Abstract
Several important pathogens cause disease by surviving and replicating within host cells. Bacterial proliferation is the product of both replication and killing undergone by the population. However, these processes are difficult to distinguish, and are usually assessed together by determination of net bacterial load. In addition, measurement of net load does not reveal heterogeneity within pathogen populations. This is particularly important in persistent infections in which slow or nongrowing bacteria are thought to have a major impact. Here we report the development of a reporter system based on fluorescence dilution that enables direct quantification of the replication dynamics of Salmonella enterica serovar Typhimurium (S. Typhimurium) in murine macrophages at both the population and single-cell level. We used this technique to demonstrate that a major S. Typhimurium virulence determinant, the Salmonella pathogenicity island 2 type III secretion system, is required for bacterial replication but does not have a major influence on resistance to killing. Furthermore, we found that, upon entry into macrophages, many bacteria do not replicate, but appear to enter a dormant-like state. These could represent an important reservoir of persistent bacteria. The approach could be extended to other pathogens to study the contribution of virulence and host resistance factors to replication and killing, and to identify and characterize nonreplicating bacteria associated with chronic or latent infections.
Keywords: Salmonella, growth, kinetics, macrophages
Salmonella enterica causes acute and chronic infections by replicating and persisting within host cells (1, 2). Intracellular bacteria express the Salmonella pathogenicity island 2 (SPI-2) type III secretion system (T3SS), which delivers more than 25 effector proteins across the vacuolar membrane separating bacteria from the host cytosol. Together these are required for bacterial proliferation in host cells including macrophages (3), and for systemic growth in host tissues (4). Bacterial proliferation is usually assessed by determination of net load, after plating cell or tissue lysates to laboratory medium and counting of colony-forming units (cfu). These are a product of both replication and killing sustained by the bacteria and the relative contributions of these processes are difficult to distinguish. Furthermore, quantitative measurements such as cfu counts do not reveal heterogeneity within the pathogen population.
Experiments using a temperature-sensitive plasmid in wild-type (WT) and mutant bacteria showed that the SPI-2 T3SS is required for replication in the mouse spleen but does not contribute significantly to the ability of the pathogen to survive in this organ (5). However, other work has indicated that the SPI-2 T3SS is involved in avoidance of host killing mechanisms (6 –8).
To provide further information on the contribution of the SPI-2 T3SS to proliferation, and to examine the dynamics of intracellular replication at the single-cell level, we have developed a reporter system based on dilution of a fluorescent protein that occurs during bacterial cell division (9). In addition to providing detailed information on these processes, we found that a surprisingly large proportion of the bacterial inoculum does not undergo any replication, but enters into a viable but nonreplicating state in macrophages, both in vitro and in vivo.
Results and Discussion
Measurement of Bacterial Replication Using Fluorescence Dilution.
To study the replication dynamics of intracellular S. Typhimurium, we constructed dual fluorescence reporter plasmids in which the production of DsRed protein is induced by the exposure of bacterial cells to arabinose, whereas that of EGFP is either constitutive (ie, pDiGc) or induced by isopropyl β-D-thiogalactoside (IPTG; ie, pDiGi; Fig. 1A). The pDiGc system enables a measure of bacterial replication by dilution of the preformed pool of DsRed proteins in egfp-expressing cells (Fig. S1). The pDiGi system extends the range of bacterial cell divisions that can be measured, by sequential removal of the two inducers (Fig. 1A). Dilution of red and green fluorescence in bacteria replicating in laboratory medium was analyzed by flow cytometry at hourly intervals, and yielded a series of overlapping, normally distributed curves (Fig. 1B and C, from right to left), reflecting uniform replication within the bacterial population. Assuming that the fluorescent proteins are partitioned equally at each cell division, and that fluorescence intensities are halved as a result, we calculated the average number of generations per time interval by using the geometric mean value of each sample. These growth curves were similar to those obtained from cfu for six generations (pDiGc, red dilution only) and approximately 10 generations (pDiGi, sequential dilution). After these times, the fluorescence intensity became too low to be measured accurately (Fig. 1D). Therefore, fluorescence dilution (FD) provides an alternative to cfu to measure more than a 1,000-fold increase in bacterial numbers.
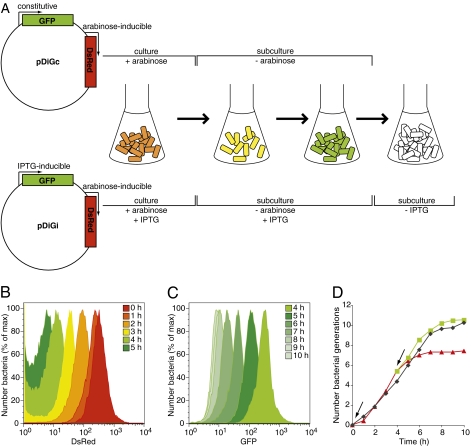
Fig. 1.
FD enables measurement of bacterial replication for up to 10 generations. (A) Structure of pDiGc and pDiGi plasmids and schematic of FD principle. With pDiGc, bacterial replication is accompanied by dilution of red fluorescence in the absence of arabinose and detection of bacteria is based on EGFP signal. With pDiGi, sequential removal of arabinose and IPTG leads to dilution of red then green fluorescence. (B and C) Flow cytometric detection of DsRed (B) or EGFP (C) fluorescence in the bacterial population (carrying pDiGi) grown in minimum liquid medium (from an OD600 of 0.05) at hourly intervals (n = 30,000 events analyzed at each time point). (D) Bacterial replication curves determined by cfu and FD analysis from one representative experiment of three. Angled arrows indicate removal of arabinose at to and IPTG at t4h. Black diamonds show cfu, red triangles red FD, and green squares green FD. The extent of replication of the population (F, fold replication) was calculated by the ratio: Yo/Yt (Y being the geometric mean of red or green fluorescence intensity of the bacterial population at a specific time). The number of generations, N, is deduced from F = 2N.
Quantitative and Qualitative Analysis of Intracellular Replication.
FD was also detected in bacteria replicating within macrophages (Movie S1 ), enabling us to compare the intracellular replication of WT S. Typhimurium and an ssaV mutant, which lacks a functional SPI-2 T3SS. Mouse primary bone marrow–derived (bm) macrophages were infected for 16 h with preinduced bacteria carrying pDiGc (Fig. 2A). Negligible replication of WT bacteria occurred in the first 6 h after uptake. However, from 6 h to 16 h, there was a 10-fold increase in bacteria, with an average cell division time of approximately 3 h. No replication of the SPI-2 mutant occurred over the same time (Fig. 2A). Therefore, the SPI-2 T3S system is absolutely required for bacterial replication in bm macrophages. Comparison of the degree of replication (calculated by FD) and net growth (calculated by counting cfu of bacteria from lysed macrophages) for WT and SPI-2 mutant bacteria showed that the difference in the replicative abilities of the two strains is sufficient to explain the difference in their overall growth at all time points (Fig. 2A). The difference between replication and net growth rates reflects the extent of killing sustained by intracellular bacteria and other processes (as detailed later) that impair their capacity to grow (ie, killing index). The killing indices of WT and SPI-2 mutant bacteria determined before (2 h to 6 h) and after (6 h to 12 h) bacterial replication had commenced (Fig. 2B) show that the SPI-2 T3SS does not contribute significantly to avoidance of host cell killing during these time periods. In view of the importance of the respiratory burst in host resistance to Salmonella (10) and evidence that the SPI-2 T3SS interferes with this (8), we compared the killing indices of the two bacterial strains in WT and phox−/− macrophages. In these experiments, bacteria were also sampled at earlier times post-uptake, when the respiratory burst is more pronounced (Fig. 2C). As expected, phox-dependent killing was detected at early (0.5 h to 6 h) but not late (6 h to 12 h) time points. However, there was no difference in the level of killing sustained by the WT and SPI-2 mutant bacteria. These results confirm that, between 0.5 h and 12 h after uptake, the SPI-2 T3SS does not have a major impact on avoidance of host cell killing.
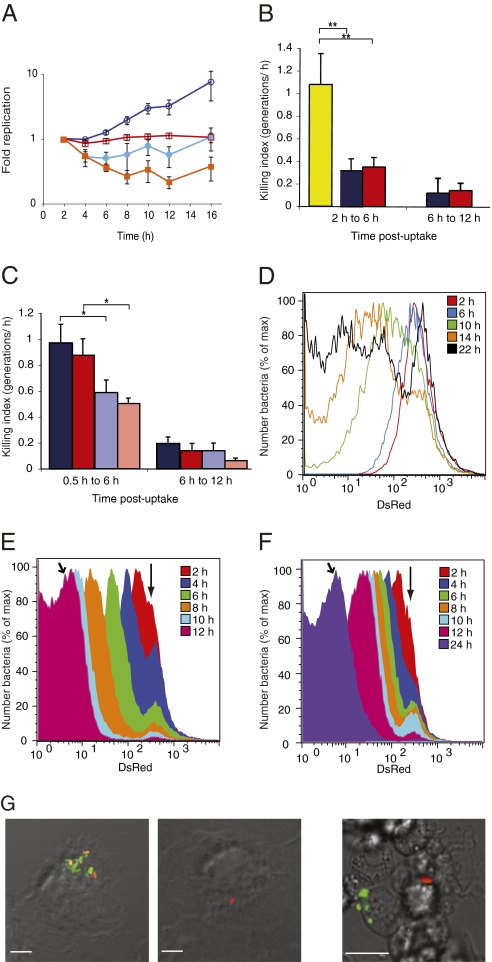
Fig. 2.
Quantification and analysis of heterogeneity of S. Typhimurium replication in macrophages with pDiGc. (A) Quantification of replication and net growth in bm macrophages. Replication kinetics of WT (dark blue, open circles) and SPI-2 mutant (red, open squares) determined by FD and net growth kinetics of WT (light blue, filled circles) and SPI-2 mutant (orange, filled squares) determined by cfu. (B and C) Killing indices of WT (dark blue) and SPI-2 mutant (red) compared with a sensitive rpoE mutant (yellow) (25) in BALB/c bm macrophages (B) and of WT and SPI-2 mutant bacteria in C57BL/6 WT bm macrophages (dark blue and dark red, respectively) or phox−/− macrophages (pale blue and pale red, respectively) (C). *P < 0.05, **P < 0.01 on paired, one-tailed Student t test. Error bars (SEM) are based on three replicate experiments. (D–F) Flow cytometric detection of DsRed fluorescence in the intracellular population at different time points (n = 50,000 events analyzed at each time point). WT bacteria released from bm macrophages (D) and WT (E) and SPI-2 mutant (F) bacteria released from RAW264.7 macrophages. Vertical arrows indicate nonreplicating subpopulation. Angled arrows indicate 12 h time point for WT (E) and 24 h time point for SPI-2 mutant bacteria (F). (G) Microscopy of infected macrophages. Left: bm macrophages infected for 24 h; one macrophage contains both replicating and nonreplicating bacteria and another contains one nonreplicating bacterium. Right: splenic macrophages isolated from BALB/c mice infected for 3 d; replicating and nonreplicating bacteria are present in different macrophages. (Scale bars: 10 μm.)
To provide a more complete description of intracellular bacterial replication, we assessed its heterogeneity by obtaining flow cytometry profiles of the distribution of the levels of red fluorescence of individual cells within the population. At 10 h, 14 h, and 22 h, the profiles of replication of the WT bacterial population did not follow normally distributed curves as observed in laboratory medium, but were spread over a much greater range, indicating highly heterogeneous replication (Fig. 2D). Microscopy revealed numerous macrophages containing bacteria that had undergone different numbers of cell division over a 22-h period (Fig. 2G). Therefore, bacteria vary in their ability to replicate, even in the same macrophage.
The histograms (Fig. 2D) and microscopy (Fig. 2G) also revealed a nonreplicating population of bacteria. Remarkably, at 22 h this represented 32.2% ± 6.4% of the total population. As FD indicated a 12-fold increase in bacterial numbers by 22 h, we estimate that as little as 15% of the initial intracellular population underwent replication. As the SPI-2 mutant failed to replicate in bm macrophages, it was possible that the nonreplicating WT bacteria were defective in a functional SPI-2 T3SS. This was tested by analyzing the profiles of replication of both strains in RAW264.7 macrophages. In contrast to bm macrophages, these cells allow moderate net growth of a SPI-2 mutant (11). Analysis of the replication profiles of WT bacteria in these macrophages revealed uniform curves, except for a subpopulation of nonreplicating cells (Fig. 2E). SPI-2 mutant bacteria replicated at a much slower rate, displaying a similar uniform FD profile at 24 h to that of WT bacteria at 12 h (Fig. 2F). Nonreplicating SPI-2 mutant bacteria were also detected (Fig. 2F). At 24 h, these accounted for 3.6% ± 1.5% of the total population, similar to the proportion of nonreplicating WT bacteria at 12 h (2.3% ± 1.16%). The strains had the same kinetics of replication for the first 6 h. Then, the SPI-2 T3SS enabled a threefold surge in the replication rate. At this time, intracellular bacteria doubled every 60 min, a rate similar to that during exponential growth in minimal medium (Fig. S2). Therefore, S. Typhimurium is very well adapted to replication within RAW264.7 macrophages. Nevertheless, as in bm macrophages, a surprisingly large proportion of the initial population did not undergo any replication, and this was independent of the SPI-2 T3SS. Nonreplicating bacteria were also detected when only EGFP was produced (as detailed later) and when a pFPV25-derived plasmid was used to express other fluorescent proteins (Fig. S3). Therefore, the nonreplicating bacteria are not an artifact of the plasmid or proteins used in these experiments.
We next examined if nonreplicating bacteria could be detected in a diseased host. WT bacteria carrying pDiGc expressing both fluorescent proteins were used to inoculate BALB/c or 129SV mice, which differ in their susceptibility to infection by S. Typhimurium (12). Splenic macrophages were recovered 2 or 3 d after inoculation. Bacteria that had not undergone any replication (from 0.5% to 27% of total bacteria) were consistently recovered from both mouse strains. These were clearly visible by microscopy and had apparently normal morphology (Fig. 2F).
Viability of Nonreplicating Bacteria.
The characteristics of nonreplicating intracellular bacteria were analyzed further by microscopy. The absolute number of nonreplicating bacteria remained constant during 3 d of infection of bm macrophages, showing that they were not degraded (Fig. S4A). Intracellular S. Typhimurium replicates within vacuoles that acquire lysosomal membrane glycoproteins such as LAMP1, but which generally avoid fusion with mature lysosomes containing hydrolytic enzymes (13). Immunofluorescence microscopy of infected macrophages revealed similar LAMP1 labeling around nonreplicating bacteria and replicating bacteria (Fig. S4B), indicating that they were contained in vacuoles. Nonreplicating bacteria from bm macrophages that had been infected for 24 h were isolated from replicating bacteria on the basis of selective tolerance to a β-lactam antibiotic (14), or after their release by cell sorting (15). They were then incubated in rich or minimal media for different periods of time. However, no resumption of growth was ever observed. To determine if these nonreplicating bacteria were viable but nonculturable or simply dead, we tested if they could synthesize protein in response to an extracellular signal (16) by using the double inducible pDiGi system as a reporter of both replication status and metabolic activity. Infection of macrophages by egfp-expressing bacteria in the absence of IPTG results in dilution of green fluorescence in the replicating but not nonreplicating population. Arabinose is then added to the cell culture medium and the production of red fluorescence indicates that bacteria have sensed the signal and responded by producing DsRed (Fig. 3A). The responses of bacteria in macrophages were first examined by time-lapse microscopy (Fig. 3 B–D). As expected, replicating bacteria that had lost their green fluorescence were revealed by the appearance of red fluorescence (Fig. 3B). Some nonreplicating (green) bacteria failed to respond to arabinose (Fig. 3C) whereas others produced red fluorescence indicating their viability (Fig. 3D). The proportions of nonreplicating but viable intracellular bacteria in bm macrophages and splenic macrophages isolated from infected mice were then quantified by FACS analysis. Despite prolonged periods within macrophages, in both cases the majority of nonreplicating bacteria produced DsRed upon addition of arabinose (Fig. 3 E–F). These experiments show that, upon uptake by macrophages, a large proportion of intracellular S. Typhimurium rapidly enters a nonreplicating but viable state.
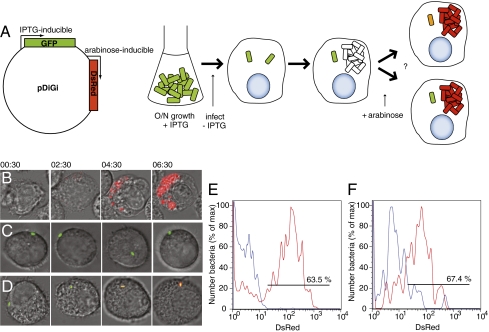
Fig. 3.
Viability of nonreplicating WT S. Typhimurium. (A) Schematic of the detection of viability of nonreplicating bacteria using the pDiGi system. (B–D) Time-lapse microscopy of infected RAW264.7 macrophages. Numbers above the fields are hours and minutes after addition of arabinose. (B) Replicating bacteria revealed by appearance of red fluorescence. (C) Nonreplicating bacterium failing to respond to induction. (D) Nonreplicating bacterium responding to arabinose by producing DsRed. (E and F) Flow cytometric detection of DsRed fluorescence in nonreplicating bacteria with (red) or without (blue) 4 h arabinose induction; (E) bm macrophages infected for 20 h (n = 800 nonreplicating bacteria); (F) splenic macrophages isolated from 4 129SV mice infected for 2 d (n = 100 nonreplicating bacteria).
Damaged or Dormant-Like Bacteria?
In bm macrophages, the proportion of responsive nonreplicating bacteria decreased from more than 40% at 24 h to 5% at 72 h (Fig. 4A). The failure of nonreplicating bacteria to regrow and the gradual decrease in their responsiveness might be a consequence of damage caused by the antimicrobial activities of macrophages (17). Alternatively, these bacteria could represent cells that have initiated an adaptive process leading to dormancy (16). To help distinguish between these possibilities, intracellular nonreplicating bacteria were removed from bm macrophages at 24 h, and incubated in saline solution for a further 48 h. Surprisingly, the rate of decrease of responsiveness of this population was very similar to that of bacteria that remained in macrophages for 72 h (Fig. 4A). This shows that, in macrophages, the process of decrease in responsiveness is initiated within the first 24 h after uptake. It is possible that damage sustained by bacteria in the first 24 h might cause a gradual loss of viability over the subsequent 48 h. Two important mediators of macrophage killing that are known to act within the first 24 h following phagocytosis are the respiratory burst (18) and effectors of IFN-γ activity (19). Therefore, we examined the proportion of responsive nonreplicating bacteria in phox−/− macrophages, or macrophages whose killing activity was stimulated with IFN-γ. In both cases, the decrease in bacterial responsiveness was not statistically different from that in control macrophages (Fig. 4A). A similar decrease was observed in nonreplicating bacteria in the more permissive RAW264.7 macrophage cell line (from 69.6% ± 9.4% at 12 h to 25.2% ± 4.4% at 24 h). Furthermore, the decrease in responsiveness was not accompanied by an increase in the proportion of nonreplicating bacteria that colocalized with the lysosomal marker cathepsin D, which remained low and constant over 72 h (Fig. 4B). Therefore, the failure to respond to inducer cannot be attributed to an increase in phagolysosomal fusion. Collectively, these results indicate that nonreplicating bacteria lose their ability to respond to the extracellular signal independently of the killing activities of macrophages, and strongly suggest that the intracellular environment signals the initiation of an adaptive response leading to dormancy.
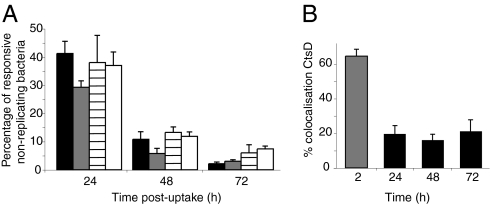
Fig. 4.
(A) Responsiveness after exposure to different conditions. Proportion of responsive nonreplicating bacteria in bm macrophages (black bars), in PBS solution after release from macrophages at 24 h (white bars), in phox−/− macrophages (gray bars), and in IFN-γ–activated macrophages (horizontal lined bars; n > 50,000 bacteria). (B) Proportion of nonreplicating bacteria colocalizing with cathepsin D (CtsD) in bm macrophages (black bars) compared with heat-killed bacteria (gray bars; n = 150–300 macrophages). Error bars (SEM) are based on three replicate experiments.
In summary, we have developed a technique based on FD that records the amount of replication undergone by individual bacteria in infected cells. This has enabled us to visualize the dynamics of intramacrophage growth of S. Typhimurium in great detail, and to measure the contribution of bacterial replication to the net growth rate. This approach could easily be applied to assess the impact of virulence and resistance genes to replication and killing of other pathogens. It could also be used to study dynamics of bacterial growth in infected hosts and nonhost environments such as biofilms. Of particular interest was the discovery of a large proportion of bacteria that enter a nonreplicating but viable state in vivo. These are tolerant to treatment by a β-lactam antibiotic and are likely to represent a hitherto unrecognized persistent or dormant reservoir of bacteria in macrophages. Fluorescent nonreplicating bacteria are amenable to further investigation and can be purified from physiologically relevant environments on the basis of antibiotic tolerance or flow cytometry. Furthermore, the approach could be used to investigate the existence of nongrowing viable cells of the pathogens associated with chronic infections, including Mycobacterium tuberculosis, Helicobacter pylori, Brucella spp., and uropathogenic Escherichia coli (20 –23).
Materials and Methods
Bacterial Strains and Plasmids.
Standard microbial techniques were used to construct strains and plasmids. Strains used in this study were S. enterica serovar typhimurium NCTC 12023 and its isogenic mutants ssaV::aphT (SPI-2 mutant) (24) and ΔrpoE::KmR (25) carrying pDiGc or pDiGi plasmids. Both plasmids encode a fluorescent-optimized DsRed protein whose expression is under the control of the arabinose-inducible P BAD promoter (26) and EGFP, either under the control of a constitutive promoter (Prpsm) in the case of pDiGc or the IPTG-inducible promoter Plac in the case of pDiGi. These plasmids were stable both in vitro and in vivo and, in agreement with a previous report (27), caused a minor increase in the bacterial cell division time in minimum medium and macrophages (by a factor of approximately 1.2). Bacteria were routinely grown in MgMES minimal medium (28) at 37 °C with aeration supplemented with carbenicillin (50 μg/mL), and appropriate sugars where indicated (0.2% arabinose, 0.5 mM IPTG) to allow production and maturation of fluorescent proteins. In the case of sequential FD, IPTG was removed by centrifugation of bacteria and replacement with fresh medium.
Cell Culture.
RAW264.7 macrophages were obtained from the European Collection of Cell Cultures (Sigma). Cells were grown in Dulbecco modified Eagle medium (DMEM; PAA Laboratories) supplemented with 10% (vol/vol) FCS (PAA Laboratories) at 37 °C in 5% (vol/vol) CO2. Primary bm macrophages were obtained from BALB/c mice, C57BL/6 WT (Charles River), or gp91phox knockout (29) mice. Cells were grown in complete medium: RPMI (Gibco) supplemented with 10% FCS, 2 mM glutamine, 10 mM Hepes, 50 μM β-mercaptoethanol, and 100 U/mL penicillin/streptomycin, and L929 cell–conditioned medium 20% (vol/vol; National Institute for Medical Research). After 3 d of culture, further fresh complete medium containing L929 cell–conditioned medium was added to the growing macrophages. On d 7, cells were washed and seeded in complete medium without antibiotic and incubated for 24 h before bacterial challenge. If needed, activation of macrophages was performed as described (19) using overnight incubation in culture medium in the presence of 103 U of recombinant mouse IFN-γ (Millipore).
Bacterial Infection of Macrophages.
Infection of macrophages was performed as described (30) using bacteria from overnight growth in minimal medium (which allowed maturation of DsRed protein), at a multiplicity of infection of 10:1. At appropriate time points, cells were washed and lysed with PBS solution containing 0.1% Triton X-100 (Sigma) to release intracellular bacteria. When required, an aliquot of released bacterial suspension was diluted in PBS solution before plating on Luria agar for cfu determination. The remaining bacterial cells were pelleted and resuspended in PBS solution. The fluorescence of the bacterial population was subsequently assessed by FACS analysis. Nonreplicating bacteria were isolated from replicating bacteria by adding 100 μg/mL cefotaxime (Sigma) to medium containing infected macrophages from 6 h after uptake to the end of the experiment. Antibiotic was then washed out three times with PBS solution before macrophage lysis. Alternatively, nonreplicating bacteria were purified by cell sorting after release from infected macrophages using a FACScan analyzer (Becton Dickinson).
Flow Cytometry Acquisition and Analysis.
Bacterial samples from laboratory culture medium or released from macrophages were resuspended in PBS. They were analyzed using a FACSCalibur cytometer (Becton Dickinson) for fluorescence intensities in FL-1 and FL-2 channels. Data were analyzed with FlowJo 8.6.3 software. In the case of red FD, the gate was set for the bacterial population based on their EGFP-positive signal. For characterization of nonreplicating bacteria, the gate was set for retained EGFP fluorescence. The extent of replication of the population (F, fold replication) was calculated by the ratio Yo/Yt, with Y being the geometric mean of red or green fluorescence intensity of the bacterial population at a specific time. The number of generations, N, is deduced from F = 2N. The killing indices were calculated by r R − r NG (with r R being replication rate and r NG being net growth rate). The estimation of the proportion of initial intracellular bacteria that undergo replication is obtained from r 0/n 0 = r/n/F (with r 0 being the proportion of initial intracellular population that will undergo replication, n 0 the proportion of initial intracellular population that does not replicate, r the proportion of replicating bacteria at time t, n the proportion of nonreplicating bacteria at time t, and F the fold replication of the population at time t).
Immunofluorescence Microscopy and Antibodies.
Cells were fixed, permeabilized, and incubated with antibodies as described (30) and observed by using a fluorescence microscope (BX50; Olympus), or a confocal laser scanning microscope (LSM510 Axiovert; Zeiss). For time-lapse microscopy using a Zeiss Axiovert 200 M microscope (Carl Zeiss), cell seeding, infection, and imaging were performed as described (31). The monoclonal rat LAMP-1 antibody 1D4B (DSHB) was used at 1:100; the polyclonal rabbit cathepsin D antibody WG (32) was a gift from S. Kornfeld (St. Louis, MO) and was used at 1:500. Secondary antibodies were obtained from Jackson Immunoresearch Laboratories: cyanine 5-conjugated donkey anti-rabbit and anti-rat antibodies were used at 1:400.
Mouse Infections.
Eight- to 12-week-old BALB/c or 129SV mice (Jackson Laboratory) were inoculated by i.p. injection of bacterial suspensions ranging from 1 × 105 to 6 × 105 cfu (BALB/c) and from 2 × 106 to 4 × 106 cfu (129SV) in a volume of 0.2 mL. This resulted in bacterial loads ranging from 1 to 3 × 106 cfu/spleen by 2 d after inoculation. Mice were killed 48 h or 72 h later. Spleens were homogenized and splenic CD11b+ cells purified using magnetic beads following manufacturer instructions (Miltenyi Biotec). In some cases, purified cells were incubated for 4 h at 37 °C with 5% CO2 in complete medium containing 0.4% arabinose. They were then lysed in PBS solution containing 0.1% Triton X-100 to release intracellular bacteria before analysis by flow cytometry.
Supplementary Material
Acknowledgments
We thank Chris Tang, Jaime Mota, Emmanuel Boucrot, and members of the Holden laboratory for helpful suggestions and critical reading of the manuscript. We thank Alan Verdie for helpful advice. This work was supported by grants from the Medical Research Council and Wellcome Trust (D.W.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000041107/DCSupplemental.
References
- 1.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 4.Hensel M, et al. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 5.Shea JE, Beuzon CR, Gleeson C, Mundy R, Holden DW. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallois A, Klein JR, Allen LA, Jones BD, Nauseef WM. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J Immunol. 2001;166:5741–5748. doi: 10.4049/jimmunol.166.9.5741. [DOI] [PubMed] [Google Scholar]
- 7.Uchiya K, et al. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 1999;18:3924–3933. doi: 10.1093/emboj/18.14.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vazquez-Torres A, et al. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 9.Roostalu J, Jõers A, Luidalepp H, Kaldalu N, Tenson T. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 2008;8:68. doi: 10.1186/1471-2180-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mastroeni P, et al. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med. 2000;192:237–248. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensel M, et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 12.Vidal SM, Pinner E, Lepage P, Gauthiers, Gros P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent from suseptible (Nramp1D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- 13.Rathman M, Barker LP, Falkow S. The unique trafficking pattern of Salmonella typhimurium-containing phagosomes in murine macrophages is independent of the mechanism of bacterial entry. Infect Immun. 1997;65:1475–1485. doi: 10.1128/iai.65.4.1475-1485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camilli A, Paynton CR, Portnoy DA. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc Natl Acad Sci USA. 1989;86:5522–5526. doi: 10.1073/pnas.86.14.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah D, et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc Natl Acad Sci USA. 2008;105:6145–6149. doi: 10.1073/pnas.0711712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyström T. Not quite dead enough: on bacterial life, culturability, senescence, and death. Arch Microbiol. 2001;176:159–164. doi: 10.1007/s002030100314. [DOI] [PubMed] [Google Scholar]
- 18.Vazquez-Torres A, Fang FC. Oxygen-dependent anti-Salmonella activity of macro-phages. Trends Microbiol. 2001;9:29–33. doi: 10.1016/s0966-842x(00)01897-7. [DOI] [PubMed] [Google Scholar]
- 19.Kagaya K, Watanabe K, Fukazawa Y. Capacity of recombinant gamma interferon to activate macrophages for Salmonella-killing activity. Infect Immun. 1989;57:609–615. doi: 10.1128/iai.57.2.609-615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehlers S. Lazy, dynamic, or minimally recrudescent? On the elusive nature and location of hte mycobacterium responsible for latent tuberculosis. Infection. 2009;37:87–95. doi: 10.1007/s15010-009-8450-7. [DOI] [PubMed] [Google Scholar]
- 21.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 22.Orme M. The latent tuberculosis bacillus (I’ll let you know if I ever meet one) Int J Tuberc Lung Dis. 2001;5:589–593. [PubMed] [Google Scholar]
- 23.Schilling JD, Lorenz RG, Hultgren SJ. Effect of trimethoprim-sulfamethoxazole on recurrent bacteriuria and bacterial persistence in mice infected with uropathogenic Escherichia coli. Infect Immun. 2002;70:7042–7049. doi: 10.1128/IAI.70.12.7042-7049.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 25.Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sörensen M, et al. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 2003;552:110–114. doi: 10.1016/s0014-5793(03)00856-1. [DOI] [PubMed] [Google Scholar]
- 27.Knodler LA, et al. Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect Immun. 2005;73:7027–7031. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu XJ, Liu M, Holden DW. SsaM and SpiC interact and regulate secretion of Salmonella pathogenicity island 2 type III secretion system effectors and translocators. Mol Microbiol. 2004;54:604–619. doi: 10.1111/j.1365-2958.2004.04297.x. [DOI] [PubMed] [Google Scholar]
- 29.Pollock JD, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 30.Beuzón CR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsden AE, Mota LJ, Münter S, Shorte SL, Holden DW. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol. 2007;9:2517–2529. doi: 10.1111/j.1462-5822.2007.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garvis SG, Beuzón CR, Holden DW. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell Microbiol. 2001;3:731–744. doi: 10.1046/j.1462-5822.2001.00153.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.