA, DiMaio D. Activation of the PDGF receptor by the bovine papillomavirus E5 transforming protein. EMBO J. 1991;10:845–855. doi: 10.1002/j.1460-2075.1991.tb08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petti L, DiMaio D. Stable association between the bovine papillomavirus E5 transforming protein and activated PDGF receptor in transformed mouse cells. P Natl Acad Sci USA. 1992;89:6736–6740. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein DJ, Andresson T, Sparkowski JJ, Schlegel R. The BPV-1 E5 protein, the 16 kDa membrane pore-forming protein and the PDGF receptor exist in a complex that is dependent on hydrophobic transmembrane interactions. EMBO J. 1992;11:4851–4859. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilson LA, DiMaio D. PDGF receptor can mediate tumorigenic transformation by the bovine papillomavirus E5 protein. Mol Cell Biol. 1993;13:4137–4145. doi: 10.1128/mcb.13.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai CC, Henningson C, DiMaio D. Bovine papillomavirus E5 protein induces oligomerization and trans-phosphorylation of the PDGF β receptor. P Natl Acad Sci USA. 1998;95:15241–15246. doi: 10.1073/pnas.95.26.15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talbert-Slagle K, DiMaio D. The bovine papillomavirus E5 protein and the PDGF β receptor: It takes two to tango. Virology. 2009;384:345–351. doi: 10.1016/j.virol.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman-Cook LL, DiMaio D. Modulation of cell function by small transmembrane proteins modeled on the bovine papillomavirus E5 protein. Oncogene. 2005;24:7756–7762. doi: 10.1038/sj.onc.1209039. [DOI] [PubMed] [Google Scholar]
- 13.Freeman-Cook LL, et al. Selection and characterization of small random transmembrane proteins that bind and activate the PDGF β receptor. J Mol Biol. 2004;338:907–920. doi: 10.1016/j.jmb.2004.03.044. [DOI] [PubMed] [Google Scholar]
- 14.Freeman-Cook LL, et al. Specific locations of hydrophilic amino acids in constructed transmembrane ligands of the PDGF β receptor. J Mol Biol. 2005;345:907–921. doi: 10.1016/j.jmb.2004.10.072. [DOI] [PubMed] [Google Scholar]
- 15.Ptacek JB, Edwards APB, Freeman-Cook LL, DiMaio D. Packing contacts can mediate highly specific interactions between artificial transmembrane proteins and the PDGF β receptor. P Natl Acad Sci USA. 2007;104:11945–11950. doi: 10.1073/pnas.0704348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talbert-Slagle K, et al. Artificial transmembrane oncoproteins smaller than the bovine papillomavirus E5 protein redefine the sequence requirements for activation of the PDGF β receptor. J Virol. 2009;83:9773–85. doi: 10.1128/JVI.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livnah O, et al. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 18.Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 19.Nilson LA, Gottlieb RW, Polack GW, DiMaio D. Mutational analysis of the interaction between the BPV E5 transforming protein and the endogenous receptor for platelet-derived growth factor in mouse C127 cells. J Virol. 1995;69:5869–5874. doi: 10.1128/jvi.69.9.5869-5874.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oates J, Hicks M, Dafforn TR, DiMaio D, Dixon AM. In vitro dimerization of the bovine papillomavirus E5 protein transmembrane domain. Biochemistry. 2008;47:8985–8992. doi: 10.1021/bi8006252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witthuhn BA, et al. JAK2 associates with the erthropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 22.Li J-P, D’Andrea AD, Lodish HF, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 23.Constantinescu SN, et al. Activation of the erythropoietin receptor by the gp55-P viral envelope protein is determined by a single amino acid in its transmembrane domain. EMBO J. 1999;18:3334–3347. doi: 10.1093/emboj/18.12.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livnah O, et al. An antagonist peptide-EPO receptor complex suggests that receptor dimerization is not sufficient for activation. Nat Struct Biol. 1998;5:993–1004. doi: 10.1038/2965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.